Abstract
The NarL response regulatory protein of Escherichia coli has been engineered by covalent modification with 1,10-phenanthroline (OP) to create a set of site-specific DNA-cleaving agents. This was accomplished by introducing single cysteine amino acid replacements at selected locations within the carboxy-terminal DNA-binding domain in or nearby the helix 8 to helix 9 region of the NarL protein using site-directed mutagenesis. Of 18 modified NarL-OP proteins made, 13 retained the ability to bind DNA as evidenced by gel mobility assays, whereas 10 of the 1,10-phenanthroline-modified proteins also exhibited specific cleavage activity for a synthetic NarL recognition sequence. These DNA-cleaving agents were divided into two groups based on the location of the cleavage sites. The first class set cleaved the DNA nearby the center of a synthetic 7–2–7 sequence composed of two NarL heptamer sites separated by a 2-bp spacer element. The second class cut the DNA at the periphery of the 7–2–7 sequence. The cleavage data are consistent with the ability of two NarL monomers to recognize and bind to the DNA in a head-to-head orientation. A second set of DNA-cleaving agents was constructed using the carboxy-terminal domain of NarL called NarLC. Similar cleavage patterns were observed whether full-length NarL or NarLC was used. The availability of 1,10-phenanthroline-modified NarL and NarLC proteins opens up the possibility to explore the position, orientation, and number of NarL recognition sites at E. coli promoters predicted to contain multiple and complex arrangements of NarL-binding sites.
Keywords: NarL response regulator, protein footprinting, copper phenanthroline, site-specific DNA cleaving agent
The NarL DNA-binding protein of Escherichia coli is a prototypical response regulatory protein involved in nitrate-dependent global control of gene expression during anaerobic cell growth conditions. NarL is representative of a large class of bacterial DNA-binding proteins called response regulators, which along with their cognate sensor transmitter proteins, make up distinct two-component regulatory systems (Parkinson and Kofoid 1992; Parkinson 1993). NarL operates along with a companion response regulator called NarP, and two sensor transmitters, NarX and NarQ (Stewart and Rabin 1995),that can independently detect the presence of submicromolar levels of nitrate if present in the cell environment (Lee et al. 1999; Wang et al. 1999). When the respiratory substrate is present, NarX and NarQ autophosphorylate, and in turn activate NarL by the transfer of phosphate to aspartic acid located at position 59. This covalent modification somehow alters the conformation of NarL to allow it to recognize and bind DNA. NarL-phosphate then acts to modulate gene expression by either the positive or negative control of transcription.
The genes targeted by NarL include those involved in anaerobic respiration to a variety of substrates, and genes needed for the fermentation of simple organic compounds (Gunsalus 1992; Gunsalus and Park 1994; Darwin and Stewart 1996). The NarL protein subfamily, also called the FixJ subfamily of response regulators, consists of ∼170 homologs across the bacterial domain. Because NarL is the only response regulator in this family for which the three-dimensional structure is known (Baikalov et al. 1996), it provides a prototype for examining the nature of these protein–DNA interactions.
The E. coli NarL response regulator binds to a poorly conserved heptameric consensus sequence defined as TACYNMT (where Y = C or T, M = A or C, and N = any nucleotide) on the basis of DNase I footprinting experiments and several genetic experiments (Tyson et al. 1994). However, the number, arrangement, and orientation of NarL molecules on the DNA at various NarL-regulated promoters varies (Li et al. 1994), and has not been extensively explored by other approaches.
