Abstract
Diverse peptide sequences recognizing the λ boxB RNA hairpin were previously isolated from a library encoding the 22-residue λ N peptide with random amino acids at positions 13–22 using mRNA display. We have statistically analyzed amino acid distributions in 65 unique sequences from rounds 11 and 12 of this selection and evaluated the resulting structural and functional predictions by alanine-scanning mutagenesis and circular dichroism spectrometry. This artificial sequence family has a consensus structure that continues the bent α helix of λ N up to position 17 when bound to λ boxB. A charge pair (E14R15) and hydrophobic patch (A21L22 or V21L22) have important functional roles in this context. Notably, amino acid covariance reveals six specific pairs of random region positions with >95% significant linkage and strong overall helical (i+1, i+3, and i+4) couplings. The covariance analysis suggests that (1) the sequence context of every residue in each insert has been optimized, (2) selected sequences are local optima on a rugged fitness landscape, and (3) it is possible to detect more subtle structural features with artificial protein sequence families than natural homologs. Our results provide a framework for investigating the structures of in vitro selected proteins by functional minimization, reselection, and covariance analysis.
Keywords: Amino acid covariance, mRNA display, in vitro protein selection, λ N peptide, boxB RNA
The methods available for the in vitro selection of novel functional proteins from random sequence pools continue to improve (Roberts 1999). The large library sizes (>1013 sequences) attainable with mRNA display have recently enabled the selection of peptides and proteins recognizing diverse biological molecules (Roberts and Szostak 1997; Cho et al. 2000). Examples include: (1) ATP-binding proteins with no detectable homology to known sequences (Keefe and Szostak 2001), (2) streptavidin-binding polypeptides containing the known HPQ sequence motif embedded in a larger unknown functional context (Wilson et al. 2001), and (3) RNA hairpin-binding peptides derived from the λ N protein with new loop-binding sequences (Barrick et al. 2001b). All of these sequences lack significant homology to structural databases of known folds and domains. Consequently, very little can be deduced about the molecular details of their functions short of pursuing a complete atomic structure by NMR or X-ray crystallography.
In contrast, structural features of novel in vitro selected nucleic acid sequences are routinely determined using a rapid combination of sequence analysis and biochemical experiments. The general procedure for characterizing a pool of novel RNA sequences begins by finding the largest deletions that preserve activity in order to identify a minimal functional domain. Different sequences containing these domains are grouped and aligned to generate artificial sequence families (Ekland and Bartel 1995). A sequence family can also be constructed by reselecting new sequences from a "doped" library where a small percentage of base changes are introduced at each position in a single functional sequence. This family forms the basis for secondary structure prediction by an additive energy model (Zuker 2000). Nucleotide covariation in sequence families of sufficient size is used to refine this model and predict tertiary contacts (Gutell 1993). Finally, the functional model is experimentally tested using a variety of biochemical techniques such as: (1) mutations disrupting and restoring putative base pairs to test secondary structure assignment, (2) alkaline hydrolysis patterns to identify single-stranded regions, and (3) nucleotide analog interference mapping to precisely define chemical groups necessary for function (Strobel and Shetty 1997). The final model from this type of analysis represents all that is currently known about some natural functional RNA families such as RNAse P (Frank and Pace 1998) and the hairpin ribozyme (Fedor 2000). The characterization of a biotin-binding RNA pseudoknot provides an unusually complete account of the sequence-directed functional analysis of an in vitro selected RNA (Wilson et al. 1998).
We have demonstrated that it is possible to efficiently isolate peptides and proteins that recognize specific RNA structures from random sequence pools with mRNA display (Liu et al. 2000; Barrick et al. 2001a). Previously, we randomized the codons for amino acids 13–22 of the 22-residue λ N peptide and selected a library of 9×1012 sequences for binding to λ boxB or two other RNA hairpins differing by a single loop base (Barrick et al. 2001b). Fifty-six of the 65 unique sequences we determined from rounds 11 and 12 of this conserved-stem selection share a consensus arginine at position 15 not present in wild-type λ N. There are only 1.2 identical amino acids on average between two sequences at the remaining nine randomized positions. Despite the sequence variety of the selected solutions, functional screening of ∼20 sequences indicated that every one specifically binds λ boxB. On this basis we hypothesized that all selected peptides adopt a common structure with shared functional contacts to recognize λ boxB.
Here we applied sequence-directed functional analysis to this artificial family of RNA-binding peptides. We examined trends in the distributions of selected amino acids on all levels: over the whole randomized region, at each individual position, and pairwise covariation between positions. To experimentally evaluate our predictions we conducted alanine-scanning mutagenesis on the winning peptide sequence and took circular dichroism spectra of selected peptides in complex with λ boxB. On the basis of these results we propose a consensus model for selected peptide binding and comment on the observed distribution of solutions in sequence space. Functional analysis of artificial sequence families reselected from perturbed libraries should prove generally useful because it can detect subtle amino acid covariance.
