Abstract
Redox reactions involving thiol groups in proteins are major participants in cellular redox regulation and antioxidant defense. Although mechanistically similar, thiol-dependent redox processes are catalyzed by structurally distinct families of enzymes, which are difficult to identify by available protein function prediction programs. Herein, we identified a functional motif, CxxS (cysteine separated from serine by two other residues), that was often conserved in redox enzymes, but rarely in other proteins. Analyses of complete Escherichia coli, Campylobacter jejuni, Methanococcus jannaschii, and Saccharomyces cerevisiae genomes revealed a high proportion of proteins known to use the CxxS motif for redox function. This allowed us to make predictions in regard to redox function and identity of redox groups for several proteins whose function previously was not known. Many proteins containing the CxxS motif had a thioredoxin fold, but other structural folds were also present, and CxxS was often located in these proteins upstream of an α-helix. Thus, a conserved CxxS sequence followed by an α-helix is typically indicative of a redox function and corresponds to thiol-dependent redox sites in proteins. The data also indicate a general approach of genome-wide identification of redox proteins by searching for simple conserved motifs within secondary structure patterns.
Keywords: CxxS motif, redox, thiol-disulfide oxidoreductase, genome, computational biology
Functional annotation of protein sequences in completely sequenced genomes is a major immediate challenge in the postgenomic era. With the increase in genomic complexity, a higher proportion of proteins is involved in cellular regulation and various signaling mechanisms. In this respect, two major mechanisms involving reversible modification of amino acid side chains to modulate protein activity are phosphorylation/dephosphorylation by kinase and phosphatase systems and reduction/oxidation by thiol-dependent enzymes. Whereas many signaling processes involving phosphorylation are well understood in terms of mechanisms and identities of participating enzymes, redox regulation of cellular processes remains a poorly characterized area. Difficulties of studying redox processes are primarily due to lack of understanding of how specificity of redox interactions is achieved, what are cellular targets of redox proteins, and what is identity of thiol-dependent redox proteins (Carmel-Harel and Storz 2000; Rhee et al. 2000; Tanaka et al. 2000; Finkel 2001; Ritz and Beckwith 2001).
Initial studies on the role of redox processes in antioxidant defense revealed a prominent role of NADPH-dependent thioredoxin and glutathione systems. In the thioredoxin system, the flow of reducing equivalents to counteract oxidative stress is as follows: NADPH → thioredoxin reductase → thioredoxin → thioredoxin peroxidase → hydrogen peroxide (Holmgren 2000). Thioredoxin interacts, in a redox manner, with a variety of other cellular proteins, such as signaling proteins (e.g., kinases and phosphatases), transcription factors, and ribonucleotide reductase and redox regulatory and antioxidant proteins (such as methionine sulfoxide reductase). The importance of these interactions is illustrated by the fact that the thioredoxin system is present in all living organisms.
In the glutathione system, the flow of reducing equivalents is similar to that of the thioredoxin system: NADPH → glutathione reductase → glutathione → glutathione peroxidase → hydrogen peroxide, or NADPH → glutathione reductase → glutathione → glutaredoxin → mixed disulfides in proteins. Glutaredoxin, a structural and functional homolog of thioredoxin, can also interact with other protein targets, such as ribonucleotide reductase, and cellular redox compounds, such as vitamin C (Cotgreave and Gerdes 1998).
Each of the protein components of the thioredoxin and glutathione systems may occur as a family of isozymes, particularly in more complex eukaryotic organisms. For example, multiple copies of thioredoxin, thioredoxin peroxidase, glutaredoxin, and glutathione peroxidase are present in the genome of Saccharomyces cerevisiae. Interestingly, these four protein families are characterized by a common fold, called thioredoxin fold, composed of a central β-sheet surrounded by α-helices and an active site involving one or two conserved cysteines (Martin 1995). In thioredoxins and glutaredoxins, a CxxC motif (two cysteines separated by two other residues) is present, whereas thiol-dependent peroxidases conserve only a single cysteine residue.
The CxxC motif in thioredoxins and glutaredoxins has been previously characterized in great detail. It is located in a partially exposed surface loop downstream of a β-strand and at the N terminus of an α-helix (Martin 1995). The identity of the amino acids separating the two cysteines in the CxxC motif and protein location influence redox properties of CxxC-containing proteins such that these proteins may serve as either reductants (e.g., thioredoxin and glutaredoxin) or oxidants (e.g., protein disulfide isomerase and DsbA; Woycechowsky and Raines 2000). In addition, it was proposed that the presence of a helix downstream of CxxC influences ionization properties of the two cysteines such that the N-terminal cysteine has reduced pKa compared to the pKa of free cysteine (Kortemme and Creighton 1995).
In the thioredoxin-catalyzed reaction, the thiolate of the low pKa cysteine attacks an intramolecular or intermolecular disulfide in a protein target to form a mixed target protein/thioredoxin intermolecular disulfide. Subsequently, the second thiol displaces the protein target thiol, resulting in the formation of a disulfide in the oxidized thioredoxin (Arner and Holmgren 2000). Thioredoxin-catalyzed reaction is a well-characterized redox reaction, but other thiol-dependent redox enzymes containing different structural folds may also use a similar reaction mechanism.
Interestingly, some thioredoxins and glutaredoxins are known that contain only the N-terminal cysteine in the CxxC motif, whereas the second cysteine is replaced with serine. This change in the active site does not appear to permit the completion of the typical thioredoxin reaction, as the CxxS-catalyzed reaction must stop at the stage of the mixed intermolecular disulfide. Indeed, CxxS mutants of thioredoxins have been previously used to obtain stable covalent complexes between thioredoxins and their target proteins for structural and functional studies and to identify physiological targets of thioredoxins (Motohashi et al. 2001; Yano et al. 2001).
Nevertheless, at least some CxxS-containing forms of thioredoxin-fold proteins retain catalytic activity. For example, mammalian thioredoxin and glutathione reductase (TGR) has a glutaredoxin-like CxxS-containing domain, which is functional in the reduction of oxidized glutathione and various mixed disulfides. Saccharomyces cerevisiae has five previously characterized glutaredoxins, including two that contain the CxxC motif and three a CxxS motif. All five proteins showed glutaredoxin activity in in vivo and in vitro assays. Another example is a recently identified protein disulfide isomerase homolog, ERp44, which assists other oxidoreductases in correct disulfide bond formation (Anelli et al. 2002).
In the present study, we asked a question of how common are CxxS motifs in proteins. Surprisingly, we found that CxxS sequences are rarely conserved in proteins and that their conservation correlates with and points to a possible redox function of a protein. This finding should help in functional characterization of redox proteins, and we present functional analyses of two bacterial, one archaeal, and one eukaryotic genomes for the presence of redox proteins containing the CxxS motif.
