Abstract
Amyloid fibrils of patients treated with regular hemodialysis essentially consists of β2-microglobulin (β2-m) and its truncated species ΔN6β2-m lacking six residues at the amino terminus. The truncated fragment has a more flexible three-dimensional structure and constitutes an excellent candidate for the analysis of a protein in the amyloidogenic conformation. The surface topology of synthetic fibrils obtained from intact β2-m and truncated ΔN6β2-m was investigated by the limited proteolysis/mass spectrometry approach that appeared particularly suited to gain insights into the structure of β2-m within the fibrillar polymer. The distribution of prefential proteolytic sites observed in both fibrils revealed that the central region of the protein, which had been easily cleaved in the full-length globular β2-m, was fully protected in the fibrillar form. In addition, the amino- and carboxy-terminal regions of β2-m became exposed to the solvent in the fibrils, whereas they were masked completely in the native protein. These data indicate that β2-m molecules in the fibrils consist of an unaccessible core comprising residues 20–87 with the strands I and VIII being not constrained in the fibrillar polymer and exposed to the proteases. Moreover, proteolytic cleavages observed in vitro at Lys 6 and Lys 19 reproduce specific cleavages that have to occur in vivo to generate the truncated forms of β2-m occuring in natural fibrils. On the basis of these data, a possible mechanism for fibril formation from native β2-m is discussed and an explanation for the occurrence of truncated protein species in natural fibrils is given.
Keywords: Amyloidosis, amyloid fibrils, β2-microglobulin, limited proteolysis, mass spectrometry
The investigation of the mechanism of amyloid fibrils formation by globular proteins is actually focused on the isolation and structural characterization of intermediates of the unfolding pathway that self aggregate in ordered fibrillar structures (Dobson 1999). In this respect, the term amyloidogenic conformation is used to indicate the conformation that a protein should acquire, even transiently, to fit properly within the fibrillar template.
Natural amyloid fibrils of patients with chronic renal failure treated with regular hemodialysis contains both full-length β2-microglobulin (β2-m) as well as some proteolitically processed fragments (Stoppini et al. 2000), with the main truncated species lacking 6 (ΔN6β2-m) and 19 (ΔN19β2-m) residues at the amino terminus, respectively. However, these shortened components were never detected in soluble form in the serum of patients but only in the fibrils. Conformational analyses of both intact β2-m and the ΔN6β2-m species carried out with a variety of experimental techniques revealed that the truncated fragment showed a more flexible structure and a greater propensity to self aggregate and to extend natural fibrils even at physiological pH (Esposito et al. 2000). On the contrary, full-length β2-m can create fibrils only at low pH both in the presence (Naiki et al. 1997) or in the absence of preformed natural fibrils (Mc Parland et al. 2000). At the low pH values required for fibril formation, full-length β2-m is partially unfolded and highly populates an intermediate state that maintains the secondary structure with residual non-native tertiary interactions. These findings are in close agreement with the current view that partial unfolding of certain globular proteins is a key step toward fibril formation. However, as mentioned above, the three-dimensional structure of these unfolding intermediates is difficult to determine due to their intrinsic dynamic properties and their tendency to self aggregate.
In this context, the behavior of ΔN6β2-m is remarkable because it still has a certain level of three-dimensional structure and shows a minimal energetic barrier for fibril formation. This species seem to be an excellent candidate for the determination of the three-dimensional structure of a protein in the amyloidogenic conformation. However, a weak point in all of the research strategies committed to the molecular characterization of amyloidogenic conformations is caused by the limits of the techniques so far available for the investigation of protein structure inside the fibrils. Atomic force microscope (Ding and Harper 1999), Fourier-transformed infra red spectroscopy (Seshadri et al. 1999), X-ray fiber diffraction (Serpell et al. 1999), and Electron Microscopy cross-sectional analysis (Serpell et al. 1995) have shed light on the overall structure of the fibril, but could not provide detailed information about intra- and intermolecular interactions of the protein monomer within the fibrils. Moreover, they were not able to describe the conformational changes occurring in the protein structure during the conversion from the native form to the fibrillar state.
The study of protein topology by limited proteolysis followed by mass spectral identification of the fragments generated was shown to be effective to probe conformational changes occurring in protein structures under different physicochemical conditions (Bianchi et al. 1999; Orrù et al. 1999; Urbani et al. 1999; Piccoli et al. 2000; Scognamiglio et al. 2001) or in the formation of protein complex or oligomerization (Scaloni et al. 1998, 1999; Atkinson et al. 2000). Recently, Wetzel and associates (Kheterpal et al. 2001) have successfully used limited proteolysis to elucidate how the A-β peptides of Alzheimer’s disease fold within the amyloid folding motif. In this study, we have used the limited proteolysis/mass spectrometry approach to investigate the surface topology of synthetic fibrils obtained from intact β2-m and the ΔN6β2-m truncated species with the aim to gain information on the structure of β2-m within the fibrillar polymer. The distribution of preferential proteolytic sites observed in fibrillar β2-m was compared with that detected on the soluble forms of the protein, leading to a description of the protein region constituting the unaccessible core of the fibrils and those still flexible and exposed to the solvent.