In many cases, protein-targeted DNA scission has been used to determine the location and orientation of protein on its DNA recognition sequences (Mack et al. 1988; Ebright et al. 1990; Pan et al. 1994; Landgraf et al. 1996;, Murakami et al. 1997). In the present study we have covalently attached a DNA-cleaving agent, 1,10-phenanthroline (OP)–copper complex, to NarL to evaluate its ability to bind and cut at a synthetic NarL recognition sequence. A complementary set of site-specific DNA-cutting reagents is defined herein, which should have application in resolving these issues of NarL site arrangements. The results of these studies should also provide useful approaches to the investigation of related response regulatory proteins of the NarL subfamily.
Results
OP modification of the NarL protein
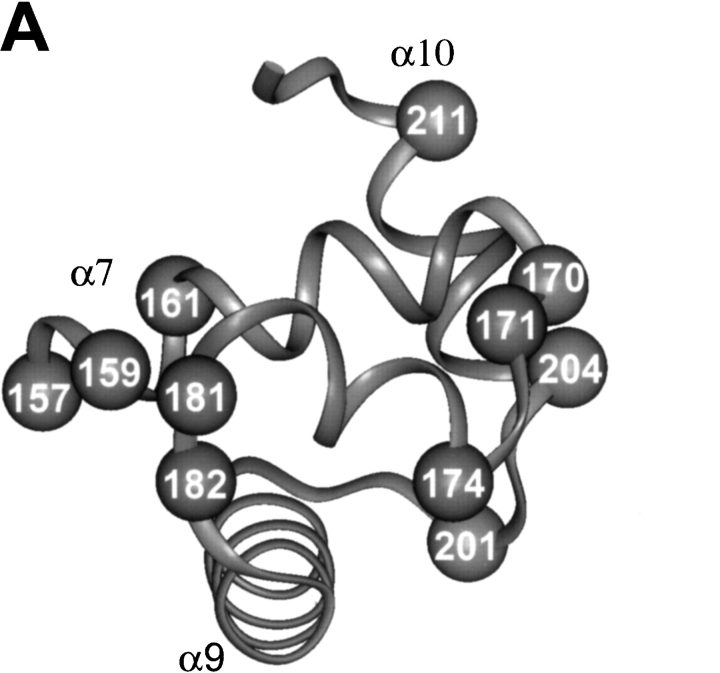
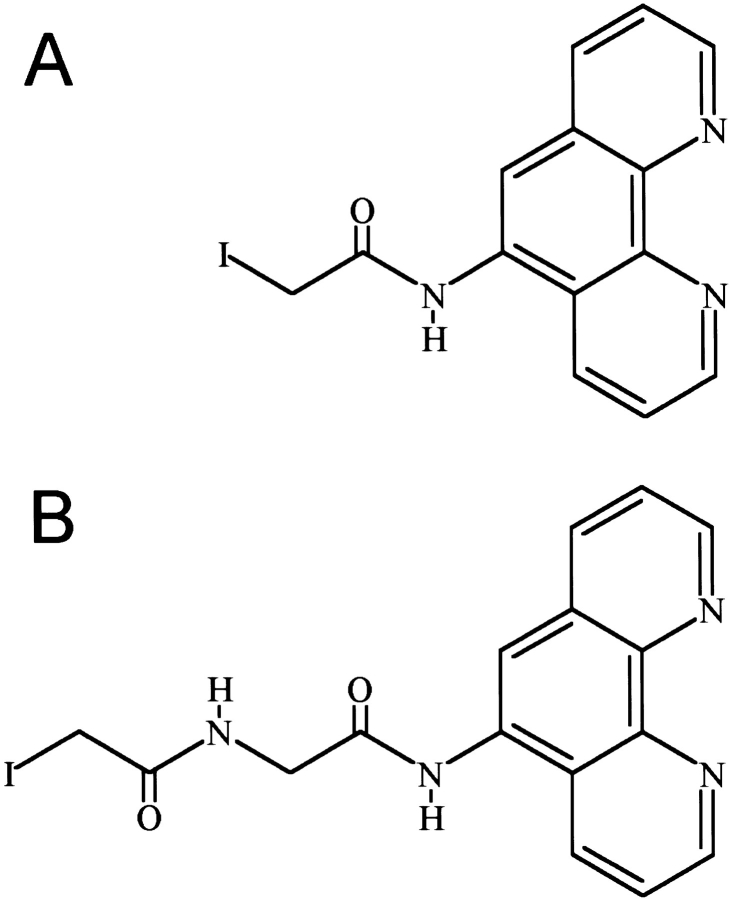
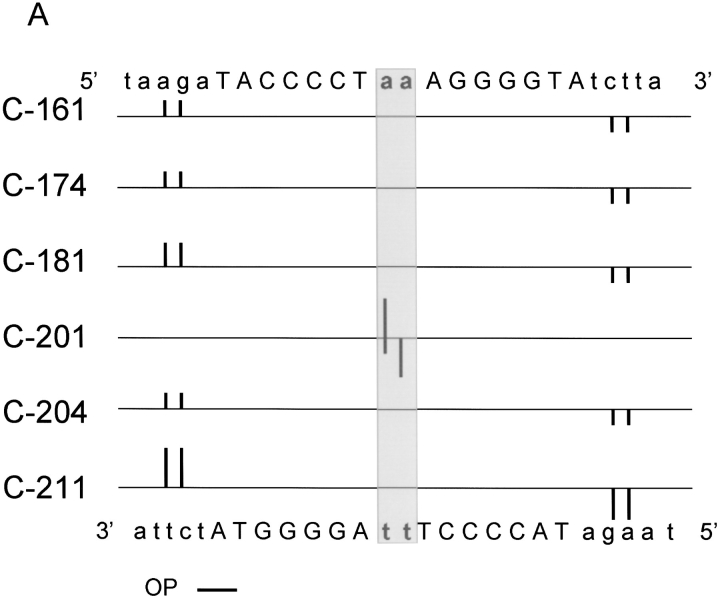
With the goal of creating NarL-OP-based DNA-cleaving agents to examine the interaction of NarL with DNA, we have used site-directed mutagenesis to introduce single cysteine replacements at selected locations in NarL to attach the OP moiety. The locations of these amino acid substitutions were based on the crystal structure of the nonphosphorylated form of NarL (i.e., inactive for DNA binding), as this is currently the only relevant structure available (Baikalov et al. 1996). For a DNA template, we used a template containing a synthetic NarL DNA 7–2–7 recognition sequence that is bound by NarL as evidenced by gel mobility shift and by DNase I footprinting methods (S. Bearson, unpubl.). This DNA contains two consensus NarL heptamer sites TACYNMT (Tyson et al. 1994) oriented in an inverted symmetrical arrangement and separated by a 2-bp spacer, TACCCCTaaAGGGGTA. NarL-phosphate and NarLC have been shown to bind this DNA with nearly equal affinity, whereas NarL binds with a 10-fold weaker affinity (S. Bearson, unpubl.). The initial chemical modification experiments were also performed using the carboxy-terminal domain of NarL (NarLC) as this protein binds DNA in a constitutive fashion (i.e., independent of the phosphorylation-dependent activation of the NarLN domain). Eleven amino acids at positions nearby or within the DNA recognition helix 8 and helix 9 within NarLC were individually replaced with cysteine (Fig. 1A ▶). After narL mutagenesis, each mutant NarLC protein was overexpressed, and purified to homogeneity (see Materials and Methods). Each intended mutation was confirmed by DNA sequence analysis (data not shown). Of the 11 modifications tested, each mutant protein was produced in a soluble form that could then be covalently modified using the 5-iodoacetylamido-1, 10-phenanthroline (IOP) or 5-iodoacetylglycylamido-1, 10-phenanthroline (IAOP) reagents depicted in Figure 2 ▶ (see Materials and Methods).
Fig. 1.
The location of individual cysteine residues in the full-length NarL and the carboxy-terminal NarL domain (NarLC) used for copper phenanthroline attachment. The amino acids chosen for replacement in A, NarLC (top) and B, NarL (bottom) are numbered relative to the amino-terminal methionine as residue number 1.
Fig. 2.
Structure of the OP-copper reagents used for protein modification: 5-iodoacetamido-1, 10-phenanthroline (IOP) and 5-iodoacetylglycylamido-1,10-phenanthroline (IAOP). Syntheses of these two compounds were described previously (Sigman et al. 1991; Shang et al. 1994).
Ability of OP-modified NarLC proteins to bind DNA
To test whether the OP-modified NarLC proteins (Fig. 1A ▶) retained the ability to bind DNA, a standard gel mobility assay was performed with the synthetic 7–2–7 DNA sequence. Of the 11 OP-modified proteins tested, 6 retained the ability to bind DNA like the wild-type NarLC protein (data not shown). They were NarLC R161C-OP, NarLC K174C-OP, NarLC D181C-OP, NarLC K201C-OP, NarLC V204C-OP, and NarLC H211C-OP. In addition, the NarLC G170C-OP, NarLC L171C-OP, and NarLC I182C-OP proteins also bound DNA but did so with somewhat weaker affinity than wild-type NarLC (data not shown). The two remaining mutant NarLC proteins, NarLC T157C-OP and NarLC R159C-OP, did not bind DNA under the standard assay conditions. Because the unmodified form of each protein (i.e., T157C and R159C) also failed to bind DNA (data not shown), the OP side group was not responsible for the observed interference in DNA binding.
