Abstract
The triple-helix is a unique secondary structural motif found primarily within the collagens. In collagen, it is a homo- or hetero-tripeptide with a repeating primary sequence of (Gly-X-Y)n, displaying characteristic peptide backbone dihedral angles. Studies of bulk collagen fibrils indicate that the triple-helix must be a highly repetitive secondary structure, with very specific constraints. Primary sequence analysis shows that most collagen molecules are primarily triple-helical; however, no high-resolution structure of any entire protein is yet available. Given the drastic morphological differences in self-assembled collagen structures with subtle changes in assembly conditions, a detailed knowledge of the relative locations of charged and sterically bulky residues in collagen is desirable. Its repetitive primary sequence and highly conserved secondary structure make collagen, and the triple-helix in general, an ideal candidate for a general parameterization for prediction of residue locations and for the use of a helical wheel in the prediction of residue orientation. Herein, a statistical analysis of the currently available high-resolution X-ray crystal structures of model triple-helical peptides is performed to produce an experimentally based parameter set for predicting peptide backbone and Cβ atom locations for the triple-helix. Unlike existing homology models, this allows easy prediction of an entire triple-helix structure based on all existing high-resolution triple-helix structures, rather than only on a single structure or on idealized parameters. Furthermore, regional differences based on the helical propensity of residues may be readily incorporated. The parameter set is validated in terms of the predicted bond lengths, backbone dihedral angles, and interchain hydrogen bonding.
Keywords: Collagen, helical wheel, triple-helix, bioinformatics, protein structure prediction
Collagen is almost unique among proteins in its use of triple-helical secondary structure. The triple-helix is composed of three polypeptide chains, each with the repeating triplet Gly-X-Y, where X and Y are frequently (∼22% occurrence of each in type I collagen) proline and 4-hydroxyproline, respectively. Since the initial high-resolution single crystal structure of a short model triple-helical peptide by Bella et al. in 1994, a total of 10 such structures have been solved (Table 1). These peptides are typically on the order of 30 amino acids in length (i.e., each peptide of the triple-helical trimer consists of 10 triplets), which is drastically shorter than the ∼1050 amino acids contained in each of the three polypeptide subunits of the typical forming collagen. We believe that an understanding of the three-dimensional orientations of charged residues and regions of high steric bulk along a collagen molecule is absolutely essential in investigating interactions of collagen with other molecules, whether collagenous or noncollagenous. This is highlighted by the distinct morphological differences observed on self-assembly of collagen with potentially quite subtle changes in assembly conditions (see Paige and Goh 2001; Paige et al. 2001). Therefore, given a triple-helical primary sequence, we would like to be able to predict both the orientations and the spatial proximity of chemically active and relevant moieties.
Table 1.
Existing high-resolution triple-helical model peptide X-ray crystal structures
| Sequencea | Residues resolved G-X-Y (triplets)b | Resolution (Å) | Reference | PDB entry |
| (P-O-G)4-P-O-A-(P-O-G)5 | 28-30-30 (26) | 1.9 | Bella et al. 1994 | 1CAG |
| (P-O-G)4-P-O-A-(P-O-G)5 | 30-30-30 (27) | 1.85 | Bella et al. 1995 | 1CGD |
| (P-P-G)10 | 7-7-7 (4) | 2.0 | Kramer et al. 1998 | 1A3I |
| 1.7 | 1A3J | |||
| (P-P-G)10 | 7-7-7 (5) | 1.9 | Nagarajan et al. 1998 | N/Ad |
| (P-O-G)10 | 7-7-7 (5) | 1.9 | Nagarajan et al. 1999 | N/Ad |
| (P-O-G)3-I-T-G-A-R-G-L-A-G-(P-O-G)4 | 18-17-18 (12)c | 2.0 | Kramer et al. 1999 | 1BKV |
| (P-O-G)4-E-K-G-(P-O-G)5 | 29-30-30 (27) | 1.75 | Kramer et al. 2000 | 1QSU |
| (P-P-G)10 | 7-7-7 (4) | 1.30 | Vitagliano et al. 2001 | 1G9W |
| (P-P-G)10 | 54-60-60 (54) | 1.30 | Berisio et al. 2002 | 1K6F |
| Total data set | 194-202-203 (168) | — | — | — |
a The one-letter code O is used for 4-hydroxyproline. Note that each structure is a homo-tripeptide.
b Number of each Gly, X and Y residue type resolved in structure and of triplets in order Gly-X-Y resolved.
c This is only the iminorich portion, as described in Materials and Methods. The middle portion we are using for aminorich statistics contains 12 of each G-X-Y and nine triplets.
d Coordinates in Protein Data Base (PDB) file format kindly provided by K. Okuyama.