Results
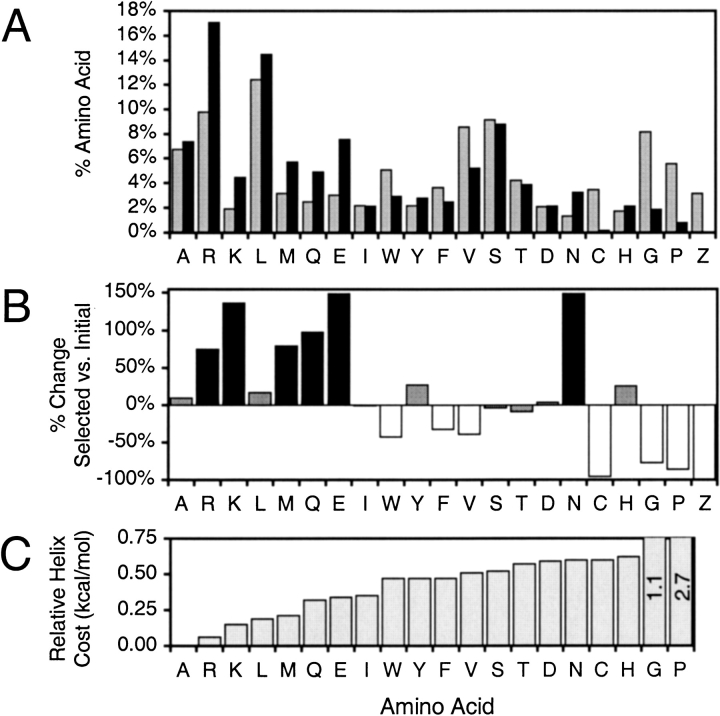
We first compared the representation of each amino acid in the 65 unique selected peptide sequences over the entire random region to its frequency in the initial random library (Fig. 1A ▶). The almost complete lack of the two helix breakers glycine and proline in selected sequences motivated us to look for a correlation between the helical propensities of amino acids and their relative representations (Figs. 1B, 1C ▶). Overrepresented amino acids (RKMQE) are generally stronger helix formers than underrepresented amino acids (WFVCGP). Asparagine is the only clear outlier to this trend. Its unexpected 1.5-fold overrepresentation in the final pool could be exaggerated by an unusually low frequency in the initial pool. Although it has exceptional hydrogen bonding potential, asparagine contacts are not especially common in natural RNA-binding protein structures (Allers and Shamoo 2001). Surprisingly, alanine and leucine have not been appreciably enriched by selection despite their high helical propensities. In general, nonpolar side chains appear to be slightly underrepresented relative to polar side chains with similar helical propensities.
Fig. 1.
Overall amino acid representation in selected randomized regions. (A) The composition in selected sequences (black) compared to the composition in the initial random library (gray). Z represents a stop codon. (B) Percent changes in amino acid representations from the initial pool to the selected pool. Shadings represent amino acids that are >30% overrepresented (black) or underrepresented (white) in final sequences. (C) Amino acids ordered by their relative intrinsic energetic costs of adopting helical dihedral angles (Munoz and Serrano 1995).
Calculating average helical stabilizations for the initial and selected pools using the intrinsic tendencies of individual amino acids to be in helical dihedral angles dramatically demonstrates the helical preference of selected peptides (Munoz and Serrano 1995). The average energetic cost of putting 10 amino acids with distributions typical of the initial pool into helical conformations is 11.5 kcal/mol. The same average cost for a selected peptide’s 10-residue random region is only 9.4 kcal/mol. For comparison, the helical cost for the corresponding residues in λ N is 8.5 kcal/mol. Seven of these 10 amino acids are part of a bent α helix when λ N is bound to λ boxB (Legault et al. 1998; Schärpf et al. 2000). The theoretical ∼2 kcal/mol stabilization of the final pool indicates similar functional helix formation by selected peptides.
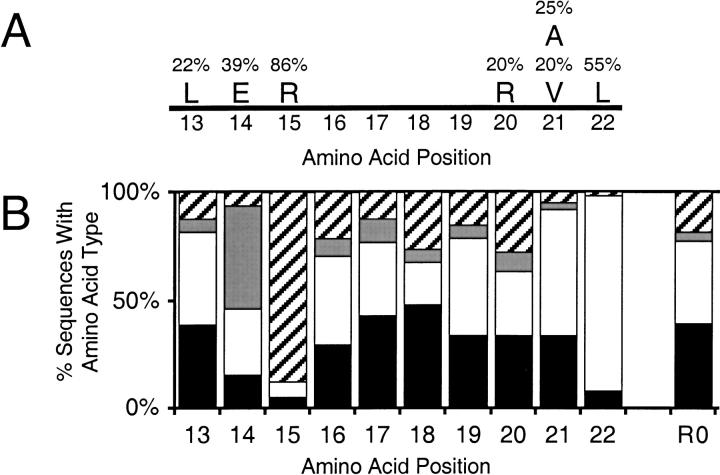
We next looked for consensus amino acids that occur in >20% of the unique selected sequences at individual positions (Fig. 2A ▶). As previously noted, there are only a few direct amino acid similarities among peptide sequences from rounds 11 and 12 of the conserved stem selection (Barrick et al. 2001b). Most importantly, arginine is present at position 15 in 86% of the selected sequences. Leucine is found in a majority of sequences at position 22, glutamate is common at position 14, and position 21 is either alanine or valine in many sequences. This analysis is unable to discern whether a specific amino acid side chain or a more general side-chain property is functionally required at a certain position.
Fig. 2.
Amino acid categorization. (A) Amino acids occurring at a randomized position in at least 20% of the unique sequences. (B) Percentage of amino acid side chains at each randomized position classified as positively charged (KR, diagonal lines), negatively charged (DE, gray), hydrophobic (GAVLIMFP, white), and polar (HNQSTCYW, black). R0 is the distribution in the initial (round zero) random pool.