Results
CxxS redox sequences
A typical functional site present in functionally characterized thiol/disulfide oxidoreductases is CxxC (two cysteines separated by two other residues). However, this motif is also present in numerous metal-binding proteins, particularly those that contain zinc, greatly complicating identification of CxxC-containing thiol/disulfide oxidoreductases. In some natural thiol/disulfide oxidoreductases, the CxxC motif is replaced with CxxS. For example, in proteins of glutaredoxin and protein disulfide isomerase families, the presence of CxxS in the active site is quite common. Several of these proteins have been functionally characterized, confirming their thiol/disulfide oxidoreductase activities.
We reasoned that CxxS, although used less frequently than the CxxC motif in thiol/disulfide oxidoreductases, would not be significantly contaminated with metal-binding proteins and therefore could potentially be used for selective enrichment of redox proteins in genome-wide searches. We also noted that the use of CxxS for redox function is not limited to one fold. For example, methionine-S-sulfoxide reductase may use CxxC or only a single cysteine, but also, less frequently, a CxxS sequence in the active site (e.g., Treponema pallidum methionine-S-sulfoxide reductase, MsrA, accession number NP_219071). Interestingly, thioredoxin-fold proteins, including thioredoxins and glutaredoxins, and proteins of the MsrA family use similar reaction mechanisms and have similar structures; however, their folds are different and their functions were acquired independently of each other (Gladyshev 2002). Another example is S. cerevisiae arsenate reductases, which are characterized by an active-site CxxS redox sequence, but have adapted a CDC25 phosphatase fold for this function (Mukhopadhyay and Rosen 2001).
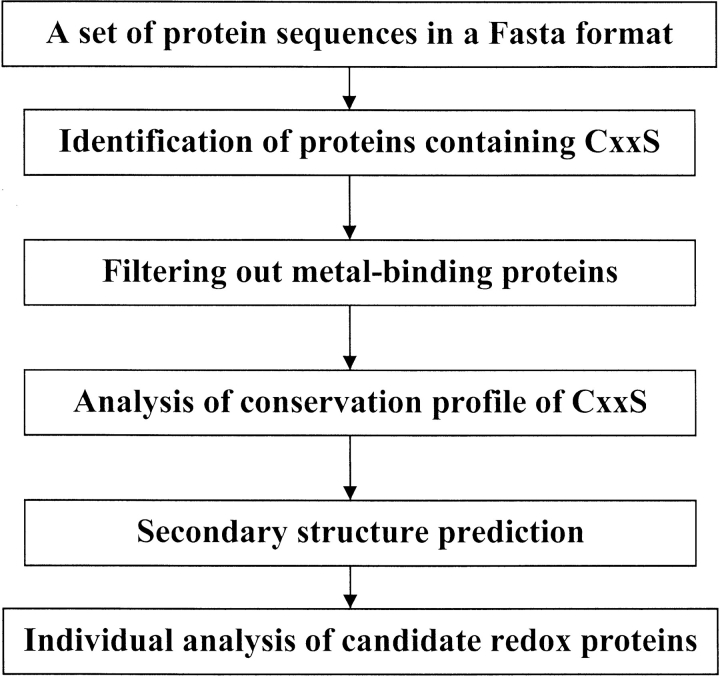
We tested how often conserved CxxS sequences were present in proteins. For this purpose, we developed a Perl script that automatically identifies conserved CxxS sequences in large sequence databases through a simple four-step algorithm (Fig. 1 ▶). In the first step, all CxxS-containing proteins were identified in a genome of interest. We found that 15%–30% of all proteins in a genome contain CxxS sequences. Therefore, this step only provided an initial crude reduction in the number of analyzed proteins. In the second step, proteins containing CxxS within metal-binding domains were filtered out using Prosite patterns. In the third step, the program searched for conservation of CxxS sequences. This step allowed a dramatic reduction in the number of analyzed proteins and was based on the observation that CxxS was always conserved in redox proteins, but rarely in other proteins. However, it should be noted that, in some redox proteins, serine in the CxxS motif was infrequently replaced with glycine, alanine, or cysteine. Therefore, these replacements were set to be tolerated by our script. The final step of the algorithm predicted secondary structure for proteins with conserved CxxS sequences. Following this automatic analysis, remaining proteins were individually analyzed for homologies to proteins of known function, known structure, or previously identified domains.
Fig. 1.
Schematic representation of the algorithm for identification of thiol/disulfide oxidoreductases containing the CxxS motif that was used in our study.
Genome-wide analysis for conserved CxxS sequences
Analysis of the complete Escherichia coli genome (4289 proteins) revealed 712 proteins that contained the CxxS sequence, and in 78 of these proteins CxxS was conserved. Subsequent manual filtering to remove buried, membrane-bound, and other contaminating sequences resulted in only 15 E. coli proteins (0.3% of the total number of proteins in the genome) that contained the conserved CxxS (Table 1).
Table 1.
Identification of proteins containing the CxxS motif in Escherichia coli, Campylobacter jejuni, Methanococcus jannaschii and Saccharomyces cerevisiae genomes
| Organism | Total proteins | Proteins with CxxS | Proteins with conserved CxxS | Proteins with CxxS, for which conservation profile is not available | Proteins with conserved CxxS flanked by an α-helix | Redox proteins (true positives/candidates) |
| Escherichia coli | 4289 | 712 | 78 (1.8%) | 49 (1.1%) | 46 (1.1%) | 15 (9/6) |
| Campylobacter jejuni | 1634 | 312 | 16 (0.97%) | 7 (0.4%) | 14 (0.86%) | 3 (1/2) |
| Methanococcus jannaschii | 1770 | 264 | 12 (0.68%) | 12 (0.7%) | 8 (0.45%) | 2 (0/2) |
| Saccharomyces cerevisiae | 6334 | 1956 | 127 (2.0%) | 285 (4.5%) | 57 (0.9%) | 24 (12/12) |
Percentages shown in parentheses indicate proportion of proteins in a genome.
Such a small number of proteins was surprising considering that the conservation profile included only two residues. To test whether a similarly small proportion of proteins contain conserved CxxS sequences in other genomes, we analyzed the bacterial genome of Campulobacter jejuni, the archaeal genome of Methanococcus jannaschii, and the eukaryotic genome of S. cerevisiae, and identified 2–24 proteins in these genomes (0.1%–0.4% of the total number of proteins; Table 1). Thus, conserved CxxS sequences were not numerous in these genomes.