DNA cleavage by OP-modified -NarLC proteins
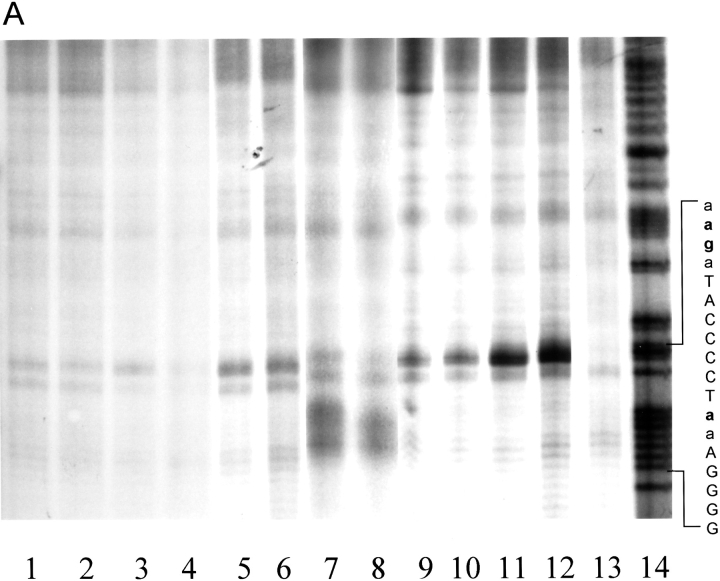
The ability of each modified NarLC protein that retained ability to bind DNA was then tested for cleavage activity using the synthetic NarL 7–2–7 sequence as a DNA substrate (Fig. 3 ▶). After gel separation, both the retarded and the free DNA bands were isolated and immersed in a standard reaction buffer. The DNA scission reaction was initiated by the addition of CuSO4, mercaptopropionic acid (MPA), and H2O2, and the reaction was allowed to proceed for 30 min at room temperature. The reaction was quenced and the 32P-labeled products were analyzed on an 8% denaturing acrylamide gel. The cleavage data for the top strand of DNA is shown in Figure 3A ▶. Two chimera NarLC proteins demonstrated strong cutting efficiency, NarLC K201C-OP and NarLC H211C-OP. Four NarLC mutant proteins, R161C-OP, K174C-OP, D181C-OP, and V204C-OP, cleaved DNA moderately relative to the above two proteins. Finally, although NarLC G170C-OP retained DNA-binding activity, it failed to cleave DNA (data not shown). Each NarLC-OP protein that exhibited cleavage activity also demonstrated site-specific cleavage: NarLC K201C-OP cleaved the DNA near the center of the 7–2–7 motif (lanes 7 and 8), whereas the other NarLC-OP chimera proteins cleaved DNA two bases upstream of the 7–2–7 motif (Fig. 3 ▶, lanes 1–6, 9–12).
Fig. 3.
Gel matrix DNA scission patterns by the NarLC mutant proteins modified with either IOP or IAOP. The 171-bp (top strand) and 180-bp (bottom strand) DNA fragments were 32P labeled on the 3′ ends as described in Materials and Methods. (A) Top strand DNA scissions. (Lane 1) NarLC R161C-OP; (lane 2) NarLC R161C-AOP; (lane 3) NarLC K174C-OP; (lane 4) NarLC K174C-AOP; (lane 5) NarLC D181C-OP; (lane 6) NarLC D181C-AOP; (lane 7) NarLC K201C-OP; (lane 8) NarLC K201C-AOP; (lane 9) NarLC V204C-OP; (lane 10) NarLC V204C-AOP; (lane 11) NarLC H211C-OP; (lane 12) NarLC H211C-AOP; (lane 13) control without protein; (lane 14) Maxam Gilbert G + A reaction. (B) Bottom strand DNA scissions. (Lane 1) NarLC R161C-OP; (lane 2) NarLC R161C-AOP; (lane 3) NarLC K174C-OP; (lane 4) NarLC K174C-AOP; (lane 5) NarLC D181C-OP; (lane 6) NarLC D181C-AOP; (lane 7) NarLC K201C-OP; (lane 8) NarLC K201C-AOP; (lane 9) NarLC V204C-OP; (lane 10) NarLC V204C-AOP; (lane 11) NarLC H211C-OP; (lane 12) NarLC H211C-AOP; (lane 13) control without protein; (lane 14) Maxam Gilbert G + A reaction. The lettering shown on the side of each panel in boldface type indicates the sites of DNA cleavage. Upper case letters indicate the heptamer sequences and the lower case letters are flanking sequences.
The ability of the OP-modified NarLC proteins to cleave the bottom strand of DNA is shown in Figure 3B ▶. As with the top strand, NarLC K201C-OP cleavage occurred near the center of the 7–2–7 DNA motif (lanes 7 and 8). As also seen with the top strand, NarLC H211C-OP cut two bases outside the heptamer sequence in the downstream flanking region on the bottom strand. The sites of DNA cleavage for the other remaining NarLC-OP chimeric proteins were also located two bases downstream from the 7–2–7 motif (Fig. 3B ▶, lanes 1–6, 9–12). Thus, the cutting patterns for both strands show an equal-spaced twofold axis of symmetry.
When the mutated NarLC proteins were modified with IAOP (see Fig. 2 ▶) rather than with IOP, a nearly identical DNA cleavage pattern was seen, except in the case of NarLC K201C where a delocalized DNA scission pattern occurred on the bottom strand (Fig. 3B ▶, lanes 7 and 8). This NarLC K201C-AOP protein possesses a 4-Å longer linker arm than does the NarLC K201C -OP protein (see Fig. 2 ▶; Pan et al. 1996). The extended reach apparently provides increased rotational freedom of the OP–Cu+ complex, and thus the ability to cut at adjacent bases on the DNA. In control experiments performed without added NarLC protein, limited scission was seen when the DNA was treated with cleavage reagents Cu++, MPA, and H2O2 (Fig. 3A,B, lane 13 ▶). These nonspecific cleavages were not included in the NarL-specific data analysis.
Ability of OP-modified full-length NarL proteins to bind DNA
To examine if the full-length form of NarL could also be converted into a DNA-cleaving agent by OP modification, seven individual cysteine mutations were introduced into narL at positions 161,174,181,192, 201,204, and 211 as was described for NarLC (see Fig. 1B ▶). All of the mutated proteins were successfully overproduced, purified, and subsequently modified with IOP as done with NarLC. The standard gel mobility-shift assay was then used to evaluate which of these full-length OP-modified proteins were able to bind DNA and what effect protein phosphorylation had on their binding ability. Protein phosphorylation was carried out in the presence of 50 mM acetyl phosphate as previously described (Schroeder et al. 1994). This procedure was demonstrated to cause phosphorylation of 90%–100% of the wild-type NarL protein. Of the seven proteins examined, four bound the DNA fragment like the wild-type NarL protein (NarL D181C-OP, NarL K201C-OP, NarL V204C-OP, and NarL H211C-OP). The three remaining proteins, NarL R161C-OP, NarL K174C-OP, and NarL K192C-OP showed loss of DNA binding under standard assay conditions. The nonphosphorylated proteins also bound DNA but migrated as slower species than the corresponding phosphorylated ones (data not shown).