Results
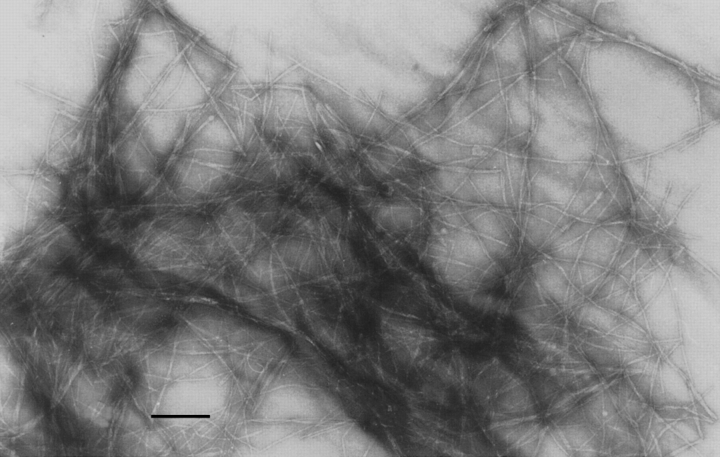
Amyloid fibrils were obtained by full-length β2-m and ΔN6β2-m at pH 4 to have identical conditions for fibril formation. As reported previously, a 15-d incubation was necessary to obtain a complete fibrillar insoluble precipitate. Both precipitates were specifically stained by Congo red and the EM analysis showed the presence of the typical fibrillar structure (Fig. 1 ▶).
Fig. 1.
Electron microscopy analysis. EM analysis of the fibrils obtained from the insoluble precipitate of ΔN6β2-m at pH 4. Space bar, 100 nm.
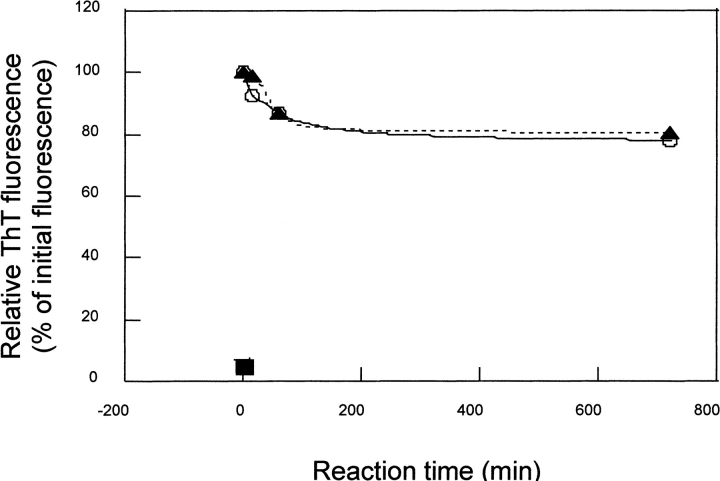
The surface topology of fibrillar β2-m and ΔN6β2-m was investigated by the limited proteolysis/mass spectrometry approach as already reported for the soluble forms (Esposito et al. 2000). These experiments aimed to investigate conformational changes affecting protein topology in the transition from the soluble state to the fibrils and to gain insight into the fibril structure by monitoring the appearance/disappearance of preferential cleavage sites (Orrù et. al. 1999; Scaloni et al. 1999). Limited proteolysis experiments were performed on the insoluble fibrils obtained from both β2-m and ΔN6β2-m using trypsin, chymotrypsin, and endoprotease Asp-N as conformational probes, as these enzymes had provided informative results on the topology of the soluble proteins. Fibrils were washed accurately before enzyme incubation to remove any possible soluble component. Fibrils recovered in the pellet after centrifugation and diluted in the working buffer of the limited proteolysis are minimally solubilized as documented by the Thioflavin assay shown in Figure 2 ▶. The partial depolimerization (10%–15%) in the first hour is mostly due to the dilution effect, because it is also evident at pH 4. In vitro fibrils obtained after 15 d of incubation and separated by centrifugation seem definitely more stable than those produced after 24 h of incubation as reported by Yamaguchi et al. (2001).
Fig. 2.
Thioflavin assay. The persistence of fibrillar structure in the proteolysis experimental conditions (pH 7.5 ▴), in β2-m fibrillogenesis conditions (pH 4 O), and in the buffer used to solubilize the proteolysed fibrils (40% CH3CN 0.4% TFA ▪ ) was monitored by thioflavine assay.
Aliquots of the reaction mixture were withdrawn at different interval times, treated with 0.4% TFA in 40% acetonitrile to solubilize fibrillar aggregates, and fractionated by HPLC. Individual fractions were then collected and the fibrils-released fragments identified by ESMS analysis.