Initial assessments of proteins selected by our program revealed that nearly 50% of them were known redox proteins. It should be pointed out that the program was designed so that all redox proteins that contained CxxS in redox active sites and were common to at least three genomes were identified. Thus, our procedure selectively identified these redox proteins (true positives) and also identified approximately an equal number of proteins, whose function was not known (candidate redox proteins).
E. coli proteins containing a conserved CxxS motif
Among 15 E. coli proteins containing the CxxS motif, 9 proteins were true positives as these proteins were previously implicated in thiol-dependent redox processes (Table 2). The other 6 proteins were hypothetical or poorly characterized proteins, and we predict that at least some of these other proteins are involved in thiol-dependent redox processes and use the CxxS motif for their function. E. coli proteins selected by the program are further discussed individually.
Table 2.
CxxS-containing proteins identified in the Escherichia coli K12 genome
| Gi number | Protein lengtha | CxxS positionb | CxxS and flanking secondary structurec | Descriptiond |
| 1787943 | 115 | 30 | β-CGFS-α | Glutaredoxin |
| 1787757 | 143 | 125 | CPVS-α | Osmotically inducible protein |
| 1789742 | 119 | 79 | α-CAAS | Hypothetical protein, YhcM |
| 1788841 | 119 | 13 | β-CSKS-α | Arsenate reductase |
| 1788991 | 104 | 14 | β-CGTS-α | Arsenate reductase |
| 1789918 | 141 | 15 | β-CGTS-α | Arsenate reductase |
| 1787577 | 322 | 41 | β-CGKS-α | Putative ATP-binding component of a transport system |
| 1788683 | 436 | 99 | β-CATS-α | Putative acyltransferase |
| 1789475 | 495 | 264 | β-CGGS-α | Altronate hydrolase |
| 1789516 | 523 | 288 | β-CGGS-α | Putative altronate hydrolase |
| 1789804 | 294 | 232 | β-CTCS-α | Disulfide bond chaperones of the HSP33 family |
| 1786351 | 114 | 108 | -CGSS- | Hypothetical protein, HesB-like |
| 1787974 | 122 | 116 | -CGES- | Hypothetical protein, HesB-like |
| 1788077 | 137 | 118 | -CVNS- | Methionine-R-sulfoxide reductase |
| 1788877 | 107 | 101 | -CGES- | HesB-like protein |
a Number of amino acids in an ORF.
b Amino acid position corresponding to Cys in the CxxS sequence.
c CxxS primary sequences and flanking secondary structure (α, α-helix; β, β-strand).
d Predicted function or information obtained by database searches.
Arsenate reductases
Three arsenate reductase homologs (gi 1788841, 1788991, and 1789918) were identified in the E. coli genome. These proteins show a structural fold not found in other proteins, and the CxxS sequence in these proteins was previously shown to be essential for catalytic activity (Shi et al. 1999). E. coli arsenate reductases catalyze arsenate reduction using glutaredoxin as a reductant, which in turn is reduced by glutathione.
Glutaredoxin
One glutaredoxin containing the CxxS motif was identified (gi 1787943), which is also a true positive identified in our genome-wide search. Although glutaredoxins containing the CxxS motif are known in other organisms, this E. coli protein has neither been described nor experimentally characterized previously.
Methionine- R -sulfoxide reductase
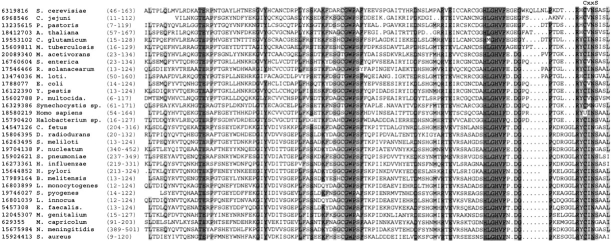
The function of this enzyme (gi 1788077; Kryukov et al. 1999) has only been revealed earlier this year (Kryukov et al. 2002). It is a small, 12–17-kD protein, present in all previously characterized genomes except those of certain obligatory parasites and hyperthermophyles. In vertebrates, some methionine-R-sulfoxide reductases contain selenocysteine residues encoded by TGA codons (Kryukov et al. 1999, 2002). During the time of our analysis, the identity of amino acids directly involved in methionine-R-sulfoxide reduction was not known. Our data indicated that the CxxS motif (or a motif containing selenocysteine in place of cysteine) located in the C-terminal portion of the protein is the actual catalytic site (Fig. 2 ▶). We recently tested this prediction by characterizing Drosophila methionine-R-sulfoxide reductase and found that mutation of either cysteine or serine in the CxxS motif disrupted thioredoxin-dependent methionine-R-sulfoxide reduction (Kumar et al., in press). A three-dimensional structure of methionine-R-sulfoxide reductase is not known, but secondary structure prediction indicated a β-strand-rich protein, the property that distinguished it from other known thiol/disulfide oxidoreductases, including arsenate reductase, glutaredoxins, and MsrAs. Thus, identification of the CxxS motif in methionine-R-sulfoxide reductase was one example where the search for the conserved motif resulted in functional predictions that were subsequently confirmed by site-directed mutagenesis and experimental characterization of mutant proteins.
Fig. 2.
Alignment of methionine-R-sulfoxide reductases. The location of the CxxS motif in C-terminal regions of the enzymes is indicated above the sequences. Residues conserved in >90% of the sequences are highlighted with dark gray. Residues conserved in >70% of the sequences are highlighted with light gray. Accession numbers (gi) for each sequence used in the alignment are indicated on the left. In vertebrate methionine-R-sulfoxide reductase, selenoprotein R, the cysteine in the CxxS motif is replaced with selenocysteine (shown as U for human enzyme).
Heat-shock protein HSP33
HSP33 (gi 1789804) is known to be regulated through disulfide bond formation, and in the reduced form, these two redox cysteines can also bind zinc (Jakob et al. 2000). The conserved CxxS motif includes one of these previously characterized redox cysteines. Thus, HSP33 is another true positive.
HesB-like proteins
HesB-like proteins, also called IscA, are involved in the formation of iron–sulfur clusters (Krebs et al. 2001; Ollagnier-de-Choudens et al. 2001). The proteins build the cluster by using conserved cysteines to coordinate iron. The thiol/disulfide function of HesB proteins is evident because of the requirement for a two-electron transfer to complete the construction of the cluster. However, the identity of iron-binding and redox cysteines in these proteins is not known. The possibility that the CxxS motif identified in HesB-like proteins is involved in thiol/disulfide reduction is very attractive, but will require further experimental verification. Three HesB-like proteins were identified in the E. coli genome (gi 1786351, 1787974, 1788877), all containing the CxxS motif.