DNA cleavage by OP-modified -NarL proteins
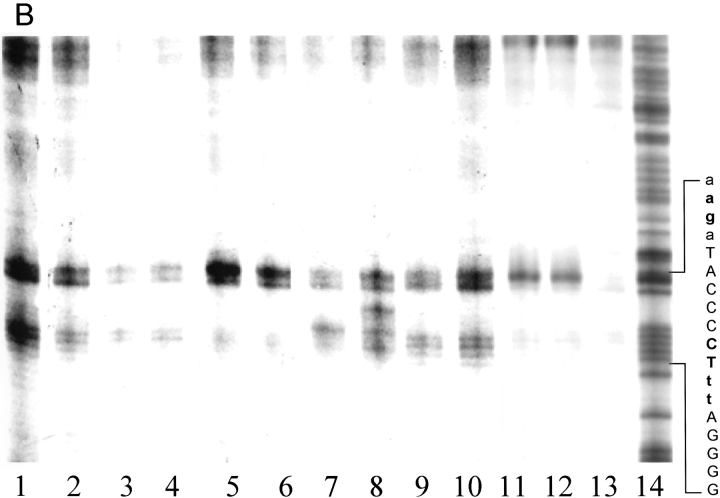
DNA scission reactions using full-length NarL proteins were performed in the same manner as those with NarLC. Figure 4A ▶ shows the cleavage data for the top strand. Protein chimeras were tested with or without phosphorylation before binding. Phosphorylated NarL K201C-OP showed significant DNA cleavage at the position located two bases downstream from the center of the 7–2–7 motif. The H211C-OP phospho-protein cleaved DNA two bases upstream of the 7–2–7 motif. Protein chimeras without phosphorylation cleaved DNA at a reduced efficiency (lanes 1 and 3). Although the cleavage pattern for NarLH211C-OP was independent of protein phosphorylation, the results differed for NarL K201C-OP (lanes 2 and 4). The nonphosphorylated K201C-OP protein cleaved at the center rather than two bases downstream from the center of the 7–2–7 sequence when the chimera protein was phosphorylated. These findings suggest that the phosphorylated and nonphosphorylated forms of the protein may recognize or bind DNA slightly differently.
Fig. 4.

Gel matrix DNA scissions by full-length mutant NarL-OP proteins. (A) Top strand DNA scissions. All proteins were phosphorylated. (Lane 1) NarL D181C-OP (+PO4); (lane 2) NarL K201C-OP (+PO4); (lane 3) NarL V204C-OP (+PO4); (lane 4) NarL H211C-OP (+PO4); (lane 5) control with no protein. (B) Bottom strand DNA scissions. The experiments using phosphorylated NarL are marked with (+PO4), and the ones lacking phosphate are marked without (+PO4). (Lane 1) NarL K201-OP; (lane 2) NarL K201C-OP (+PO4); (lane 3) NarL H211C-OP; (lane 4) NarL H211C-OP (+PO4); (lane 5) control with no protein. Proteins were phosphorylated as described in Materials and Methods as is the DNA cleavage procedure. The boldface letters at the side of the DNA ladder indicate the sites of cleavage.
The DNA cleavage data for the bottom strand is shown in Figure 4B ▶. Two proteins, NarL K201C-OP and NarL H211C-OP, exhibited strong DNA-cutting efficiency. Two other OP-modified proteins, NarL D181C-OP and NarL V204C-OP, cut the DNA weakly. The cleavage patterns from full-length K201C-OP and H211C-OP are almost identical to those from the carboxyl terminus, with K201C-OP cleaving at the center and H211C-OP cleaving at two bases downstream from the side of the 7–2–7 motif.
Summary of DNA cleavage patterns for OP-modified NarLC and NarL proteins
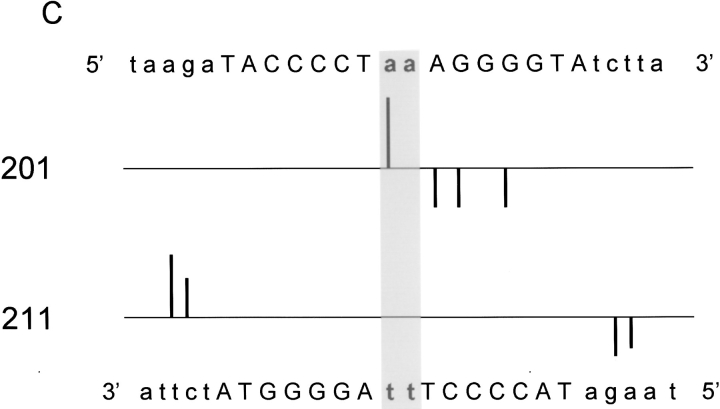
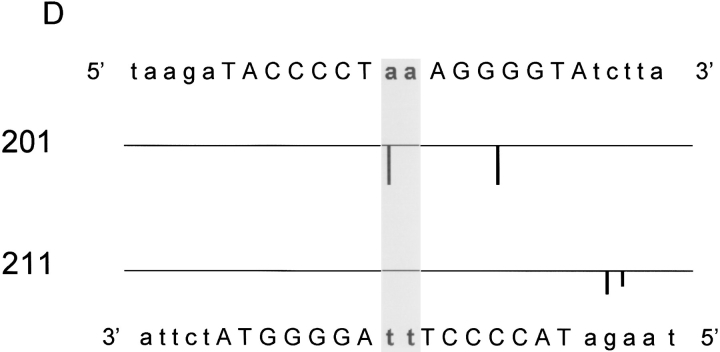
The DNA scission patterns for both the NarLC-OP and NarLC-AOP proteins (see Fig. 3 ▶) and for the full-length NarL-OP proteins (Fig. 4 ▶) are depicted as histograms in panels A, B, C, and D of Figure 5 ▶, respectively. The patterns are highly conserved among the various protein reagents (discussed below).
Fig. 5.
(Legend on facing page.)