To discriminate between these possibilities, we categorized the amino acids at each position based on their physicochemical properties into positively charged (KR), negatively charged (DE), hydrophobic (GAVLIMFP), and uncharged polar (HNQSTCYW) groups and compared the distributions to those of the initial round zero random pool (Fig. 2B ▶). Positions 13 and 16–20 have overall distributions of amino acid types close to the initial pool: 18% positively charged (+), 5% negatively charged (−), 38% hydrophobic (H), and 39% uncharged polar (P). Still, there are distinct preferences at each position for certain types of amino acids. The best contrast is drawn between the polar and positive charge proclivities of position 18 (26.2% +; 6.2% −; 20.0% H; 47.7% P) and hydrophobic predilection of position 19 (15.4% +; 6.2% −; 44.6% H; 33.8% P). At positions 14, 15, 21, and 22 this categorization trivially emphasizes that one or two amino acids dominate the distribution. If the side chains appearing in more than 20% of the sequences are reduced to their unselected levels, then the categorized distribution becomes close to that of the initial pool at positions 14, 15, and 21. This implies that these positions require a specific amino acid side chain rather than a more relaxed category of side chains. At position 22, after deemphasizing leucine, 80% of the remaining side chains are hydrophobic. Thus, the requirement here is for a nonpolar side chain, although leucine is definitely preferred. In fact, only six of the 65 sequences have polar side chains at position 22, and four of these are the bulky aromatics tryptophan and tyrosine.
In highly optimized protein sequences, the identity of an amino acid at one position can significantly influence what amino acids have high fitnesses at functionally related positions. In families of such sequences this coupling can be detected by looking for covariance in the distributions of amino acids at two positions. To investigate amino acid covariance in our selected peptides, we calculated the mutual information of each pair of positions in the random region and determined the significance of the resulting values by comparing them to randomized data sets. Selected peptides have an average pairwise linkage significance of 62.2%. This is elevated from the average 50% significance within randomly shuffled sequence sets with 99.9% confidence. Therefore, these 65 unique selected sequences appear to individually optimize a common set of positional interactions.
The specific pairs of amino acids that are most interdependent offer a higher-resolution statistical perspective on the peptides’ consensus structure. There are three pairs of positions with >99.9% significant linkage (Fig. 3A ▶). Positions 14 and 15 show dramatic charge exclusion. Every occurrence of negatively charged glutamate and aspartate side chains at position 14 occurs when position 15 is arginine. Conversely, only one in 56 sequences with Arg15 has a positively charged side chain at position 14. The amino acid distribution at position 18 is also highly dependent on the presence of arginine at position 15. Tryptophan and tyrosine are found at position 18 in only one of the 56 sequences with Arg15. However, position 18 is one of these aromatic amino acids in half (4/8) of the sequences without Arg15. Trp18 stacking by λ N on the exposed final base of the λ boxB loop is an important hydrophobic contact in the wild-type structure (Su et al. 1997). Apparently, Arg15 enforces a bound peptide structure where this interaction is not as favorable. Positions 21 and 22 form a hydrophobic patch. They contain a remarkable number of AL and VL pairs: 24 compared to the 16.1 expected if there was no covariance. Unfortunately, it is difficult to draw simple structural conclusions from the six other pairs of positions with >90% significant linkage.
Fig. 3.

Pairwise amino acid covariance. (A) Pairwise covariance between randomized positions. Connectors are labeled with the overall covariance significance between the two positions. (B) Pairwise covariance periodicity. The sum of significance scores for all amino acid pairs separated by a certain distance across the whole random region (gray circles) is compared to the average linkage in random sequences (black squares) and the maximum possible values (black line). (C) Average amino acid helical propensities at each position. The three positions with the least favorable amino acid distributions are shaded black. The line represents the average propensity of the initial pool distribution.
However, covariance does provide further evidence for helicity that is independent of individual amino acid tendencies. Plotting the strength of interactions with different periodicities clearly shows that i+1, i+3, and i+4 interactions are greatly overrepresented (Fig. 3B ▶). These side chains are in close proximity in an α helix: within a turn for i+1 and between adjacent turns of the helix for i+3 and i+4. Other periodicities, such as i+2 interactions of an extended β strand configuration, appear at roughly the frequencies expected by chance. If we assume that a completely helical consensus has 100% significances for all i+3 and i+4 pairs and that random 50% significances correspond to no helicity, then we interpolate 4.2 and 5.3 helical residues for selected random regions from i+3 and i+4 pairs, respectively. This calculation does not take into account possible biases in sequence sampling, the distribution of significances across the random region, or the incompleteness of measured helical covariances due to flanking constant regions. Despite these simplifications, the average value of 4.7 helical residues agrees well with the value of 4.9 obtained from a similar ad hoc interpolation based on the average helical stabilization of the selected pool. For this calculation, zero helicity corresponds to the helical stabilization of the initial pool. The lower energy of putting residues 13–22 in helical dihedral angles for wild-type λ N is equated to seven helical residues based on its structure in complex with λ boxB (Legault et al. 1998; Schärpf et al. 2000).