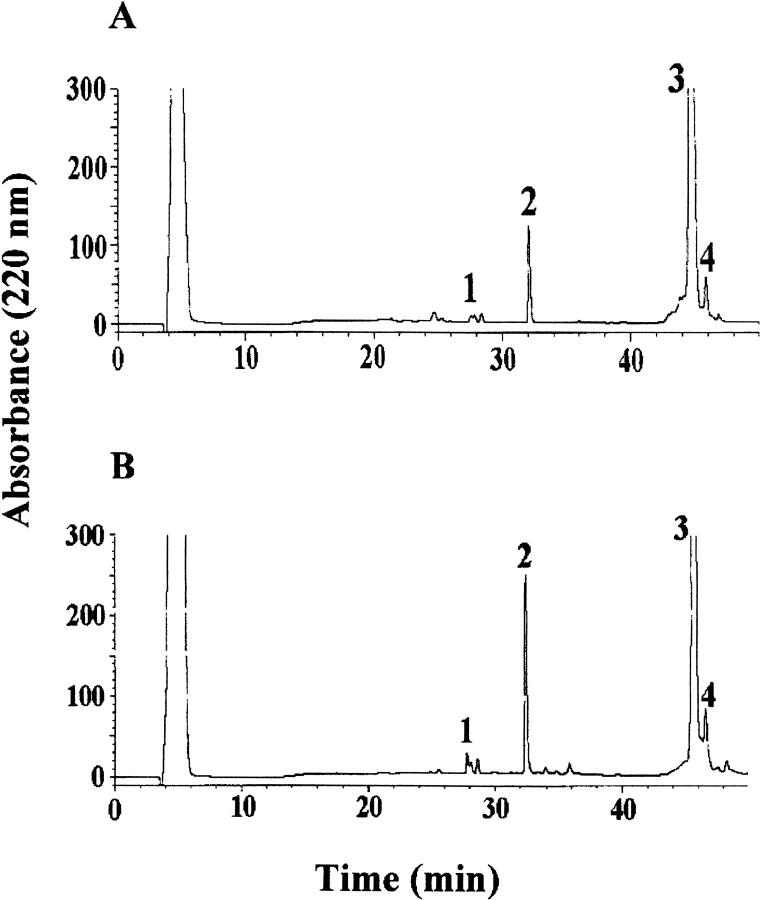
Figure 3 ▶ shows the HPLC analysis of the aliquots withdrawn following 15 min of trypsin digestion of the fibrils from β2-m (A) and ΔN6β2-m (B). Almost identical chromatographic profiles were obtained that essentially showed the presence of three peaks in addition to the undigested proteins (fraction 3). Intact β2-m was immediately cleaved at Lys 6, releasing the fragment 7–99, which coeluted with the untouched protein. In both chromatograms, peak 2 was identified as the peptide 92–99, indicating the occurrence of a preferential cleavage site at Lys 91; this interpretation was confirmed by the presence of the complementary peptide 7–91 coeluting with other species in fraction 3. Finally, the identification of the complementary peptide pairs 7–19 and 20–99 in peaks 1 and 4, respectively, indicated the occurrence of a secondary cleavage site at Lys 19.
Fig. 3.
Limited proteolysis experiment with trypsin. HPLC analysis of the aliquot withdrawn following 15 min of trypsin incubation of the fibrils from β2-m (A) and ΔN6β2-m (B). The indicated fractions were manually collected and the eluted peptides were identified by ESMS.
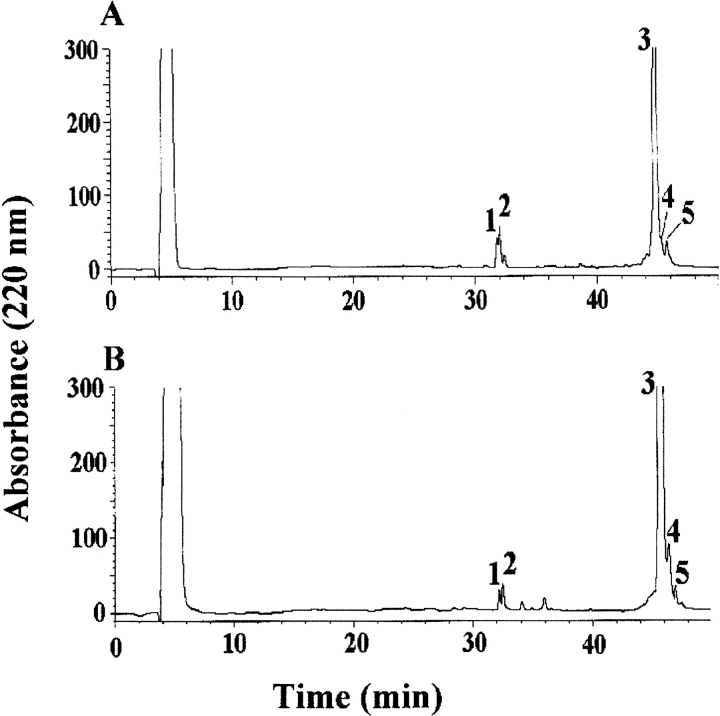
Similar results were obtained when the proteolytic experiments were carried out with endoprotease Asp-N and chymotrypsin. Both proteins showed identical accessibility in the carboxy-terminal region as shown by the preferential cleavage sites occurring at Asp 96 and Asp 98 following AspN digestion. Figure 4 ▶ shows the HPLC chromatograms of the samples withdrawn after 15 min of chymotrypsin digestion of the fibrils from intact β2-m (A) and ΔN6β2-m (B). Mass spectral analysis of the individual fractions revealed the presence of the complementary fragments 1–10 (or 7–10 in the case of ΔN6β2-m) and 11–99 in peaks 1 and 3, respectively, identifying Tyr 10 as a preferential proteolytic site. A secondary cleavage site was inferred at Leu 87 by the occurrence of the complementary peptide pairs 88–99 and 11–87, eluting in fractions 2 and 5, respectively. Finally, peak 4 contained the fragment 11–95, suggesting the presence of a further secondary cleavage site at Trp 95.