Osmotically induced protein
Osmotically induced protein (OsmC; gi 1787757) is a putative envelope protein of unknown function that is required for long-term survival in stationary phase and resistance to organic peroxides (Mongkolsuk et al. 1998; Atichartpongkul et al. 2001). Mutations in this protein lead, directly or indirectly, to increased sensitivity to oxidative stress. E. coli OsmC contains a conserved CxxS motif in the C-terminal portion, and, in addition, it has a conserved cysteine residue in the middle of the protein sequence (Fig. 3 ▶). Our data indicate that OsmC is directly involved in redox function and that it uses the CxxS motif for this purpose. A possibility is also that the CxxS motif is involved in the formation of a disulfide bond with the second conserved cysteine. The CxxS motif in OsmC is followed by an α-helix, a feature that is common for thiol-disulfide oxidoreductases (see below).
Fig. 3.
Alignment of proteins of the OsmC family. The location of the CxxS motif in C-terminal regions of the protein is indicated above the sequences. Residues conserved in >90% of the sequences are highlighted with dark gray. Residues conserved in >60% of the sequences are highlighted with light gray. Accession numbers (gi) for each sequence used in the alignment are indicated on the left.
Hypothetical protein YheM
YheM (gi 1789742) is a small protein of unknown function and is another attractive candidate for being a redox protein that uses a thiol/disulfide function. In Chromatum vinosum, it is a part of the locus involved in sulfur oxidation (Pott and Dahl 1998). In E. coli, the gene for this protein is located in an operon with the following gene order: YheO (has one conserved Cys), YheN (has a conserved CxxC site; in some members of the family the second Cys is replaced with alanine), YheM, and YheL (no conserved cysteines are present in this protein). A similar gene organization (at least three genes are in the same order) is also seen in Buchnera sp. APS, Pseudomonas aeruginosa, Vibrio cholerae, Haemophilus influenzae, Pasteurella multocida, and Thermotoga maritima. These data indicate a functional linkage between these four gene products, but none of them has been functionally characterized. The CxxS motif in YheM is located in the C-terminal portion of the protein (Fig. 4 ▶), and we suggest that it is directly involved in redox function.
Fig. 4.
Alignment of proteins of the Yhe family. The location of the CxxS motif in C-terminal regions of the proteins is indicated above the sequences. Residues conserved in >80% of the sequences are highlighted with dark gray. Residues conserved in >60% of the sequences are highlighted with light gray. Accession numbers (gi) for each sequence used in the alignment are indicated on the left.
Putative altronate hydrolases
D-Altronate hydrolases (gi 1789575 and 1789516) belong to a class of Fe2+-requiring enzymes, but the function of iron in these enzymes is not known (Dreyer 1987). Mannonic and altronic hydrolases act, respectively, on mannonate, the intermediate aldonate of the glucuronate branch, and altronate, the intermediate aldonate of the galacturonate branch of the hexuronate pathway, yielding 2-keto-3-deoxy-gluconate. Thiol/disulfide function and/or regulation have not been described for these poorly characterized enzymes. Our data indicate that the identified CxxS sequence is possibly involved in a redox process.
C. jejuni proteins containing the CxxS motif
Methionine- R -sulfoxide reductase
In addition to the E. coli genome (Fig. 2 ▶), this enzyme (gi 6968546) was found in the C. jejuni genome (Table 3). As discussed above, our analyses resulted in functional predictions for this protein, which were subsequently experimentally confirmed.
Table 3.
CxxS-containing proteins identified in the Campylobacter jejuni genome
| Gi number | Protein lengtha | CxxS positionb | CxxS and flanking secondary structurec | Descriptiond |
| 6967954 | 390 | 156 | β-CGGS-α | Putative altronate hydrolase |
| 6968729 | 435 | 194 | β-CHPS-α | Hypothetical protein Cj 1295 |
| 6968546 | 119 | 106 | -CVNS- | Methionine-R-sulfoxide reductase |
a Number of amino acids in an ORF.
b Amino acid position corresponding to Cys in the CxxS sequence.
c CxxS primary sequences and flanking secondary structure (α, α-helix; β, β-strand).
d Predicted function or information obtained by database searches.
Hypothetical protein Cj1295
This is a putative aminopeptidase (gi 6968729). Thiol/disulfide function or redox regulation has not been previously described for proteins of this family.
Predicted altronate hydrolase
This enzyme (gi 6967954) is described above for the E. coli genome.
M. jannaschii proteins containing the CxxS motif
Predicted coding region
This protein (gi 1591280; Table 4) is a homolog of the E. coli OsmC protein (Fig. 3 ▶; Mongkolsuk et al. 1998; Atichartpongkul et al. 2001). As discussed above, this is an excellent candidate for being a redox protein that uses CxxS for redox function.
Table 4.
CxxS-containing proteins identified in the Methanococcus jannaschii genome
| Gi number | Protein lengtha | CxxS positionb | CxxS and flanking secondary structurec | Descriptiond |
| 1591280 | 189 | 172 | α-CLIS | Predicted coding region |
| 1499579 | 275 | 235 | β-CELS-α | Conserved hypothetical protein |
a Number of amino acids in an ORF.
b Amino acid position corresponding to Cys in the CxxS sequence.
c CxxS primary sequences and flanking secondary structure (α, α-helix; β, β-strand).
d Predicted function or information obtained by database searches.
Conserved hypothetical protein
This is a small protein (gi 1499579) conserved in archaea and containing a DsrE-like domain. The Chromatium vinosum cluster dsrABEFHCMK encodes sulfite oxidase polypeptides DsrA and DsrB, four small soluble proteins (DsrE, DsrF, DsrH and DsrC), a transmembrane protein (DsrM) with similarity to heme-b-binding polypeptides, and a soluble protein (DsrK) resembling [4Fe–4S]-cluster-containing heterodisulfide reductase from methanogenic archaea. The cluster was found to be involved in oxidation of sulfur, but specific functions for the proteins involved are not known. Interestingly, approximately half of DsrE-like homologs in archaea contain a CxxC motif in place of CxxS. We predict that the DsrE-like protein identified in our searches participates in a thiol/disulfide redox reaction through its CxxS motif (Fig. 5 ▶).
Fig. 5.
Alignment of DsrE-like proteins. The location of the CxxS motif in C-terminal regions of the proteins is indicated above the sequences. Residues conserved in 100% of the sequences are highlighted with dark gray. Residues conserved in >60% of the sequences are highlighted with light gray. Accession numbers (gi) for each sequence used in the alignment are indicated on the left.
S. cerevisiae proteins containing conserved CxxS: True positives
Glutaredoxins
Five glutaredoxins that contain the CxxS motifs were present in the yeast genome (gi 6319488, 6320193, 6320303, 6321022, and 6325198; Table 5). Three of them have previously been identified and characterized, but the remaining two proteins are newly discovered glutaredoxins. Glutaredoxins show a thioredoxin fold and use CxxC or CxxS motifs and glutathione for thiol/disulfide function (Martin 1995).