The location of these ∼5 helical residues can be predicted from several sequence characteristics. First, examining helical stabilization at each position separately shows that positions 19–21 have significantly less stabilization than the rest of the random region (Fig. 3C ▶). This location agrees with the simpler observation that helix-breaking glycine and proline residues are confined to positions 19–22 except in a single peptide. Prediction of helix location based on i+3 and i+4 coupling strengths is limited by the inability to calculate covariances with constant amino acids on the periphery of the random region. Still, it is notable that position 17 has very weak helical couplings to positions 20 and 21 that are indicative of an amino acid in the final turn of a helix (Fig. 3A ▶). Position 18 has weak helical linkages in both directions, and the i+1 covariance between positions 17 and 18 is the least significant in the entire random region. This evidence strongly suggests that on average the bent α helix of the constant λ N stem continues up to position 17 in selected peptides.
The first 13 codons of the λ N encoding DNA library sequence accumulated mutations from the PCR and reverse-transcription steps of selection because primers did not overlap this region. Favorable mutations outside the random region could potentially cause a population bottleneck and decrease the effective pool complexity. Most observed mutations are silent changes in the third, wobble base of a codon. Most notably, the Ala3 GCC codon has T substituted for the final C in 68% of the selected sequences, and the AGG codon of Arg7 has its final G changed to A in 38% of the sequences. The library used the same codons for Ala12 and Arg11, but there is only one sequence with the corresponding C to T transition and not a single sequence has the G to A change. This pattern of silent mutations could result from unintended selection for increased in vitro translation or reverse-transcription efficiency if they alter the secondary structure of the mRNA. Only one DNA sequence mutation causing a change in the encoded amino acid sequence occurs in more than two sequences. Eleven of the 87 total sequences have the initial G of the GAC codon of Asp2 changed to A so that they instead encode asparagines. This is a very conservative amino acid change that is actually present in the sequence of the N peptide from phage P22 (Tan et al. 1993). It is unlikely that the incompletely penetrant substitutions in our selected sequences significantly affected the sequence composition of the pool.
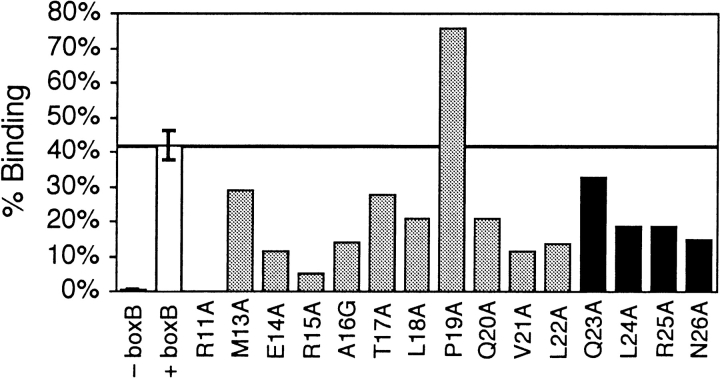
We used alanine-scanning mutagenesis of the most common selected peptide to verify the amino acid side-chain requirements deduced from sequence analysis (Fig. 4 ▶). Clone 11–36 was one of ten sequences coding for the random region MERATLPQVL. We extended mutagenesis of 11–36 (1–29) into the C-terminal region past position 22 to determine whether selected peptides utilized contacts to the constant amino acids coded by the reverse transcription primer (QLRNSCA) during selection and included the R11A substitution as a control known to decrease affinity by >50-fold (Su et al. 1997). The one native alanine residue was mutated to glycine. Because alanine is the strongest helix former and contains only a side-chain methyl group, a decrease in mutant peptide binding must be due to removing side-chain interactions rather than inhibiting helix formation. On the other hand, an increase in affinity could be due to the extra stabilization of a helix by alanine outweighing any stabilization the side chain contributes.
Fig. 4.
Alanine-scanning mutagenesis of 11–36. White bars represent native 11–36 binding with and without immobilized target. Black bars indicate that the mutated residue was encoded by the C-terminal primer region kept constant during selection. (Two measurements for native 11–36 bindings and one measurement for all mutants).
Every point mutation except P19A decreases the amount of peptide binding to λ boxB. As expected, the greatest losses of binding from alanine substitutions in the random region occur at positions with an amino acid consensus: E14, R15, V21, and L22. The ubiquitous R15 side chain makes the strongest contribution to binding of any random region residue. Apparently, some amino acids in the constant region participate in binding as much as random region side chains. Still, compared to R11 the roughly 1.5- to 8-fold stabilization provided by any side chain at positions 13–26 is marginal. The single sequence closely related to 11–36 has a random region of MERAMLPRVL. Interestingly, this less-abundant relative has fairly conservative T17M and Q20R changes at the two positions where alanine substitutions decrease binding least. The pairwise covariance between positions 17 and 20 is also one of the least significant couplings.
A threefold decrease in binding for the helix-destabilizing A16G substitution shows that helix formation is important for binding. The helix-destabilizing P19A substitution causes an opposite almost twofold increase in binding. Why wasn’t the P19A 11–36 mutant isolated during selection when it binds more tightly than 11–36? The selection incorporated 10-fold higher concentrations of nonspecific competitor tRNA compared to the assay used here, so a decrease in specificity could be confused with an increase in affinity. We believe the distinctive proline of 11–36 decreases its affinity for λ boxB but grants much greater specificity against binding noncognate RNAs. In fact, the 11–36 (1–22) peptide discriminates against binding the other two RNA hairpins used during selection better than all of the other selected peptides we have tested, although it does not have the highest affinity for λ boxB (Barrick et al. 2001b). Sensing changes in helical propensities from residues outside the predicted α helix is not surprising in a short peptide where flanking residues can greatly aid helix formation.