Summary of the DNA scission patterns for the 7–2–7 NarL recognition site by NarL-OP and NarLC -OP proteins. The DNA histograms show the cleavage of the top and bottom strands of the DNA. The sequence of the top strand is shown on the top of each panel, and the sequence of the bottom strand is shown on the bottom of each panel. The mutant proteins used for the cleavage experiments are indicated on the left. The upward bars indicate the cleavage pattern for the top strand and the downward ones indicate the cleavage pattern for the bottom strand. The bar size reflects the intensity of the cleavage. (A) Histogram showing cleavage by NarLC-OP. (B) Histogram showing cleavage by NarLC-AOP. (C) Histogram showing cleavage by NarL-OP-phosphate. (D) Histogram showing cleavage by NarL-OP (nonphosphorylated form).
Discussion
Previous studies have demonstrated that many DNA-binding proteins can be readily converted into site-specific DNA-cleaving agents (Chen et al. 2001). This conversion is achieved by linking OP to a native cysteine within a DNA-binding protein, or to an introduced cysteine. In the presence of CuSO4 and mercaptopropionic acid, these modified DNA-binding proteins cleave DNA by oxidatively attacking the deoxyribose moiety. This strategy has been successfully used to study the orientation of protein-binding sites on DNA. The proteins studied using this approach include Fis (Pan et al., 1994, 1996), TrpR (Sutton et al. 1993; Landgraf et al. 1996), Engrailed (Pan et al. 1995), Cro (Bruice et al. 1991), CAP (Ebright et al. 1990; Pendergrast et al. 1994), OmpR (Harrison-McMonagle et al. 1999), Msx-1 (Shang et al. 1994), and LexA (Dumoulin et al. 1996). In this study we were able to successfully construct a family of complementary protein reagents for NarL and NarLC that recognize and bind specifically to DNA target sequences composed of two NarL heptamers organized in a 7–2–7 arrangement. The DNA cleavage data are consistent with two molecules of NarL (or NarLC) protein bound in a head-to-head arrangement on the synthetic 7–2–7 NarL recognition sequence (Fig. 5 ▶). Although the binding of full-length NarL to DNA normally occurs in a phosphorylation-dependent fashion, the present studies were done at elevated protein levels where nonphosphorylated NarL also binds DNA (S. Bearson, unpubl.). This was necessary due to the requirements of the OP Cu+ assay.
DNA-binding affinities of NarL mutant proteins
Full-length NarL protein consists of a carboxy-terminal domain (9 kD) that recognizes and binds DNA and an amino-terminal domain (15 kD) that can be activated through phosphorylation at aspartic acid residue 59. As shown previously by gel retardation assay, the binding abilities of wild-type NarLC and full-length NarL protein upon phosphorylation to DNA were equivalent. Not all OP-modified NarL mutants behaved the same; although NarLC R161C-OP and NarLC K174C-OP maintain their binding ability, the corresponding full-length NarL mutants were unable to bind DNA. This suggests a subtle difference of conformation of the carboxy-terminal domain relative to the amino-terminal domain in the full-length NarL protein. NarL K192C is located on α helix 9, the recognition helix where the protein and DNA are proposed to contact (Baikalov et al. 1996). This protein also failed to bind. It is also noted that although the nonphosphorylated NarL mutant proteins were able to bind DNA, the mobility of their protein–DNA complexes is noticeably slower than that of the phosphorylated samples. The difference in mobility could be due to (1) increase of negative charge in the phosphorylated protein, or (2) conformation changes in NarL induced by phosphorylation of D59.
Analysis of DNA cleavage
The substrate DNA used in this study is a synthetic sequence containing two NarL consensus heptamers organized in a 7–2–7 arrangement similar to that found in the nirB promoter (cgTACCCATtaATGGGTAcg) (Tyson et al. 1993; Wu et al. 1998). Previous studies have shown that the binding of NarL to DNA containing a half-site is significantly weaker by ∼8- to 10-fold (S. Bearson, unpubl.), which suggests that NarL assembles onto the DNA to form a homodimer. Although a head-to-head arrangement of monomers in the dimeric complex is suggested by DNase I footprinting, there is no direct evidence to substantiate this arrangement. The targeted scission data generated by NarL-OP conjugates provides a means to confirm this proposal.
Among the chimeric NarL mutant proteins examined (i.e., either full-length or the carboxy-terminal domain), the K201C-OP and H211C-OP reagents are the most efficient DNA cutters. This indicates that amino acid 201 on the α helix 9 to α helix 10 loop and amino acid 211 on the carboxy-terminal end of α helix 10 where OP is tethered must be properly positioned and accessible to the deoxyribose bond where DNA cleavage occurs.
In general, the overall cleavage patterns generated by NarLC-OP, NarL-OP(+PO4), or NarL-OP are similar to one another (Figs. 3 and 4 ▶ ▶). Only the NarL mutant K201C-OP shows cleavage in the center and also on both strands of the DNA. Cleavage patterns from the rest of mutant-OP proteins show two clusters of cleavage on specific strands of the DNA, equally spaced from the center of twofold symmetry. The only scenario that can account for this result is that two NarL monomers positioned on the DNA in a head-to-head fashion with a dyad symmetry.
A closer examination of the cleavage patterns among NarLC-OP, NarL-OP with phosphorylation, and NarL-OP without phosphorylation revealed some minor differences. NarL K201C-OP cleaves the bottom strand DNA with two different features from NarLC K201C-OP where (1) the cleavage site is shifted two nucleotides downstream from the center where NarLC-OP cuts, and (2) an additional cleavage site is seen at four nucleotides downstream from the dyad center. No cutting at this site occurs in the NarLCK201C-OP cleavage pattern. This difference in cutting ability could be due to the positioning of the 15 kD amino-terminal domain (NarLN) in the full-length NarL mutant. As noted by Baikalov, the NarLN and NarLC domains of NarL must adopt a different conformation relative to each other when bound to DNA versus when in the uncomplexed state (Baikalov et al. 1996). A distinct conformation for NarL versus NarL-phosphate on the DNA may account for the slightly different cutting patterns where the monomers are orientated somewhat differently (Fig. 5 ▶). Finally, both monomers on the DNA are apparently each phosphorylated, as the cutting patterns for NarL versus NarL-PO4 were distinct.