Table 5.
CxxS-containing proteins identified in the Saccharomyces cerevisiae genome
| Gi number | Protein lengtha | CxxS positionb | CxxS and flanking secondary structurec | Descriptiond |
| 6319488 | 203 | 108 | β-CPYS-α | Glutaredoxin |
| 6320193 | 231 | 136 | β-CSYS-α | Glutaredoxin |
| 6320303 | 285 | 211 | β-CGFS-α | Glutaredoxin |
| 6321022 | 244 | 171 | β-CGFS-α | Glutaredoxin |
| 6325198 | 150 | 60 | β-CGFS-α | Glutaredoxin |
| 6320726 | 517 | 62 | β-CLHS-α | Protein thiol-disulfide isomerase, ER- |
| 405 | β-CIHS-α | resident protein | ||
| 6321642 | 148 | 90 | β-CMLS-α | Required for arsenate resistance |
| 6325458 | 130 | 76 | β-CTGS-α | Required for arsenate resistance |
| 6320882 | 449 | 198 | CRES-α | S-adenosyl-L-homocysteine hydrolase |
| 6324041 | 373 | 181 | α-CVAS-α | Hypothetical ORF; Caf40p |
| 6324588 | 757 | 583 | α-CISS-α | B-type regulatory subunit of protein phosphatase 2A |
| 6320878 | 72 | 24 | α-CASS | Hypothetical ORF; Yer039c-ap |
| 6321792 | 472 | 3 | CTVS-α | Component of pheromone response pathway |
| 6319497 | 1165 | 448 | CIAS-α | Required for chitin synthesis |
| 6321005 | 801 | 565 | α-CIVS-α | Secretion (Golgi retention) deficient |
| 6322745 | 717 | 405 | β-CGTS-α | Required for biosynthesis of cell wall |
| 6322811 | 256 | 199 | β-CSSS-α | NifU-like protein |
| 6324247 | 904 | 848 | α-CRSS-α | Required for mismatch repair in mitosis and meiosis |
| 6325025 | 1887 | 1305 | CATS-α | Trifunctional enzyme |
| 6319816 | 168 | 157 | -CVNS- | Methionine-R-sulfoxide reductase |
| 14318476 | 674 | 239 | -CFNS- | Hypothetical ORF |
| 6323001 | 250 | 244 | -CGES- | HesB-like protein |
| 6324521 | 396 | 284 | -CGYS- | S-Adenosylmethionine decarboxylase |
| 6325324 | 185 | 177 | -CGSS- | HesB-like protein |
a Number of amino acids in an ORF.
b Amino acid position corresponding to Cys in the CxxS sequence.
c CxxS primary sequences and flanking secondary structure (α, α-helix; β, β-strand).
d Predicted function or information obtained by database searches.
Protein disulfide isomerase
Protein disulfide isomerases are endoplasmic reticulum-resident thioredoxin-fold enzymes that catalyze thiol/disulfide interchange reactions, resulting in formation or rearrangement of protein disulfide bonds. These reactions are important for proper folding and disulfide bonding of proteins. The enzyme identified by our searches (gi 6320726) has two thioredoxin domains, each containing the CxxS motif. This protein has been previously experimentally characterized (Anelli et al. 2002). Interestingly, replacement of CxxS with CxxC resulted in increased disulfide-bond formation. It was proposed that the CxxS motif in this protein participates in redox reactions other than those forming disulfide bonds (Anelli et al. 2002).
Methionine- R -sulfoxide reductase
This true positive (gi 6319816) is discussed above (Fig. 2 ▶).
NifU-like protein
The function of this protein (gi 6322811) is not known. Clusters of Othologous Groups (COG) predict that this protein has a thioredoxin fold and that CxxS corresponds to the thiol/disulfide active center of thioredoxins. Although we also propose the redox function for this protein, secondary structure predictions indicate that it is unlikely to have a thioredoxin fold (data not shown). The cysteine of the CxxS motif may also be considered as part of the upstream CxxC motif. NifU proteins are involved in the formation or repair of iron–sulfur clusters (Olson et al. 2000; Seidler et al. 2001). As discussed above for HesB proteins, iron–sulfur cluster formation requires a redox step, and the CxxS motif is an attractive candidate for this function (Krebs et al. 2001; Ollagnier-de-Choudens et al. 2001).
HesB-like proteins
HesB-like proteins (gi 6323001 and 6325324) were also identified in the E. coli genome (Krebs et al. 2001; Ollagnier-de-Choudens et al. 2001).
Arsenate reductases
Two arsenate reductase homologs (gi 6321642 and 6325458) were identified in the S. cerevisiae genome. These proteins are members of the CDC25A superfamily of protein phosphotyrosyl phosphatases (PTPases), containing a phosphatase signature sequence HCxxxxxR. These proteins require glutathione and glutaredoxin as a source of reducing equivalents (Mukhopadhyay and Rosen 2001). Mutation of the cysteine in the CxxS motif resulted in the loss of arsenate resistance. It was suggested that the CxxS motif, as part of the phosphatase-like Cys–X5–Arg motif, is the catalytic center of the enzyme involved in the reduction of arsenate.
S. cerevisiae proteins: Candidate redox proteins
B-type regulatory subunit of protein phosphatase 2A (B56 family)
This protein (gi 6324588) is an attractive candidate redox protein that may be relevant to both redox and signaling pathways. Serine/threonine phosphatase 2A (PP2A) is a major intracellular protein phosphatase that regulates multiple aspects of cell growth and metabolism. This protein contains highly conserved structural (A) and catalytic (C) subunits. The ability of this widely distributed heterotrimeric enzyme to act on a diverse array of substrates is controlled by a highly variable regulatory and targeting B subunit. At least three distinct gene families encoding B subunits are known: B/B55/CDC55, B`/B56/RTS1, and B"/PR72/130. No homology has been identified among the B families, and little is known about how these B subunits interact with the PP2A A and C subunits (McCright and Virshup 1995; Li and Virshup 2002).
Component of pheromone response pathway
This protein (gi 6321792) is a putative G-protein α-subunit, but the function of this protein has not been characterized in detail. G proteins couple receptors of extracellular signals to intracellular signaling pathways, and their α subunits are weak GTPases. A peptide pheromone can bind to a G-protein transmembrane receptor, leading to activation of a MAP kinase cascade that results in transcriptional induction of genes necessary for the mating process (Dietzel and Kurjan 1987; Boman and Kahn 1995; Coleman and Sprang 1996; Saito et al. 1998).