Fig. 4.
Limited proteolysis experiment with chymotrypsin. HPLC analysis of the aliquot withdrawn following 15 min of chymotrypsin incubation of the fibrils from β2-m (A) and ΔN6β2-m (B). The indicated fractions were collected manually, and the eluted peptides were identified by ESMS.
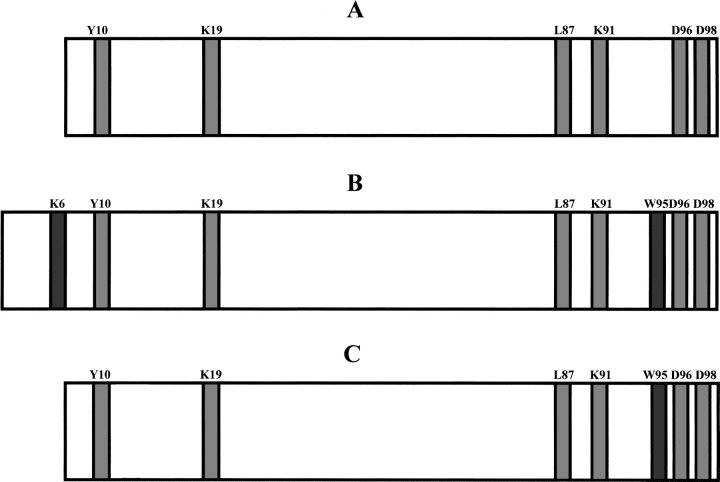
The distribution of preferential cleavage sites observed on the fibrils originated from intact β2-m and ΔN6β2-m is reported in Figure 5 ▶, in which previous data obtained on the two soluble proteins are also reported for comparison (Esposito et al. 2000). These data clearly indicated that truncated ΔN6β2-m exhibited an identical distribution of preferential proteolytic sites both in solution and in the fibrillar form, with cleavages occurring at positions 10, 19, 87, 91, 96, and 98. On the contrary, a totally different pattern of proteolytically accessible sites was identified in the fibrillar full-length β2-m as compared with the soluble form. Once inserted in the fibrillar polymer, a large portion of the central region of β2-m became protected against protease action, whereas a much higher accessibility was observed at the amino- and carboxy-termini. In particular, the region encompassing residues 53–61, which, in the soluble protein, had been easily cleaved at numerous sites (Asp 53, Lys 58, Asp 59, Trp 60, and Ser 61; Esposito et al. 2000), was found to be completely resistant to proteolysis.
Fig. 5.
Schematic representation of the results obtained from limited proteolysis experiments. The preferential cleavage sites common to the soluble ΔN6β2-m (A) and to the fibrils originated from both intact β2-m (B) and ΔN6β2-m (C) are highlighted in light gray; those observed on intact β2-m (B) and ΔN6β2-m (C) at the fibrillar state only are highlighted in dark gray.
This region corresponds to the loop connecting strands V and VI; the terminal end of strand V and possibly part of the connecting loop to strand VI had been proposed previously to be involved in intermolecular β-sheet pairings in the truncated β2-m species (Esposito et al. 2000). The absence of proteolytic accessibility in the same region in the fibrillar form of the full-length β2-m strongly indicates that this portion of the protein molecule is involved in the formation of the fibril core.
The higher accessibility of the amino- and carboxy-terminal ends of intact β2-m was shown by the occurrence of preferential proteolytic cleavages at Lys 6, Tyr 10, and Lys 19 in the amino-terminal region, and at Asp 96 and Asp 98 at the carboxyl terminus. It should be noted that these cleavages were never observed in the full-length β2-m in solution (Esposito et al. 2000).
Finally, when these results were compared with those obtained on the truncated ΔN6β2-m species both in solution and in the fibrils, a nearly coincident distribution of preferential proteolytic sites was clearly identified.