Its supercoiled nature has led the triple-helix to be linked with the α-helical coiled coil, which has enjoyed a great deal of success with structural prediction (Skolnick et al. 1999; Kajava 2001), including parameterization (Harbury et al. 1995) and statistical analysis of common features in crystal structure (Yang et al. 1999). The collagen triple-helix should be more straightforward for parameterization and prediction than is the α-helical coiled-coil, in that the packing of multiple helices side-by-side and the prediction of the relative orientations of helices are not issues. To provide an experimental basis for the prediction of the three-dimensional layout of residues in a collagen-scale triple-helix, we have performed a statistical analysis of all existing high-resolution model peptide structures. Although models of the triple-helix have been developed that successfully describe some of the features of X-ray diffraction experiments and single crystal structures (Brodsky and Shah 1995; Mayo 1996; Beck and Brodsky 1998) or during the course of molecular dynamics studies (Klein and Huang 1999), we have decided to avoid any preconceptions of the structural motif and instead simply perform a statistical analysis using all presently available structures.
Results and Discussion
Two factors must be specified to fix residues within an α-helix: the number of residues per 360° turn and the translation along the length of the helix per residue. This is insufficient for a triple-helix, however, because the residues do not all fall the same distance from the middle of the helix. Hence, a third parameter must also be included: the distance from the center-line of the helix. Unlike an α-helix, in which the rise between residues is constant, each residue in a Gly-X-Y triplet will be translated differently along the length of the helix. As a final wrinkle in comparison to the α-helical wheel, the tripeptide nature of the triple-helix structure means that the relative locations of the three polypeptide chains must also be part of our parameter set.
Taking the Gly-X-Y triplets as independent subunits within each model peptide structure and considering each triple-helical peptide structure in a cylindrical frame of reference, we have statistically determined values of translation along the long-axis of the helix (Δz), the angular stagger (Δθ), and the radius from the helix center (rh). The following five sets of statistics are sufficient to locate the backbone and Cβ atoms in a triple-helix:
rh at each triplet position for each backbone atom (N, Cα, C‘ and O) and for Cβ;
Δz and Δθ for triplets, determined from Glyn → Glyn+1, Xn → Xn+1, and Yn → Yn+1 using all backbone atoms;
Δz and Δθ between Cα atoms for Gly → X and X → Y, and Δz for Y → Gly;
Δz and Δθ of N, C‘, O, and Cβ relative to Cα at each triplet position; and
the chain-to-chain Δz and Δθ.
As is apparent from Table 1, the currently available peptide structures are almost exclusively Gly-imino-imino in all cases, except Protein Data Bank (PDB) entry 1BKV. (Analysis was performed separately for the iminodeficient region in the middle of 1BKV.) Rather than excluding specific triplets from the data set on the basis of conformational differences or anomalies, we have chosen to perform a statistical filter on the data set, as described in Materials and Methods.
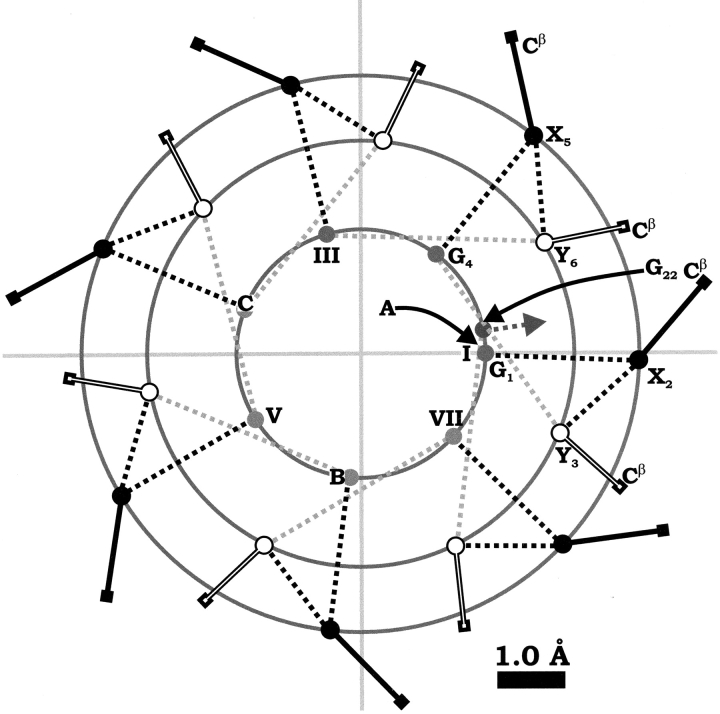