S-adenosyl-L-homocysteine hydrolase
S-Adenosylhomocysteine (AdoHcy) hydrolase (gi 6320882) catalyzes the reversible hydrolysis of AdoHcy to adenosine and homocysteine, playing an essential role in modulating the cellular homocysteine levels and regulating activities of a host of methyltransferases in eukaryotic cells. Thiol-disulfide function for this protein has not been previously considered, but the levels of homocysteine are known to be regulated through a redox pathway (Turner et al. 2000).
Required for biosynthesis of cell wall
This protein (gi 6322745) catalyzes the first step in the hexosamine pathway, which is required for biosynthesis of cell wall precursors. It contains amidotransferase and phosphosugar isomerase domains that are found in proteins that regulate the expression of genes involved in synthesis of phosphosugars (Bateman 1999).
The remaining yeast proteins identified in our searches either have defined nonredox function, are poorly characterized, or are hypothetical proteins with no known structure. These hypothetical proteins include (1) protein required for mismatch repair in mitosis and meiosis (gi 6324247; DNA mismatch repair enzyme/predicted ATPase); (2) trifunctional enzyme (gi 6325025; contains an ACPS-like domain of the 4`-phosphopantetheinyl transferase superfamily, a ketoacyl-synthase-like domain, and a short chain dehydrogenase domain); (3) hypothetical ORF (gi 14318476; putative membrane-associated glucosyltransferase); (4) S-adenosylmethionine decarboxylase (gi 6324521; involved in the biosynthesis of polyamines); (5) a protein required for chitin synthesis (gi 6319497; probable chitin synthase/glycosyltransferase involved in cell wall biogenesis); (6) hypothetical ORF Caf40p (gi 6324041; homology to different cell differentiation proteins); (7) hypothetical ORF Yer039c-ap (gi 6320878); and (8) secretion-deficient Golgi transport protein (gi 6321005). Further studies are needed to determine which of these proteins are involved in redox processes through their conserved CxxS sequences.
Most CxxS-containing redox proteins contain a downstream α-helix
Analysis of secondary structures for true positive and candidate redox proteins from the four genomes listed in Tables 2–5 revealed that the majority (31 out of 44, or 72%) contained a predicted α-helix immediately downstream of the CxxS motif. This secondary structure pattern was present independently of the structural fold in redox proteins. For example, glutaredoxins, E. coli arsenate reductases, yeast arsenate reductases, OsmC, and HSP33 all showed different folds but had a downstream α-helix flanking the CxxS motif.
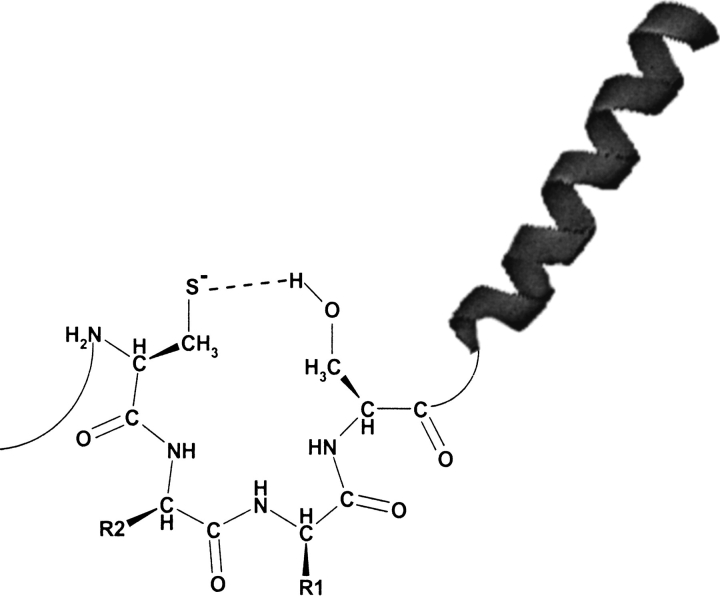
Only 13 proteins identified in the four genomes in our searches did not contain a predicted α-helix downstream of the CxxS motif. Eight of these were HesB and methionine-R-sulfoxide reductases, and three proteins had a predicted α-helix upstream of the CxxS. These data indicate that the presence of an α-helix may assist in some way in redox function provided by the CxxS motif (Fig. 6 ▶). Generation of a dipole that could stabilize the cysteine thiolate in these proteins is a possible explanation of this phenomenon. The presence of the helix could also be a structural feature that preferentially flanks surface-exposed active-site loops in redox enzymes. Independently of the reason for the presence of this helix, a conserved CxxS–α-helix motif would be a strong predictor of thiol-dependent redox function.
Fig. 6.
A model for the involvement of serine and the α-helix in stabilization of cysteine thiolate involved in redox reactions. The presence of an α-helix downstream of the CxxS sequence could also be owing to frequent occurrence of these structures near catalytic surface-exposed loops.
Discussion
The major outcome of our study is identification of a new functional redox motif, CxxS, that is used by structurally distinct families for redox function. The specificity and selectivity of our searches were such that this simple redox motif, when conserved and present in the context of a simple secondary structure pattern, could be used as a predictor of both redox function and location of redox sites in proteins. Some of our predictions are illustrated in Figures 2–5 ▶ ▶ ▶ ▶, and several other predictions are discussed above.
Identification of redox function in proteins is difficult because of the multiplicity of families of redox enzymes and the extreme divergence of their members. Even within the most studied family of thiol-dependent enzymes, thioredoxin-fold proteins, intergroup homology analyses can rarely identify functional relationships (Martin 1995).
To assist in homology-based analyses, various function prediction approaches have been used for identification of redox function, but these are restricted to specific subsets of redox proteins. For example, a sequence—to structure—to function paradigm has been used to identify thiol-disulfide oxidoreductases by threading sequences to a fuzzy functional form descriptor (Fetrow et al. 1998, 2001). However, this approach was limited to thioredoxin-fold proteins containing a conserved proline residue located in close proximity to the active-site cysteine(s).
In contrast to homology- and threading-based algorithms, our approach is not limited to a specific structural fold; rather, it identifies redox function in structurally unrelated proteins. Interestingly, the CxxS groups were typically located in these proteins downstream of an α-helix, implying that this structural element assists in redox function (Fig. 6 ▶). The molecular basis behind the flanking occurrence of an α-helix is not known, but may involve generation of a dipole and stabilization of a cysteine thiolate, or simply be indicative of preferential presence of helices downstream of enzyme active sites.