Discussion
Protein structure and dynamics can be probed by the effect of proteolytic enzymes, as the main property of the sites in which limited proteolysis occurs in a globular protein consists of an enhanced chain flexibility (Zappacosta et al. 1996; Polverino de Laureto et al. 1999). These sites are generally devoid of a stable secondary structure and are not embedded in rigid hydrogen-bonding constrains. Local unfolding is most likely required for the proteolytic cleavage of a peptide bond, because a large conformational change is needed to accommodate a peptide segment within the active site of a protease (Hubbard et al. 1998).
In several amyloid proteins, specific proteolytic cleavages seem to constitute a key step along the transition pathway from the soluble to the fibrillar form, as a large number of these proteins accumulate in natural fibrils in truncated forms. Following protease action, partial unfolding of the polypeptide chain can occur to generate the amyloidogenic conformation, giving rise to the fibrillogenic phenomenon. In some cases, like APP (Hooper et al. 2000), the proteolytic event is an absolute requirement for fibrils formation, whereas in others the schedule of the proteolytic digestion, whether occurring before or after the fibrillogenic event, is largely unknown. However, it is clear that the natural proteolytic phenomena occurring in amyloidogenic proteins spares the polypeptides regions involved in the formation of the fibril core. On the contrary, cleavages occurring outside of the protein core minimize the energetic barrier that protects several globular proteins from the transition toward the fibrillar aggregate (Wetzel 1997).
β2-m can be isolated from natural amyloid fibrils in intact and amino-terminal truncated forms (Linke et al. 1989; Bellotti et al. 1998; Stoppini et al. 2000). Conformational analyses carried out using a variety of different techniques such as NMR, limited proteolysis/mass spectrometry, CD, H/D exchange monitored by mass spectrometry, and definition of the thermodynamic parameters of the unfolding showed clearly that significative structural differences exist between the two forms in solution (Esposito et al. 2000). However, in the definition of a molecular mechanism for β2-m fibrils formation, two questions still remained unsolved. First, the description of the amyloidogenic conformation of β2-m, that is, the stable, partly folded intermediate that initiates the aggregation process and, second, the role of the proteolytic processing that leads to the truncated forms of the protein, that is, whether it is the release of the six amino-terminal residues that originates the amyloidogenic conformation, or the proteolysis occurs as a repair mechanism only after the fibrillogenic process had already started.
The surface topology of synthetic fibrils obtained in vitro from intact β2-m and truncated ΔN6β2-m was studied by the limited proteolysis/mass spectrometry approach that appeared particularly suited to gain insights into the structure of β2-m within the fibrillar polymer. The distribution of prefential proteolytic sites observed in both fibrils revealed that the central region of the protein spanning residues 53–60, which had easily been cleaved in the full-length globular β2-m, was fully protected in the fibrillar form. In addition, the amino- and carboxy-terminal regions of β2-m became more flexible and exposed to the solvent in the fibrils, whereas they were completely masked in the native protein.
These findings depict a scenario in which β2-m molecules in the fibrils consist of an unaccessible core comprising residues from 20 to 87 with the strands I and VIII being unconstrained in the fibril quaternary structure and exposed to the proteases. Protection of strands V and VI, rather susceptible to proteases in the soluble species, confirms our hypothesis (Esposito et al. 2000) that this region is endowed with less conformational stability and is most likely involved in intermolecular β sheet pairings, originating the fibril core.
In certain diseases not associated with amyloidosis, a complement-mediated cleavage at Lys 58 (at the beginning of strand V) is detectable in plasmatic β2-m (Nissen et al. 1990). The role of this cleavage in β2-m amyloidogenesis was recently re-examined (Heegaard et al. 2002). Such a cleavage induces in vitro significant conformational modifications of β2-m that acquires the capacity to bind Congo Red and heparin, both properties suggesting a higher amyloidogenic potential of this conformer. However, β2-m isolated from amyloid fibrils in patients only consists of intact, ΔN6- and ΔN19-truncated protein forms (Bellotti et al. 1998; Stoppini et al. 2000), thus excluding the occurrence of Lys 58–β2m in natural fibrils.
It is remarkable that we have been able, for the first time, to obtain rather detailed structural information on the fibrils structure by defining the protein region constituting the core of the fibrils and involved in the polymerization of the protein, and those exposed to the solvent and still endowed with conformational flexibility. Moreover, proteolytic cleavages observed in vitro at Lys 6 and Lys 19 reproduce specific cleavages that have to occur in vivo to generate the truncated forms of β2-m isolated from natural fibrils (Linke et al. 1989; Stoppini et al. 2000). These cleavages were neither observed in globular full-length β2-m, not even as secondary cleavage sites (Esposito et al. 2000,) nor were they obtained in vitro by use of a truncated species of β2-m, in which the first three residues had been removed (Δ3β2-m) as substrate (M. Monti, S. Principe, S. Giorgetti, V. Bellotti, A. Amoresano, and P. Pucci, unpubl.). As a whole, these results show that in vivo, the proteolytic processing leading to the amino-terminal truncated forms occurs only after the β2-m protein is constrained within the fibril structure.