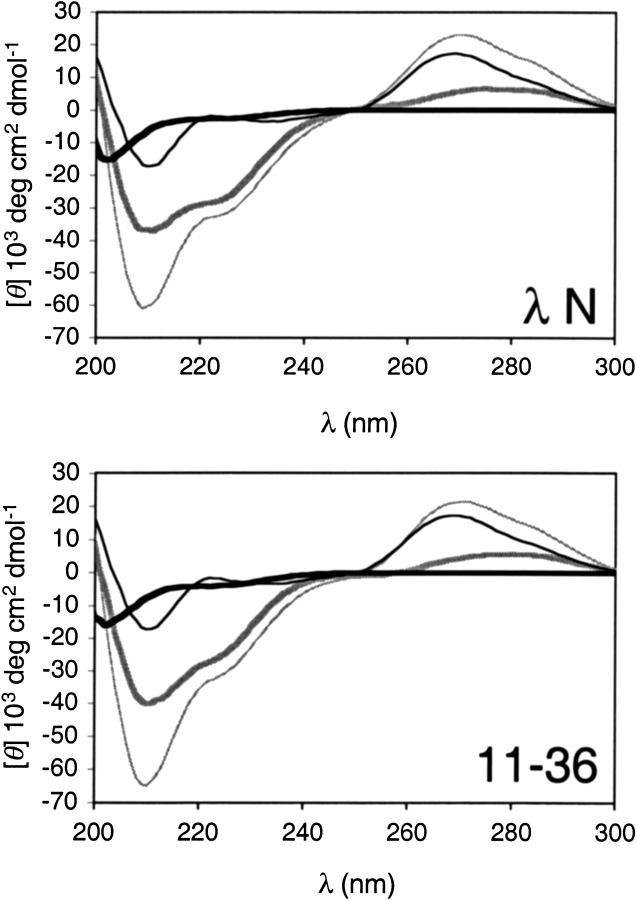
We experimentally tested our prediction that the random region adopts an α helical secondary strutcture when bound to λ boxB using circular dichroism spectometry on truncated 22 residue versions of several selected peptides (Fig. 5 ▶). λ N peptide is unstructured in solution. Upon binding λ boxB RNA, it folds into an α helix whose length can be calculated from the induced ellipticity (Su et al. 1997). We used the same procedure to calculate the number of helical amino acids in selected peptides bound to λ boxB (Table 1). Each selected peptide exhibits <15% helicity when free in solution. Our calculation of 16 total helical amino acids for bound λ N (1-22) agrees with the NMR structure that shows a bent α helix extending from Q4 to K19 (Legault et al. 1998; Schärpf et al. 2000). If we assume that positions 1–12 contribute the same 9 helical amino acids to the total signal in selected peptides, as they do in wild-type λ N, then the five peptides tested have 4–6 helical residues within positions 13–22. The average value of 4.6 agrees surprisingly well with our prediction of ∼5 helical residues. Peptide truncation to position 22 would destabilize helical structure in the C terminal residues of the random region. Thus, the observed helicity must also reside at the beginning of the random region. Apparently the end of the λ N stem initiated α helix always extends at least one residue past the conserved arginine to position 16 and can continue as far as position 18 in individual selected peptides. The heterogeneity of helix lengths hints that there are several idiosyncratic ways to extend the λ N stem mode of binding.
Fig. 5.
Circular dichroism spectra of wild-type λ N and 11–36 peptides bound to λ boxB RNA. Free peptide (thick black line), free boxB RNA (thin black line), and peptide-RNA complex (thin gray line) spectra are shown. The difference spectrum of bound peptide (thick gray line) is the peptide-RNA complex spectrum minus the free boxB RNA and free peptide spectra. The positive peak in the bound peptide spectrum around 280 nm is induced RNA structure (Su et al. 1997). [θ] is the molar ellipticity per amino acid residue in the peptide for all spectra, including free RNA. Bound peptide spectra of 11–10, 12–39, 12–47, and 12–50 have the same shape as λ N and 11–36 (data not shown).
Table 1.
Helicity of selected peptides bound to λ boxB RNAa
| Peptide sequence | −[θ]222 | #AA | |
| λ N | MDAQTRRRERRAEKQAQWKAAN | 25,800 | 16.0 |
| 11–10 | ––––––––––––ALRNEKFWVV | 21,100 | 13.1 |
| 11–36 | ––––––––––––MERATLPQVL | 20,600 | 12.8 |
| 12–39 | ––––––––––––LQRSRARHAL | 24,100 | 14.9 |
| 12–47 | ––––––––––––LERTKLEKAL | 21,000 | 13.0 |
| 12–50 | ––––––––––––NMRMYRSLVI | 22,700 | 14.1 |
a The first 12 amino acids are identical in all peptides. [θ]222 is the molar ellipticity per amino acid residue in deg cm2 dmol−1 from the bound peptide difference spectrum. #AA is the number of helical amino acids calculated from [θ]222. We estimate a <5% error in the measurement of [θ]222. There may be as high as a 15% error in #AA for λ N, 11–10, and 12–50 due to aromatic side chain contributions to [θ]222.