In any event, the finding that α-helices are frequently located next to cysteine-based redox motifs may be very useful for identification of thiol/disulfide proteins, including those that do not contain the CxxS motif. For example, searches for CxxC-containing thiol/disulfide oxidoreductases are difficult because of the presence of a large number of zinc-containing proteins. However, most of these do not have α-helices downstream of the CxxC, which may allow efficient filtering of these proteins during the searches.
The finding of CxxS motifs in thioredoxin-fold proteins that were located in place of CxxC motifs resulted in considerable debate within the field in regard to the ability of CxxS proteins to catalyze redox reactions that require two cysteines. Recent studies illustrate that at least glutaredoxins and protein disulfide isomerases of this family are active even when only one cysteine is present (Anelli et al. 2002).
We showed that a search for the conserved CxxS motif allows prediction of redox function on a genome-wide basis. It will be of interest to determine how many such proteins are present in the human genome. Considering our analyses of bacterial, archaeal, and eukaryotic genomes, a large number of human CxxS-containing redox proteins may be expected. Further studies are needed to identify such proteins and characterize their functions.
Although our method can be used to identify both redox proteins and redox sites in proteins, it would be incorrect to view these predictions as a proof of identification of thiol-dependent processes. Because the CxxS motif involves only two residues, these can be conserved in some proteins for various reasons. The presence of conserved CxxS sequences upstream of an α-helix is a strong indication of redox function, but further studies may be needed to confirm such predictions.
Finally, our study illustrated that a large number of reactions use CxxS motifs, either with or without the help of other redox cysteines. CxxS motifs can form intermolecular disulfide bonds that may help to retain target proteins in a specific compartment or to serve as intermediates in disulfide-bond formation, isomerization, or reduction (Cotgreave and Gerdes 1998; Carmel-Harel and Storz 2000; Holmgren 2000; Rhee et al. 2000; Tanaka et al. 2000; Finkel 2001; Ritz and Beckwith 2001). Protein disulfide isomerases are examples of such proteins. The CxxS-containing proteins may also provide antioxidant defense using small redox molecules for reduction of various oxidized compounds or proteins. Glutaredoxins that use glutathione as an electron donor are examples of this function (Cotgreave and Gerdes 1998; Carmel-Harel and Storz 2000; Holmgren 2000; Ritz and Beckwith 2001). CxxS may also directly attack certain other forms of oxidized sulfur, such as sulfoxides, with formation of sulfenic acid intermediates, and methionine sulfoxide reductases are an example of such function (Kryukov et al. 2002). Furthermore, in certain proteins, CxxS may be used for both redox function and to coordinate metals, as evident from the analysis of HSP33 proteins containing CxxS and CxxC motifs (Jakob et. al. 2000). Further experimental analyses will undoubtedly provide additional important roles for CxxS, a redox motif present in structurally distinct proteins that was defined in our study.
Materials and methods
The script that we developed for identifying potential redox proteins containing the CxxS motif involves four steps: (1) identification of all proteins containing CxxS; (2) filtering out proteins that use cysteine in the CxxS sequences to bind metals; (3) analysis of a conservation profile of CxxS sequences among homologous proteins; and (4) prediction and analysis of secondary structure for proteins containing the conserved CxxS motif (Fig. 1 ▶).
Analyses were carried under Red Hat Linux 7.2. Perl script was used in steps 1 and 2 for identification of CxxS sequences and filtering out metal-binding proteins, and in steps 3 and 4 for interaction between external programs and in output preparation and visualization. To identify metal-binding proteins, Prosite patterns (release April 2002) were used (Hofmann et al. 1999).
The analysis of conservation profiles was performed using the output file of Position-Specific Iterated BLAST (Altschul et al. 1997). A database of known thioredoxin-fold proteins (>500 proteins) was constructed from the NCBI nonredundant database and was used to optimize the parameters of conservation profile searches. For subsequent genomic searches, these parameters were set as follows: expectation value, 0.0001; optimal number of iterations, 3; and conservation cutoff limit, 65% identity of both cysteine and serine (the CxxS motif) among homologs present in the NCBI nonredundant database. Only the following replacements were allowed: Cys to Ser, Cys to Ala, Cys to Gly, Ser to Cys, Ser to Ala, and Ser to Gly. Proteins that did not satisfy these requirements were automatically removed from further analysis.
For secondary structure prediction we chose PSIPRED (Jones 1999) as a simple, fast, and reliable secondary structure prediction method. The program incorporates two feed-forward neural networks, which perform analysis in an output obtained from PSI-BLAST. Again, a database composed of known thioredoxin-fold proteins was analyzed by PSIPRED to optimize the parameters of secondary structure prediction. The following parameters were then selected: β-strand, 3 or more amino acid residues between −1 and −10 residues (with respect to cysteine in the CxxS motif); and α-helix, 7 or more consecutive amino acid residues between +1 and +20 residues. The accuracy of predicting β-strands and α-helices was ∼78%–80%.
Following the analysis of secondary structures in proteins containing a conserved CxxS motif, these proteins were manually analyzed for sequence homology to proteins with known function (PSI-BLAST), occurrence in completely sequenced genomes, and gene context (Clusters of Orthologous Groups, COG), solvent accessibility of CxxS sequences (for proteins with known structures), and identity of protein domains (DART, Pfam, and COG).
All protein sequences that were used in this work were obtained from GenBank with the following release dates: C. jejuni (June 2001), M. jannaschii (April 2001), E. coli K12 (April 2001), and S. cerevisiae (February 2002).