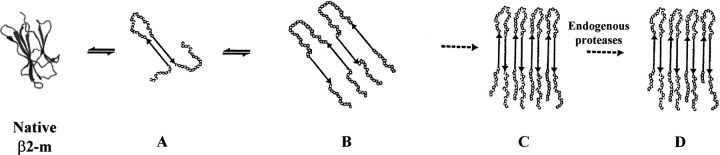
On the basis of these results, a molecular mechanism for fibril formation in amyloidosis occurring during long-term hemodialysis can be proposed (Fig. 6 ▶). β2-m must lose certain tertiary interactions to self aggregate in a well-ordered fibrillar polymer and a significant conformation transition is required. Partial unfolding of full-length protein could be caused by pH reduction, interaction with dialytical membranes, or hydrostatic pressure created in the dialytical system. These conformational changes generate the partly unfolded intermediate (A) characterized by an amyloidogenic conformation whose overall topologic structure resembles that of the truncated ΔN6β2-m form. The transient component A is more susceptible to amyloid deposition and originates the accumulation of insoluble material, which, in turn, drives the entire equilibrium toward the formation of fibrils. Once the amyloid deposit tends to accumulate, the extracellular proteases attempt to digest the insoluble material as a repair mechanism. However, the tightly packed structure of the fibril core prevents proteolysis and addresses protease action toward the amino-and carboxy-terminal ends, thus explaining the presence of the amino-terminal truncated forms of β2-m in natural fibrils.
Fig. 6.
Possible fibrillogenic pathway of β2-m. Native β2-m undergoes conformational changes to generate the partly unfolded intermediate (A) characterized by an amyloidogenic conformation whose overall topologic structure resembles that of the truncated ΔN6β2-m form. The transient component (A) originates the accumulation of soluble aggregates (B), which, in turn, drives the entire equilibrium toward the formation of fibrils (C). When the extracellular proteases attempt to digest the insoluble material, the tightly packed structure of the fibril core prevents proteolysis and addresses protease action toward the amino-and carboxy-terminal ends (D).
The proposed mechanism is in good agreement with recent results reported on the folding process of β2-m. The refolding pathway of full-length β2-m does not follow a simple two-state transition, but a more complex mechanism with two partly folded intermediates accumulating along the pathway (Chiti et al. 2001a). In particular, the species I2 has a long lifetime due to its very slow interconversion into the native protein. This species is significantly populated under physiological conditions and is able to aggregate in the presence of preformed fibrils (Chiti et al. 2001b). Spectroscopic data suggest that a close similarity exists between the ΔN6β2m (Esposito et al. 2000) and the I2 species of full-length β2-m (Chiti et al. 2001b), leading to the hypothesis that this metastable species share the same amyloidogenic conformation with ΔN6β2-m and intermediate A. According to the data obtained on I2, intermediate A might then favor aggregation either directly by exposing hydrophobic residues, thus enabling the establishment of intermolecular interactions, or by favoring equilibria with other less-structured conformations. Surface topology studies on these species are currently in progress to study whether the kinetic intermediate I2, and the truncated ΔN6β2-m form (the amyloidogenic component A) might be conformationally coincident.
Materials and methods
Samples preparation
Fibrils from both intact and Δ6β2-m were prepared as described previously (Esposito et a., 2000) at pH 4 and separated from soluble β2-m species by centrifugation in ultracentrifuge at 10,000 rpm. The pellet was washed extensively and finally resuspended in the optimum buffer to perform the limited proteolysis experiments.
Limited proteolysis experiments
Limited proteolysis experiments were carried out by incubating β2-m and ΔN6β2-m fibrils with trypsin, chymotrypsin, and endoprotease AspN. Enzymatic digestions were all performed in 100 mM sodium phosphate (pH 7.5) at 37°C using enzyme-to-substrate ratios ranging between 1:100 and 1:1000 (w/w). The persistence of fibrils at this pH in the interval of proteolytic digestion was monitored by the Thioflavin assay. No differences in specific fluorescence of the thioflavin–fibrils complex was found in buffer at pH 4 or 7.5. The extent of the reaction was monitored on a time-course basis by sampling the incubation mixture at different time intervals and by dissolving the remaining aggregates in 40% acetonitrile containing 0.4% TFA.
Chromatographic separation of peptides
Peptide mixtures from the different proteolysis experiments were fractionated by reverse-phase HPLC on a Phenomenex Jupiter C18 column (250 × 2.1 mm, 300 Å pore size) with a linear gradient from 5% to 50% acetonitrile in 0.1% trifluoroacetic acid (TFA) over 40 min, at a flow rate of 200 μL/min; elution was monitored at 220. Individual fractions were collected manually and identified by ESMS.
Mass spectrometry
Proteolytic fragments were analyzed by electrospray MS using an API-100 single quadrupole instrument (Applied Biosystem). Samples were injected directly into the ion source by a Harward syringe pump at a flow rate of 3 μL/min. Data were acquired and processed by the Biomultiviewer software provided by the manufacturer. The instrument was calibrated by a separate injection of horse heart myoglobin (average molecular mass 16,951.5 Da); all masses are reported as average mass.