Discussion
Our sequence-directed biochemical analysis has provided a rough structural model for how the R15 peptide family recognizes the λ boxB RNA hairpin. On average, the bent α helix of the wild-type λ N peptide stem continues halfway through the random region to position 17 in selected peptides. Arg15 is within this helix in every individual sequence we tested and clearly makes the most important RNA contact of any random region side chain. It probably recognizes a specific pocket between negatively charged phosphates of the λ boxB RNA loop backbone, as do Arg8 and Arg11 of the λ N stem (Legault et al. 1998; Schärpf et al. 2000). The adjacent negatively charged Glu14 side chain probably prevents nonspecific binding by patterning the electrostatic surface of the peptide rather than directly participating in RNA recognition. The hydrophobic AL or VL pair at positions 21 and 22 may directly pack against exposed bases in the RNA loop or cooperate with the functional side chains in the C-terminal constant region.
We previously demonstrated that adding high concentrations of tRNA during binding to λ boxB effectively competes away peptide sequences that have nonspecific RNA affinity (Barrick et al. 2001a). These conditions dramatically increase the enrichment of specific RNA-binding peptides during a round of selection, presumably because they make the fitness landscape more rugged by increasing the steepness of its optima. Many of our new observations support this conclusion. (1) Site-directed mutagenesis shows that every single alanine mutant in the random region of 11–36 changes the amount of peptide binding by at least 1.5-fold. A smooth fitness landscape would have neutral mutations. (2) Our initial library encoded almost all of the 1013 possible 10-amino acid sequences, but there was only a single pair of unique random regions that shared >50% identity out of 87 total sequences. If the landscape were smooth, we would expect to isolate considerably more related sequences relative to unique optimal sequences. (3) Strong pairwise covariance scores between random region positions indicate that the function of a selected amino acid is highly dependent on its sequence context. Simulations of NK fitness landscape models have established that greater interdependence between individual amino acid contributions to fitness creates a more rugged landscape (Kauffman 1993).
Serial selection is a stringent optimization method: A sequence with only 1.5-fold better performance than another will be represented ∼130 times as often in the pool after 12 rounds of selection. With the known ruggedness of the fitness landscape, we therefore expect almost every peptide remaining in the selected pool to be a local optimum in sequence space. The dearth of close sequence relatives in the selected pool and decrease in binding accompanying all alanine substitutions in 11–36 (except P19A for the reasons discussed) agree with this hypothesis. Selected R15 peptides are clustered in sequence space by shared side chains at four positions. Within these general constraints, they appear to reach their individual fitness peaks by optimizing idiosyncratic coupled "weak interactions". The apparent similarity of the absolute fitness at each local optimum is exaggerated: Selection conditions were not stringent enough to efficiently separate peptides past a certain specificity/affinity threshold. After further rounds of selection with increased magnesium concentration and decreased λ boxB concentration, a rarer sequence consensus without R15 grows to dominate the pool (J. Barrick and R. Roberts, in prep.).
The methods detailed here provide a general framework for a sequence-directed approach to investigating the structure and function of novel proteins. The common binding mode of the selected peptides and enforced alignment of their short random regions by conserved flanking sequences made our final RNA-binding peptide pool a suitable sequence family for immediate analysis. Generally, functional minimization followed by reselection from a "doped" library based on a single selected sequence would be used to generate an artificial sequence family. This is precisely the nucleic acid paradigm (Ellington and Szostak 1990; Bartel et al. 1991). The greater conformational flexibility and chemical diversity of amino acids did not hopelessly complicate an analogous prediction and experimental evaluation of model structures for our peptides. However, de novo and similarity-based structure prediction algorithms for longer protein sequences are not as reliable or straightforward as those available for nucleic acids. Consequently, information from amino acid covariance in the artificial sequence family is critical for creating an initial structural model.
Covariance analysis has been applied to independently folded proteins in several contexts. Nonadditivity in thermodynamic mutant cycles constructed by site-directed mutagenesis has been used to determine pairs of interacting residues at protein-protein interfaces in the Shaker K+ channel/scorpion toxin (Hidalgo and MacKinnon 1995) and barnase/barstar complexes (Schreiber and Fersht 1995). Covariance has been examined in a number of natural protein families. Originally, it was used to assess functional linkage in the highly variable V3 loop of the HIV-1 envelope protein (Korber et al. 1993). It was subsequently applied to protein families with known structures, revealing salt bridges and coupled amino acids involved in DNA binding in homeodomains (Clarke 1995), clusters of coupled residues in the hydrophobic core of RNA recognition motifs (Crowder et al. 2001), and long-range energetic coupling through the core of a PDZ domain family (Lockless and Ranganathan 1999). The predictive power of covariance in these data sets is limited by the number of sequences that can be constructed by site-directed mutagenesis or the quality of alignment and structural conservation within an evolutionarily related sequence family.