Model of NarL–DNA complex
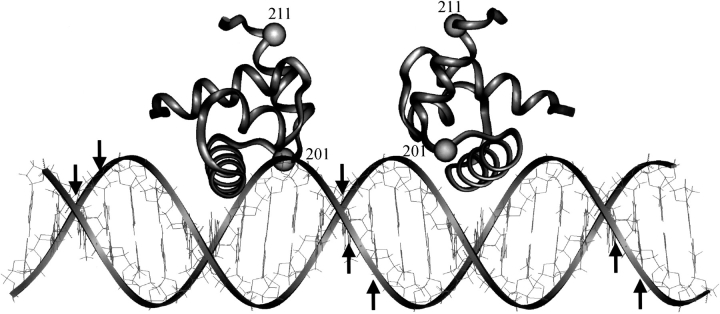
A model shown in Figure 6 ▶ was constructed using linear B form DNA, and the NarLC domain coordinates obtained from the protein data bank (file #1RNL) to illustrate the OP–Cu+ cleavage data. Two NarLC monomers are placed on the DNA in a symmetrical arrangement. The NarL α helix 9, one of the two DNA recognition helices, is placed close to the major groove of the DNA, with residue 192 contacting the DNA. This arrangement would be in agreement with the loss of DNA binding when NarL residue 192 is mutated from lysine to cysteine. Each monomer is positioned such that residue 201 is located toward the minor groove and on either side of the 7–2–7 dyad axis. They are also juxtaposed with the resulting distances between the cysteine of residue 201 and the C1′ atoms of the DNA cleavage sites ∼13–14 Å. This distance is well within the range of the length of OP linker segment, thus allowing the cleavage of deoxyribose to occur.
Fig. 6.
Model of the NarLC–DNA complex showing the locations of residues 201 and 211 in each NarLC monomer. Two molecules of NarLC are positioned above the major grooves of linear DNA at the 7–2–7 motif. The down arrows indicate the positions of DNA cleavage on the top strand and the up arrows indicate cleavage on the bottom strand. The positions of amino acids 201 and 211 are indicated on the ribbon structure by the small spheres.
The crystal structure of a NarLC–DNA complex indicates that α helix 10 may serve a role in NarL dimerization (A. Maris, pers. comm.). This model would position residue 211 of the NarL protein too far from the observed DNA cleavage sites. For the cleavage to occur, DNA flanking the core region has to wrap around a NarL homo-dimer. Due to possible structural differences between the NarL crystalline and liquid states, and the length of DNA used, a significant bending within the DNA recognition sequence beyond that observed in the crystalline state may occur. In fact, DNA cleavage by NarL H211C-OP on the top strand DNA occurs only upstream from the core binding site, whereas on the bottom strand cleavage occurs only downstream from the core. The difference in the chemical reactivity toward the top and bottom strand of DNA may be attributed to this bending of DNA, or to the narrowing of minor groove, which makes the deoxyribose on only one strand of DNA accessible to OP–Cu+.
In conclusion, tethering OP–Cu+ at amino acid residues 201 and 211 in the NarL protein yield efficient and specific DNA cleavage reagents. Relatively minor differences were observed in the cleavage patterns whether full-length NarL or the carboxy-terminal domain of NarL was used. This suggests that the geometry of the NarL molecule orientation on the DNA is essentially identical. DNase I footprinting studies support this proposal (S. Bearson, unpubl.). Thus, the carboxy-terminal domain NarL proteins K201C-OP and H211C-OP should be attractive reagents to study the interactions of NarL-regulated promoters where the NarL consensus recognition sequences are partially defined (e.g., narK, narG, nirB, frfA, napF, and frdA (Li et al. 1994), or for those where no data are currently available. The number, position, and orientation of NarL monomers on these promoters can then be experimentally addressed. The approach described herein for NarL may also have application for other NarL subfamily response regulators, including UhpA, BvgA, DegU, and ComA (Parkinson 1993).
Materials and methods
Materials
Oligonucleotides used for mutagenesis, subcloning, and sequencing were purchased from GIBCO-BRL (Gaithersburg, MD.). HiTrap Chelating HP column (1 mL) was purchased from Amersham Pharmacia Biotech. All restriction endonucleases, polymerases, and other enzymes were purchased from New England Biolabs (Cambridge, MA). 5-iodoacetylamido-1,10-phenanthroline (IOP) (Sigman et al. 1991) and 5-iodoacetylglycylamido-1,10-phenanthroline (IAOP) (Shang et al. 1994) were synthesized according to the published procedure.
Bacterial strains and plasmids
E. coli strain JM109 and plasmid pQE9 (Qiagen, Chatsworth, CA) were used for protein expression experiments.
Site-directed mutagenesis and subcloning
The Kunkel mutagenesis method with the M13MK1 template was used for site-directed mutagenesis as described previously (Kunkel 1985). Double-stranded M13MK1 containing the mutations were amplified using PCR with two primers, GX41 (CCATCACGGA TCCAGTAATCAGGAACCGGCTA) and GX42 (CTGGGAAA AGCTTTCAGAAAATGCGCTCCTGAT), for the full-length NarL mutants. For the NarL carboxy-terminal domain mutants starting from residue A147, the primer GX47 (CTGGGAAAA GCTTCCGTAATCAGAAAATGCGCTCC) instead of GX41 was used. The amplified fragments were digested with BamHI and HindIII restriction enzymes, and then subcloned into the vector pQE9 digested with the same enzymes. Thus, each overexpressed NarL or NarLC protein contains the MRGSHHHHHHGS sequence at the amino terminus. The host strain JM109 was used for overexpression of the NarL mutant proteins. The intended mutagenesis and subcloning events were confirmed by DNA sequencing.