When necessary, identification of disulphide-bridged peptides was carried out by reduction of the samples with 10 mM dithiothreitol in 50 mM Tris HCl, 1 mM EDTA (pH 7.5), containing 6 M GdnHCl, for 2 h at 37°C under nitrogen, followed by MALDI mass spectrometry (MALDI/MS) analysis using a Voyager DE instrument (Applied Biosystem). Typically, 1 μL of analyte solution was mixed with 1 μL of α-cyano-4-hydroxycinammic acid 10 mg/mL in acetonitrile/0.2% TFA, 70:30 v/v, containing 250 fmole of bovine insulin. The mixture was applied onto the metallic sample plate and air dried. Mass calibration was performed using the quasi-molecular ions of insulin at m/z 5734.5 and a matrix peak at m/z 379.1. All mass values are reported as average masses.
Acknowledgments
This study was supported by MURST (PRIN project Protein folding and misfolding), COFIN 2000 prot MM05221899 and Ministero della Sanità: ricerca finalizzata sulla Malattia di Alzheimer. We thank Dr. Fabrizio Chiti for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0206902.
References
- Atkinson, R.A., Joseph, C., Dal Piaz, F., Birolo, L., Stier, G., Pucci, P., and Pastore A. 2000. Binding of α-actinin to titin: Implications for Z-disk assembly.Biochemistry 39 5255–5264. [DOI] [PubMed] [Google Scholar]
- Bellotti, V., Stoppini, M., Mangione, P., Sunde, M., Robinson, C.V., Asti, L., Brancaccio, D., and Ferri, G. 1998. β2-microglobulin can be refolded into a native state from ex vivo amyloid fibrils.Eur. J. Biochem. 258 355–560. [DOI] [PubMed] [Google Scholar]
- Bianchi, E., Orru, S., Dal Piaz, F., Ingenito, R., Casbarra, A., Biasiol, G., Koch, U., Pucci, P., and Pessi, A. 1999. Conformational changes in human hepatitis C virus NS3 protease upon binding of product-based inhibitors.Biochemistry. 38 13844–13852. [DOI] [PubMed] [Google Scholar]
- Chiti, F., Mangione, P., Andreola, A., Giorgetti, S., Stefani, M., Dobson, C.M., Bellotti, V., and Taddei, N. 2001a. Detection of two partially structured species in the folding process of the amyloidogenic protein β 2-microglobulin.J. Mol. Biol.. 307 379–391. [DOI] [PubMed] [Google Scholar]
- Chiti, F., De Lorenzi, E., Grossi, S., Mangione, P., Giorgetti, S., Caccialanza, G., Dobson, C.M., Merlini, G., Ramponi, G., and Bellotti V. 2001b. A partially structured species of β 2-microglobulin is significantly populated under physiological conditions and involved in fibrillogenesis.J. Biol. Chem. 276 46714–46721. [DOI] [PubMed] [Google Scholar]
- Ding, T.T. and Harper, J.D. 1999. Analysis of amyloid-β assemblies using tapping mode atomic force microscopy under ambient conditions.Methods Enzymol. 309 510–525. [DOI] [PubMed] [Google Scholar]
- Dobson, C.M. 1999. Protein misfolding, evolution and diseaseTrends Biochem. Sci. 24 329–332. [DOI] [PubMed] [Google Scholar]
- Esposito, G., Michelutti, R., Verdone, G., Viglino, P., Hernandez, H., Robinson, C.V., Amoresano, A., Dal Piaz, F., Monti, M., Pucci, P., et al. 2000. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation.Protein Sci. 9 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heegaard, N.H., Roepstorff, P., Melberg, S.G., and Nissen, M.H. 2002. Cleaved {β};{sub2}-microglobulin partially attains a conformation that has amyloidogenic features.J. Biol. Chem. 277 11186–11189 [DOI] [PubMed] [Google Scholar]
- Hooper, N.M., Trew, A.J., Parkin, E.T., and Turner, A.J. 2000. The role of proteolysis in Alzheimer’s disease.Adv. Exp. Med. Biol. 477 379–390. [DOI] [PubMed] [Google Scholar]
- Hubbard, S.J., Beynon, R.J., and Thornton, J.M. 1998. Assessment of conformational parameters as predictors of limited proteolytic sites in native protein structures.Protein Eng. 11 349–359. [DOI] [PubMed] [Google Scholar]
- Kheterpal, I., Williams, A., Murphy, C., Bledsoe, B., and Wetzel, R. 2001. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis.Biochemistry. 40 11757–11767. [DOI] [PubMed] [Google Scholar]
- Linke, R.P., Hampl, H., Lobeck, H., Ritz, E., Bommer, J., Waldherr, R., and Eulitz, M. 1989. Lysine-specific cleavage of β 2-microglobulin in amyloid deposits associated with hemodialysis.Kidney Int. 36 675–681. [DOI] [PubMed] [Google Scholar]