An optimal randomized library for detecting covariance will maximize its information content by introducing as many mutations as possible while ensuring that its sequences remain functionally equivalent. One strategy, selection of functional sequences from alanine-doped libraries, has already proven useful for combinatorial alanine-scanning mutagenesis (Weiss et al. 2000). Although these perturbed libraries have been used primarily to determine the statistical energies of many single alanine mutants simultaneously, energetic nonadditivity in pairs of mutations indicating functional linkage has been noted in selected sequences (Gregoret and Sauer 1993). Because artificial sequence families are unambiguously aligned and structurally homogeneous by construction, there is much less statistical noise in their covariance scores compared to natural sequence families. Our observation of secondary structure linkages for the first time hints at the more subtle features they can detect. Theoretically, the predictive power of covariance analysis in reselected sequence families is only limited by the initial library complexity and number of clones that can be reasonably sequenced. Covariance analysis cannot replace the wealth of information generated by a full structure determination, but it can provide a rapid way of garnering some structural information that can be validated by simpler experimental techniques. Artificial protein sequence families may additionally prove useful for cheaply improving the coverage of sequence-to-structure databases and investigating the evolutionary relationships of protein folds.
Materials and methods
Basic sequence analysis
Of 87 individual sequenced clones from the round 11 and 12 pools of the selection, 65 unique sequences with different amino acids in at least three of the 10 randomized positions were chosen to calculate all statistics. Only one sequence (clone 12–136) with a unique random region was excluded by this procedure. The initial pool had NNS codons (N = A, T, C, G; S = C, G) for the 10 randomized amino acids (13–22). The empirical distribution of bases at N and S positions was determined by sequencing 14 members of the initial pool. We extrapolated the representation of each amino acid in the initial pool from NNS codons with these distributions (N = 17.9% A, 30.0% T, 23.6% C, 28.6% G; S = 40.7% C, 59.3% G) and the standard mammalian genetic code. The intrinsic helical propensities of the amino acids tabulated by Munoz and Serrano (1995) were used to evaluate helical trends within selected sequences. Secondary structure prediction algorithms such as GORIV (Garnier et al. 1996) were not useful for investigating the bound conformation of selected peptides as they typically predict the entire random region to be helical.
Pairwise covariance analysis
Covariance between pairs of positions in the randomized region of the 65 unique selected peptide sequences was calculated according to the standard definition of mutual information (Korber et al. 1993). It quantifies interdependence between two positions as a log-odds score relating the number of times an amino acid pair is actually observed at two positions to the number of times it would be expected to randomly appear if the amino acid distributions at each position were independent. To determine the significance of the resulting values, scores were also calculated for 100,000 random sets of 65 sequences generated by shuffling amino acids between sequences while preserving their position within the random region. The significance of a linkage score was determined from its rank order in these lists of random scores.
Alanine-scanning mutagenesis of 11–36
Mutant peptides were synthesized through standard PCR, in vitro transcription with T7 RNA polymerase, and in vitro translation in rabbit reticulocyte lysate (Barrick et al. 2001a). To construct site-directed mutants, the 3′ primer 21.108 was replaced with a primer encoding the four codons before the substituted amino acid, the mutated codon (GCC for alanine and GGC glycine), and the entire C-terminal peptide sequence. Quantitation of affinity-precipitated peptides was as previously described for library 3 peptides using biotinylated λ boxB (Barrick et al. 2001b). Here we used 6 μL of crude in vitro translation and included 5 × 500 μL washes with N binding buffer.
Circular dichroism spectrometry
λ boxB RNA (5′- GCCCUGAAAAAGGGCC) was prepared by T7 transcription. Fmoc or Boc chemistry was used to chemically synthesize truncated selected peptides (aa 1–22) with free acid and amine termini. Peptides were purified to a single peak by reversed-phase HPLC, and their identities were confirmed by mass spectrometry. Circular dichroism spectra were recorded with an Aviv 62 DS CD spectrophotometer at 25°C. Free peptide (10 μM–20 μM), free RNA (5 μM–7 μM), and RNA-peptide complex spectra (5μM–7 μM RNA with 1.2× peptide) were taken in 10 mM potassium phosphate buffer (pH 7.8). A bound peptide’s difference spectrum was determined by subtracting the spectra of an equivalent amount of free RNA and free peptide from the spectrum of the RNA-peptide complex (Su et al. 1997). The fractional helicities of free and bound peptides were calculated from measurements of their ellipticity at 222 nm (Chakrabartty et al. 1994). Following this treatment, we used [θ]222 values of 0 and −35,500 deg cm2 dmol−1 per amino acid residue for 0% and 100% helicity. Aromatic Phe, Tyr and Trp residues make additional contributions to [θ]222 when they are in α helical model peptides (Chakrabartty et al. 1993). We did not correct our calculations of the number of helical amino acids for this effect because we are unsure of its magnitude in the context of λ N and selected peptides. Attempting to correct for Trp and Tyr by assuming a linear relation between a peptide’s induced aromatic signal and fractional helicity (Chakrabartty et al. 1994) did not change the calculated average extent of helicity into the random region (data not shown).
Electronic supplemental material
Supplemental material includes tables of the 87 clone sequences analyzed and covariance significance scores for every amino acid pair in the random region.
Acknowledgments
J.E.B. is an HHMI predoctoral fellow. This work was supported by NSF career grant 9876246 to R.W.R. We thank S.L. Mayo for suggesting we examine helical covariance and T.T. Takahashi for his comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0208902.