Overexpression and purification of (His)6-tagged NarL mutant proteins
E. coli JM109 cells containing the indicated overexpression plasmid were grown in 10 mL of LB broth with 100 μg/mL ampicillin overnight at 37°C. The cells were transferred to 500 mL of LB broth containing 100 μg/mL ampicillin and grown at 37°C until an OD600 of 0.7–1.0. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 2 mM. The cultures were incubated for 3 additional h, chilled to 4°C, and cells were harvested by centrifugation at 4000 rpm. Cell pellets were stored at −70°C, or directly resuspended for protein purification. Cell material (∼1.5 g wet weight) was resuspended with 12 mL of resuspension buffer (20 mM phosphate buffer at pH 7.8, 10 mM imidazole, 1 M NaCl, 2 mM MgCl2) containing 14 μL of DNase I and RNase A solution (10 mg/mL RNase A, 5 mg/mL DNase I), and then passed through a French Pressure Cell four times at 14,000 psi. Cell disruption was monitored microscopically to confirm >90% cell breakage. The cell suspension was centrifuged at 15,000 rpm for 0.5 h at 4°C, and the supernatant fraction was decanted and applied to a Pharmacia HiTrap Chelating HP column (1 mL). For FPLC chromatography, all buffers were filtered and then degassed for 15 min immediately before use. The Hi-Trap chelating column (1 mL) was washed and eluted at a flow rate of 1 mL/ min. Nickel sulfate or nickel chloride was preloaded onto the column before protein samples were applied. The wash buffer was 20 mM phosphate buffer at pH 7.8, 10 mM imidazole, 1 M NaCl, and the elution buffer was 20 mM phosphate buffer at pH 7.8, 500 mM imidazole, 0.1 M NaCl. The column was washed with 20% elution buffer until the OD280 was restored to baseline. The (His)6-tagged NarL proteins were typically eluted at a 60%-to-40% ratio of wash-to-elution buffer. The NarL carboxy-terminal domain protein was concentrated using a Centricon-3 filter (Amicon), whereas the full-length NarL protein was concentrated using a Centricon-10 filter. The protein was dialyzed with storage buffer (20 mM Tris-HCl at pH 7.0, 500 mM NaCl, 10% glycerol), and quickly frozen in a dry ice/ethanol bath.
Phosphorylation of NarL proteins
NarL and mutant proteins were phosphorylated by acetyl phosphate at a ratio of 1,280:1 acetyl phosphate/protein in 50 mM MOPS at pH 7.0, 50 mM CaCl2, 0.5 mM EDTA, and 20 mM MgCl2 as previously described (Schroeder et al. 1994). The protein concentration was typically 200 μM.
Preparation of DNA
The 263-bp DNA fragment containing the synthetic NarL 7–2–7 sequence was obtained by PCR of plasmid pIS69 (S. Bearson, unpubl.). The 3′-labeling of the top strand was achieved by restricting the PCR product with HindIII, and then end-filled with [α-32P]dATP to give a 171-bp DNA fragment. To 3′-label the bottom strand, the PCR fragment was restricted with EcoRI, and then end-filled with [α-32P]-labeled dATP to give a 180-bp DNA fragment. Both labeled fragments were subsequently purified by 8% nondenaturing acrylamide gel.
Derivatization of NarL mutant proteins
For standard derivatization (Sutton et al. 1993) of both full-length and carboxy-terminal NarL mutant proteins (NarLC), 20 μL of protein (200 μM) in 20 mM Tris at pH 8.0, 1 mM EDTA, 500 mM NaCl, 10% glycerol, and 1 mM DTT was incubated with 5-iodoacetamido-1,10 phenanthroline (IOP) or 5-iodoacetylglycylamido-1,10 phenanthroline (IAOP) in dimethyl formamide (20 mM) at 4°C overnight. The amount of OP reagent used was 10 times the amount of protein used plus 40 nmoles for DTT. Derivatized protein was analyzed by mass spectrometry: the OP-labeling efficiency was demonstrated to be 95% to 100% with one OP molecule per protein monomer.
Gel shift assay for DNA binding
The 32P-labeled DNA fragment containing the 7–2–7 NarL binding site (∼40,000–50,000 cpm) was dissolved in 12.5 μL of buffer with a final concentration of 10 mM Tris at pH 7.5, 50 mM KCl, 7 mM CaCl2, 500 ng of poly[d(I-C)] and 10% glycerol (v/v). The indicated NarLC-OP (2.5 μL or 0.12 μg of protein), or the NarL-OP (2.5 μL or 0.18 μg of protein) in either the phosphorylated or nonphosphorylated protein in 50 mM MOPS at pH 7.0, 50 mM CaCl2, 0.5 mM EDTA, and 20 mM MgCl2 was then added. The final protein concentration was 1 μM for NarLC, and 500 nM for full-length NarL. After incubating at room temperature for 10 min, the protein–DNA mixture was loaded onto a native 6%, 0.5× TB polyacrylamide gel with 2% glycerol. The EDTA was omitted from both the gel and gel-running buffer to prevent chelating the copper, which would inhibit DNA cleavage by NarL-OP in subsequent steps. The gel was run at 250 V at 4°C.
DNA cleavage
The cleavage reactions were carried out within the gel matrix as described (Sutton et al. 1993). After gel separation, slices containing the DNA–protein complex were immersed in 100 μL of buffer (20 mM Tris-Cl at pH 7.5, 100 mM KCl, 14 mM CaCl2). The OP-Cu+ scission reaction was initiated with the addition of 1 μL of 9 mM CuSO4, 10 μL of 58 mM 3-mercatopropionic acid, and 1μL of 0.92 M H2O2. The reaction proceeded for 30 min at room temperature. The scission products were analyzed on an 8% denaturing polyacrylamide gel.
Molecular modeling
The modeling of the NarLC–DNA complex was performed using the Insight II software (Molecular Simulations). The same DNA sequence used for gel shift and DNA cutting experiments was used for the linear B DNA fragment.
Acknowledgments
We thank Imke Schroeder for the construction of plasmid pIS69, Jenney Yang and Yandong Zhao for technical assistance, and Ben Wong for his assistance in constructing the protein–DNA model. We also acknowledge the expert services of the UCLA Mass spectrometer facility for NarL-OP analysis.
This work was supported in part by grants from the National Institutes of Health, AI21678 (RG) and GM21199 (DS).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
NarL, NarL response regulator protein
NarLc, NarL carboxy-terminal domain
NarLN , NarL amino-terminal domain
OP, 1,10-ortho-phenanthroline
IOP, 5-iodoacetylamido-1,10-phenanthroline
IAOP, 5-iodoacetylglycylamido-1,10-phenanthroline
MPA, mercaptopropionic acid
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0212502.
References
- Baikalov, I., Schroder, I., Kaczor-Grzeskowiak, M., Grzeskowiak, K., Gunsalus, R.P., and Dickerson, R.E. 1996. Structure of the Escherichia coli response regulator NarL.Biochemistry 35 11053–11061. [DOI] [PubMed] [Google Scholar]
- Bruice, T.W., Wise, J.G., Rosser, D.S., and Sigman, D.S. 1991. Conversion of lphage Cro into an Operator-specific nuclease.J. Am. Chem. Soc. 113 5446–5447. [Google Scholar]
- Chen, C-H.B., Milne, L., Landgraf, R., Perrin, D.M., and Sigman, D.S. 2001. Artificial nucleases.Chembiochem 2 735–740. [DOI] [PubMed] [Google Scholar]
- Darwin, A.J., and Stewart, V. 1996. The nar modulon systems: Nitrate and nitrite regulation anaerobic gene expression. In Regulation of Gene Expression in Escherichia coli (eds. E.C.C. Lin and A.S. Lynch), pp. 343–360. R.G. Landes Company & Hall, New York.