- McParland, V.J., Kad, N.M., Kalverda, A.P., Brown, A., Kirwin-Jones, P., Hunter, M.G., Sunde, M., and Radford, S.E. 2000. Partially unfolded states of β(2)-microglobulin and amyloid formation in vitro.Biochemistry 39 8735–8746. [DOI] [PubMed] [Google Scholar]
- Naiki, H., Hascimoto, N., Suzuki, S., Kimura, H., Nakakuki, K., and Gejyo, F. 1997. Establishment of a Kinetic model of dialysis-related amyloid fibril extention in vitro.Amyloid 4 223–232. [Google Scholar]
- Nissen, M.H., Roepstorff, P., Thim, L., Dunbar, B., and Fothergill, J.E. 1990. Limited proteolysis of β 2-microglobulin at Lys-58 by complement component C1s.Eur. J. Biochem. 189 423–429. [DOI] [PubMed] [Google Scholar]
- Orrù, S., Dal Piaz, F., Casbarra, A., Biasiol, G., De Francesco, R., Steinkuhler, C., and Pucci, P. 1999. Conformational changes in the NS3 protease from hepatitis C virus strain BK monitored by limited proteolysis and mass spectrometry.Protein Sci. 8 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli, R., De Lorenzo, C., Dal Piaz, F., Pucci, P., and D’Alessio, G. 2000. Trypsin sheds light on the singular case of seminal RNase, a dimer with two quaternary conformations.J. Biol. Chem. 275 8000–8006. [DOI] [PubMed] [Google Scholar]
- Polverino de Laureto, P., Scaramella, E., Frigo, M., Wondrich, F.G., De Filippis, V., Zambonin, M., and Fontana, A. 1999. Limited proteolysis of bovine α-lactalbumin: Isolation and characterization of protein domains.Protein Sci. 8 2290–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaloni, A., Miraglia, N., Orrù, S., Amodeo, P., Motta, A., Marino, G., and Pucci, P. 1998. Topology of the Calmodulin-Melittin complex.J. Mol. Biol. 277 945–958. [DOI] [PubMed] [Google Scholar]
- Scaloni, A., Monti, M., Acquaviva, R., Tell, G., Damante, G., Formisano, S., and Pucci, P. 1999. Topology of the thyroid transcription factor 1 homeodomain-DNA complex.Biochemistry 38 64–72. [DOI] [PubMed] [Google Scholar]
- Scognamiglio, R., Notomista, E., Barbieri, P., Pucci, P., Dal Piaz, F., Tramontano, A., and Di Donato, A. 2001. Conformational analysis of putative regulatory subunit D of the toluene/o-xylene-monooxygenase complex from Pseudomonas stutzeri OX1.Protein Sci. 10 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpell, L.C., Sunde, M., Fraser, P.E., Luther, P.K., Morris, E.P., Sangren, O., Lundgren, E., and Blake, C.C. 1995. Examination of the structure of the transthyretin amyloid fibril by image reconstruction from electron micrographs.J. Mol. Biol. 254 113–118. [DOI] [PubMed] [Google Scholar]
- Serpell, L.C., Fraser, P.E., and Sunde, M.1999. X-ray fiber diffraction of amyloid fibrils.Methods Enzymol. 309 526–536. [DOI] [PubMed] [Google Scholar]
- Seshadri, S., Khurana, R., and Fink, A.L. 1999. Fourier transform infrared spectroscopy in analysis of protein depositsMethods Enzymol. 309 559–576. [DOI] [PubMed] [Google Scholar]
- Stoppini, M., Arcidiaco, P., Mangione, P., Giorgetti, S., Brancaccio, D., and Bellotti, V. 2000. Detection of fragments of β2-microglobulin in amyloid fibrils.Kidney Int. 57 349–350. [DOI] [PubMed] [Google Scholar]
- Urbani, A., Biasiol, G., Brunetti, M., Orrù, S., Dal Piaz, F., Casbarra, A., Pucci, P., Nardi, C., Gallinari, P., De Francesco, R., et al. 1999. Multiple determinants influence complex formation of the hepatitis C virus NS3 protease domain with its NS4A cofactor peptide.Biochemistry 38 5206–5215. [DOI] [PubMed] [Google Scholar]
- Wetzel, R. 1997. Immunoglobulin deposition disorderAdv. Protein Chem. 50 183–242. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, I., Hasegawa, K., Takahashi, N., Gejyo, F., and Naiki, H. 2001.. Apolipoprotein E inhibits the depolymerization of β 2-microglobulin-related amyloid fibrils at a neutral pH.Biochemistry. 40 8499–8507. [DOI] [PubMed] [Google Scholar]
- Zappacosta, F., Pessi, A., Bianchi, E., Venturini, S., Sollazzo, M., Tramontano, A., Marino, G., and Pucci, P. 1996. Probing the tertiary structure of proteins by limited proteolysis and mass spectrometry: The case of Miniboby.Protein Sci. 5 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]