References
- Allers, J. and Shamoo, Y. 2001. Structure-based analysis of Protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 311 75–86. [DOI] [PubMed] [Google Scholar]
- Barrick, J.E., Takahashi, T.T., Balakin, A., and Roberts, R.W. 2001a. Selection of RNA-binding peptides using mRNA-peptide fusions. Methods 23 287–293. [DOI] [PubMed] [Google Scholar]
- Barrick, J.E., Takahashi, T.T., Ren, J.S., Xia, T.B., and Roberts, R.W. 2001b. Large libraries reveal diverse solutions to an RNA recognition problem. Proc. Natl. Acad. Sci. 98 12374–12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P., Zapp, M.L., Green, M.R., and Szostak, J.W. 1991. Hiv-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell 67 529–536. [DOI] [PubMed] [Google Scholar]
- Chakrabartty, A., Kortemme, T., Padmanabhan, S., and Baldwin, R.L. 1993. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effects on measurement of helix propensities. Biochemistry 32 5560–5565. [DOI] [PubMed] [Google Scholar]
- Chakrabartty, A., Kortemme, T., and Baldwin, R.L. 1994. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 3 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, G., Keefe, A.D., Liu, R.H., Wilson, D.S., and Szostak, J.W. 2000. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J. Mol. Biol. 297 309–319. [DOI] [PubMed] [Google Scholar]
- Clarke, N.D. 1995. Covariation of residues in the homeodomain sequence family. Protein Sci. 4 2269–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder, S., Holton, J., and Alber, T. 2001. Covariance analysis of RNA recognition motifs identifies functionally linked amino acids. J. Mol. Biol. 310 793–800. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1995. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 23 3231–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, A.D. and Szostak, J.W. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346 818–822. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. 2000. Structure and function of the hairpin ribozyme. J. Mol. Biol. 297 269–291. [DOI] [PubMed] [Google Scholar]
- Frank, D.N. and Pace, N.R. 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67 153–180. [DOI] [PubMed] [Google Scholar]
- Garnier, J., Gibrat, J.F., and Robson, B. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266 540–553. [DOI] [PubMed] [Google Scholar]
- Gregoret, L.M. and Sauer, R.T. 1993. Additivity of mutant effects assessed by binomial mutagenesis. Proc. Natl. Acad. Sci. 90 4246–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell, R.R. 1993. Comparative studies of RNA: Inferring higher-order structure from patterns of sequence variation. Curr. Opin. Struct. Biol. 3 313–322. [Google Scholar]
- Hidalgo, P. and MacKinnon, R. 1995. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science 268 307–310. [DOI] [PubMed] [Google Scholar]
- Kauffman, S.A. 1993. The structure of rugged fitness landscapes. In The origins of order: Self-organization and selection in evolution, pp. 33-67. Oxford UP, New York.
- Keefe, A.D. and Szostak, J.W. 2001. Functional proteins from a randomsequence library. Nature 410 715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B.T.M., Farber, R.M., Wolpert, D.H., and Lapedes, A.S. 1993. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: An information theoretic analysis. Proc. Natl. Acad. Sci. 90 7176–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault, P., Li, J., Mogridge, J., Kay, L.E., and Greenblatt, J. 1998. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell 93 289–299. [DOI] [PubMed] [Google Scholar]
- Liu, R.H., Barrick, J.E., Szostak, J.W., and Roberts, R.W. 2000. Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 318 268–293. [DOI] [PubMed] [Google Scholar]
- Lockless, S.W. and Ranganathan, R. 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286 295–299. [DOI] [PubMed] [Google Scholar]
- Munoz, V. and Serrano, L. 1995. Elucidating the folding problem of helical peptides using empirical parameters. II. Helix macrodipole effects and rational modification of the helical content of natural peptides. J. Mol. Biol. 245 275–296. [DOI] [PubMed] [Google Scholar]
- Roberts, R.W. 1999. Totally in vitro protein selection using mRNA-protein fusions and ribosome display. Curr. Opin. Chem. Biol. 3 268–273. [DOI] [PubMed] [Google Scholar]
- Roberts, R.W. and Szostak, J.W. 1997. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. 94 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärpf, M., Sticht, H., Schweimer, K., Boehm, M., Hoffmann, S., and Rösch, P. 2000. Antitermination in bacteriophage λ: The structure of the N36 peptide-boxB RNA complex. Eur. J. Biochem. 267 2397–2408. [DOI] [PubMed] [Google Scholar]
- Schreiber, G. and Fersht, A.R. 1995. Energetics of protein-protein interactions: Analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248 478–486. [DOI] [PubMed] [Google Scholar]
- Strobel, S.A. and Shetty, K. 1997. Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. Proc. Natl. Acad. Sci. 94 2903–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, L., Radek, J.T., Hallenga, K., Hermanto, P., Chan, G., Labeots, L.A., and Weiss, M.A. 1997. RNA recognition by a bent α-helix regulates transcriptional antitermination in phage l. Biochemistry 36 12722–12732. [DOI] [PubMed] [Google Scholar]
- Tan, R.Y., Chen, L., Buettner, J.A., Hudson, D., and Frankel, A.D. 1993. RNA recognition by an isolated α helix. Cell 73 1031–1040. [DOI] [PubMed] [Google Scholar]
- Weiss, G.A., Watanabe, C.K., Zhong, A., Goddard, A., and Sidhu, S.S. 2000. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. 97 8950–8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C., Nix, J., and Szostak, J. 1998. Functional requirements for specific ligand recognition by a biotin-binding RNA pseudoknot. Biochemistry 37 14410–14419. [DOI] [PubMed] [Google Scholar]
- Wilson, D.S., Keefe, A.D., and Szostak, J.W. 2001. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. 98 3750–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2000. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 10 303–310. [DOI] [PubMed] [Google Scholar]