- Dumoulin, P., Ebright, R.H., Knegtel, R., Kaptein, R., Granger-Schnarr, M., and Schnarr, M. 1996. Structure of the LexA repressor–DNA complex probed by affinity cleavage and affinity photo-cross-linking.Biochemistry 35 4279–4286. [DOI] [PubMed] [Google Scholar]
- Ebright, R.E., Ebright, Y.W., Pendergrast, P.S., and Gunasekera, A. 1990. Conversion of a helix-turn-helix motif sequence-specific DNA binding protein into a site-specific DNA cleavage agent.Proc. Natl. Acad. Sci. 87 2882–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus, R.P. 1992. Control of electron flow in Escherichia coli: Coordinated transcription of respiratory pathway genes.J. Bacteriol. 174 7069–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus, R.P. and Park, S.J. 1994. Aerobic–anaerobic gene regulation in Escherichia coli: Control by the ArcAB and Fnr regulons.Res. Microbiol. 145 437–450. [DOI] [PubMed] [Google Scholar]
- Harrison-McMonagle, P., Denissova, N., Marinez-Hackert, E., Ebright, R.H., and Stock, A.M. 1999. Orientation of OmpR monomers within an OmpR: DNA complex determined by DNA affinity cleaving.J. Mol. Biol. 285 555–566. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection.Proc. Natl. Acad. Sci. 82 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf, R., Pan, C., Sutton, C., Pearson, L., and Sigman, D.S. 1996. Engineering of DNA binding proteins into a site-specific cutters: Reactivity of Trp repressor-1,10-phenanthroline chimeras.Protein Engineering 9 603–610. [DOI] [PubMed] [Google Scholar]
- Lee, A.I., Delgado, A., and Gunsalus, R.P. 1999. Signal-dependent phosphorylation of the membrane bound NarX two-component sensor transmitter protein of Escherichia coli: Nitrate elicits a superior anion ligand response compared to nitrite.J. Bacteriol. 181 5309–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Kustu, S., and Stewart, V. 1994. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12.J. Mol. Biol. 241 150–165. [DOI] [PubMed] [Google Scholar]
- Mack, D.P., Iverson, B.L., and Dervan, P.B. 1988. Design and chemical synthesis of a sequence-specific DNA-cleaving protein.J. Am. Chem. Soc. 110 7572–7574. [Google Scholar]
- Murakami, K., Owens, J.T., Belvyaena, T.A., Meares, C.F., Busby, S.J., and Ishiama, A. 1997. Positioning of two α subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors.Proc. Natl. Acad. Sci. 94 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C.Q., Feng, J., Finkel, S.E., Landgraf, R., Sigman, D.S., and Johnson, R.C. 1994. Structure of the E. coli Fis–DNA complex probed by protein conjugated with 1,10-phenanthroline Copper(I) complex.Proc. Natl. Acad. Sci. 91 1721–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C.Q., Landgraf, R., and Sigman, D.S. 1995. Drosophila engrailed-1,10-phenanthroline Chimeras as probes of homeodomain–DNA complexes.Protein Sci. 4 2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C.Q., Finkel, S.E., Crampton, S.E., Feng, J.A., Sigman, D.S., and Johnson, R.C. 1996. Variable structures of fis–DNA complexes determined by flanking DNA–protein contacts.J. Mol. Biol. 264 675–695. [DOI] [PubMed] [Google Scholar]
- Parkinson, J.S. 1993. Signal transduction schemes of bacteria.Cell 73 857–871. [DOI] [PubMed] [Google Scholar]
- Parkinson, J.S. and Kofoid, E.C. 1992. Communication modules in bacterial signaling proteins.Annu. Rev. Genet. 26 71–112. [DOI] [PubMed] [Google Scholar]
- Pendergrast, P.S., Ebright, Y.W., and Ebright, R.H. 1994. High-specificity DNA cleavage agent: Design and application to kilobase and megabase DNA substrates.Science 265 959–962. [DOI] [PubMed] [Google Scholar]
- Schroeder, I., Wolin, C.D., Cavicchioli, R., and Gunsalus, R.P. 1994. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli.J. Bacteriol. 176 4985–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Z., Ebright, Y.W., Ilerr, N., Pendergrast, P.S., Echelard, Y., McMaahon, A.P., Ebright, R.H., and Abate, C. 1994. DNA affinity cleaving analysis of homeodomain–DNA interaction: Identification of homeodomain consensus sites in genomic DNA.Proc. Natl. Acad. Sci. 91 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman, D.S., Kuwabara, M.D., Chen, C.-H.B., and Bruice, T.W. 1991. Nuclease activity of 1, 10-phenanthroline-copper in the study of protein-DNA interactions.Methods Enzymol. 208 408–433. [DOI] [PubMed] [Google Scholar]
- Stewart, V. and Rabin, R.S. Dual sensors and dual response regulations interact to control nitrate-and nitrite-responsive gene expression in Escherichia coli. In Two-Component Signal Transduction (eds. J.A. Hoch and T.J. Silhavy), pp. 233–252. American Society for Microbiology, Washington, D.C.
- Sutton, C.L., Mazumder, A., Chen, C.B., and Sigman, D.S. 1993. Transforming the E. coli Trp repressor into a site-specific nuclease.Biochemistry 32 4225–4230. [DOI] [PubMed] [Google Scholar]
- Tyson, K.L., Bell, A.I., Cole, J.A., and Busby, S.J.W. 1993. Definition of nitrate and nitrite response elements at the anaerobically inducible Escherichia coli nirB promoter: Interactions between FNR and NarL.Mol. Microbiol. 7 151–157. [DOI] [PubMed] [Google Scholar]
- ———. 1994. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: Identification of common target heptamers for both NarP- and NarL-dependent regulation.Mol. Microbiol. 5 353–360. [DOI] [PubMed] [Google Scholar]
- Wang, H., Tseng, C-P., and Gunsalus, R.P. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to sub-micromolar concentrations of nitrate but not nitrite.J. Bacteriol. 181 5303–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.C., Tyson, K.L., Cole, J.A., and Busby, S.J.W. 1998. Regulation of transcription initiation at the Escherichia coli nir operon promoter: A new mechanism to account for co-dependence on two transcription factors.Mol. Microbiol. 27 493–505. [DOI] [PubMed] [Google Scholar]