Acknowledgments
We thank Gregory Kryukov for helpful suggestions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0218302.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli, T., Alessio, M., Mezghrani, A., Simmen, T., Talamo, F., Bachi, A., and Sitia, R. 2002. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 21 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner, E.S. and Holmgren, A. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267 6102–6109. [DOI] [PubMed] [Google Scholar]
- Atichartpongkul, S., Loprasert, S., Vattanaviboon, P., Whangsuk, W., Helmann, J.D., and Mongkolsuk, S. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147 1775–1782. [DOI] [PubMed] [Google Scholar]
- Bateman, A. 1999. The SIS domain: A phosphosugar-binding domain. Trends. Biochem. Sci. 24 94–95. [DOI] [PubMed] [Google Scholar]
- Boman, A.L. and Kahn, R.A. 1995. Arf proteins: The membrane traffic police? Trends. Biochem. Sci. 20 147–150. [DOI] [PubMed] [Google Scholar]
- Carmel-Harel, O. and Storz, G. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54 439–461. [DOI] [PubMed] [Google Scholar]
- Coleman, D.E. and Sprang, S.R. 1996. How G proteins work: A continuing story. Trends Biochem. Sci. 21 41–44. [PubMed] [Google Scholar]
- Cotgreave, I.A. and Gerdes, R.G. 1998. Recent trends in glutathione biochemistry—glutathione–protein interactions: A molecular link between oxidative stress and cell proliferation? Biochem. Biophys. Res. Commun. 242 1–9. [DOI] [PubMed] [Google Scholar]
- Dietzel, C. and Kurjan, J. 1987. The yeast SCG1 gene: A G α-like protein implicated in the a- and α-factor response pathway. Cell 50 1001–1010. [DOI] [PubMed] [Google Scholar]
- Dreyer, J.L. 1987. The role of iron in the activation of mannonic and altronic acid hydratases, two Fe-requiring hydro-lyases. Eur. J. Biochem. 166 623–630. [DOI] [PubMed] [Google Scholar]
- Fetrow, J.S., Godzik, A., and Skolnick, J. 1998. Functional analysis of the Escherichia coli genome using the sequence-to-structure-to-function paradigm: Identification of proteins exhibiting the glutaredoxin/thioredoxin disulfide oxidoreductase activity. J. Mol. Biol. 282 703–711. [DOI] [PubMed] [Google Scholar]
- Fetrow, J.S., Siew, N., Di Gennaro, J.A., Martinez-Yamout, M., Dyson, H.J., and Skolnick, J. 2001. Genomic-scale comparison of sequence- and structure-based methods of function prediction: Does structure provide additional insight? Protein Sci. 10 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, T. 2001. Reactive oxygen species and signal transduction. IUBMB Life 52 3–6. [DOI] [PubMed] [Google Scholar]
- Gladyshev, V.N. 2002. Thioredoxin and peptide methionine sulfoxide reductase: Convergence of similar structure and function in distinct structural folds. Proteins 46 149–152. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., Bucher, P., Falquet, L., and Bairoch, A. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren, A. 2000. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox. Signal. 2 811–820. [DOI] [PubMed] [Google Scholar]
- Jakob, U., Eser, M., and Bardwell, J.C. 2000. Redox switch of hsp33 has a novel zinc-binding motif. J. Biol. Chem. 275 38302–38310. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Kumar, R.A., Koc, A., Cerny, R.L., Gladyshev, V.N. Reaction mechanism, evolutionary analysis and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 2002 (in press). [DOI] [PubMed]
- Kortemme, T. and Creighton, T.E. 1995. Ionisation of cysteine residues at the termini of model α-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 253 799–812. [DOI] [PubMed] [Google Scholar]
- Krebs, C., Agar, J.N., Smith, A.D., Frazzon, J., Dean, D.R., Huynh, B.H., and Johnson, M.K. 2001. IscA, an alternate scaffold for Fe–S cluster biosynthesis. Biochemistry 40 14069–14080. [DOI] [PubMed] [Google Scholar]
- Kryukov, G.V., Kryukov, V.M., and Gladyshev, V.N. 1999. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 274 33888–33897. [DOI] [PubMed] [Google Scholar]
- Kryukov, G.V., Kumar, R.A., Koc, A., Sun, Z., and Gladyshev, V.N. 2002. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. 99 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. and Virshup, D.M. 2002. Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur. J. Biochem. 269 546–552. [DOI] [PubMed] [Google Scholar]
- Martin, J.L. 1995. Thioredoxin—A fold for all reasons. Structure 3 245–250. [DOI] [PubMed] [Google Scholar]
- McCright, B. and Virshup, D.M. 1995. Identification of a new family of protein phosphatase 2A regulatory subunits. J. Biol. Chem. 270 26123–26128. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk, S., Praituan, W., Loprasert, S., Fuangthong, M., and Chamnongpol, S. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. Phaseoli. J. Bacteriol. 180 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, K., Kondoh, A., Stumpp, M.T., and Hisabori, T. 2001. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. 98 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, R. and Rosen, B.P. 2001. The phosphatase C(X)5R motif is required for catalytic activity of the Saccharomyces cerevisiae Acr2p arsenate reductase. J. Biol. Chem. 276 34738–34742. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens, S., Mattioli, T., Takahashi, Y., and Fontecave, M. 2001. Iron–sulfur cluster assembly: Characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem. 276 22604–22607. [DOI] [PubMed] [Google Scholar]
- Olson, J.W., Agar, J.N., Johnson, M.K., and Maier, R.J. 2000. Characterization of the NifU and NifS Fe–S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry 39 16213–16219. [DOI] [PubMed] [Google Scholar]
- Pott, A.S. and Dahl, C. 1998. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144 1881–1894. [DOI] [PubMed] [Google Scholar]
- Rhee, S.G., Bae, Y.S., Lee, S.R., and Kwon, J. 2000. Hydrogen peroxide: A key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE 53 PE1. [DOI] [PubMed] [Google Scholar]
- Ritz, D. and Beckwith, J. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55 21–48. [DOI] [PubMed] [Google Scholar]
- Saito, Y., Kimura, K., Oka, T., and Nakano, A. 1998. Activities of mutant Sar1 proteins in guanine nucleotide binding, GTP hydrolysis, and cell-free transport from the endoplasmic reticulum to the Golgi apparatus. J. Biochem. (Tokyo) 124 816–823. [DOI] [PubMed] [Google Scholar]
- Seidler, A., Jaschkowitz, K., and Wollenberg, M. 2001. Incorporation of iron–sulphur clusters in membrane-bound proteins. Biochem. Soc. Trans. 29 418–421. [DOI] [PubMed] [Google Scholar]
- Shi, J., Vlamis-Gardikas, A., Aslund, F., Holmgren, A., and Rosen, B.P. 1999. Reactivity of glutaredoxins 1, 2, and 3 from Escherichia coli shows that glutaredoxin 2 is the primary hydrogen donor to ArsC-catalyzed arsenate reduction. J. Biol. Chem. 274 36039–36042. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Nakamura, H., Nishiyama, A., Hosoi, F., Masutani, H., Wada, H., and Yodoi, J. 2000. Redox regulation by thioredoxin superfamily; protection against oxidative stress and aging. Free Radic. Res. 33 851–855. [DOI] [PubMed] [Google Scholar]
- Turner, M.A., Yang, X., Yin, D., Kuczera, K., Borchardt, R.T., and Howell, P.L. 2000. Structure and function of S-adenosylhomocysteine hydrolase. Cell Biochem. Biophys. 33 101–125. [DOI] [PubMed] [Google Scholar]
- Woycechowsky, K.J. and Raines, R.T. 2000. Native disulfide bond formation in proteins. Curr. Opin. Chem. Biol. 4 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, H., Wong, J.H., Lee, Y.M., Cho, M.J., and Buchanan, B.B. 2001. A strategy for the identification of proteins targeted by thioredoxin. Proc. Natl. Acad. Sci. 98 4794–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]