This analysis allows generation of the Cα and Cβ trace (looking down the helical long-axis) shown in Figure 1 ▶ in what we propose to be a useful helical-wheel format for the triple-helix, along the lines of the α-helical wheel of Schiffer and Edmundson (1967). The well-known left-handed triplet-to-triplet helicity (in which each triplet itself winds in a right-handed manner) and right-handed chain-to-chain helicity are readily apparent in Figure 1 ▶. Along with this projection down the helical long-axis, we produce the corresponding position of each atom along the long-axis. All parameters required for production of a Cα trace of a triple-helix backbone are detailed in Table 2. Although the Cα trace, perhaps of the X and Y residues only, may be the most desirable manner to represent a triple-helix with upwards of 3000 residues, applications such as side-chain prediction may require the entire backbone. Given the highly characteristic dihedral angles of the triple-helix (Brodsky and Shah 1995; Mayo 1996; Beck and Brodsky 1998), prediction of the Cα locations should also allow the locations of the other backbone atoms to be predicted. Statistical analysis of the set of model peptide crystal structures shows this to indeed be true. Parameters allowing incorporation of the remainder of the backbone and of Cβ atoms are provided in Table 3. PDB format model structure files, using each parameter set, for the simple sequence [(G-A-A)10]3 are available in the Supplementary Material. Triple-helix structures generated using the statistically derived parameters in Tables 2 and 3 show very close agreement with expected bond lengths, as based on the work of Engh and Huber (1991). The backbone dihedral angles are also in excellent agreement with those of the data sets used. These validations are shown in detail in Table 4 for both the iminorich and iminodeficient parameter sets. Further validation of the parameter set is provided through analysis of the hydrogen-bonding patterns, using the DSSP program of Kabsch and Sander (1983). Both of the model PDB structures display the expected Gly-to-X hydrogen-bond patterns, with calculated strengths of −1.9 to −2.0 kcal/mole for the iminorich parameter set and −2.8 to −2.9 kcal/mole for the parameters based on the iminodeficient middle portion of 1BKV. As more high-resolution structures are solved, the statistically derived parameters will certainly improve.
Fig. 1.
Helical wheel for idealized triple-helical (G-X-Y)7-G sequence in region containing high imino acid content in X and Y positions. Circles indicate Cα positions for G (grey), X (black), and Y (white) residues (first six are labeled) for chain A. The odd-numbered triplets are labeled I–VII. Cβ positions are indicated for X and Y. The locations of Cα of the first G residue of chains B and C are shown by letters on the G radius circle. Note that triplet I is N-terminal, and that every residue after G1 is translated further C-terminal along the length of the helix (i.e., into the page). Numerical parameters are given in Tables 2 and 3.
Table 2.
Statistics required for Cα trace of triple-helix derived from all available high-resolution triple-helical model peptide crystal structures (Table 1)
| Iminorich regions | Aminorich region in 1BKV | |||
| Statistic | Ne | μ (σ)e | Ne | μ (σ)e |
| Triplet to triplet | ||||
| Δθ (°)a,b | 1836 | 53 (7.3) | 86 | 40 (5.6) |
| Δz (Å)a | 1890 | 8.53 (0.16) | 95 | 8.65 (0.14)h |
| Chain to chain | ||||
| Δθ (°)a,b,c | 2168 | −102 (8.8) | 120 | −107 (8.7) |
| Δz (Å)a,c | 1479 | 2.8433… (0.17) | 96 | 2.8833… (0.15) |
| Cα | ||||
| rH (Å) G | 184 | 1.83 (0.27) | 12 | 1.72 (0.30) |
| X | 184 | 4.10 (0.33) | 12 | 4.06 (0.27) |
| Y | 191 | 3.14 (0.28) | 12 | 3.15 (0.22) |
| Δθ (°)a,d G → X | 154 | −1.4 (5.0)f | 9 | −1.1 (3.6) |
| G → Y | 150 | −22 (6.4) | 9 | −25 (5.4) |
| Δz (Å)a,d G → X | 158 | 3.03 (0.087)g | 9 | 3.03 (0.08) |
| X → Y | 178 | 3.46 (0.10) | 12 | 3.34 (0.08) |
| Y → G | 183 | 2.04 (0.16) | 12 | 2.28 (0.20) |
a A positive Δθ is a counter-clockwise rotation. A positive Δz is a translation along the helical long-axis from N-terminal to C-terminal.
b A chain-to-chain Δθ of 102° corresponds to 3.5 residues per turn of 360°, when considered with a triplet-to-triplet Δθ of 53°, implying 6.8 triplets per turn; this is extremely close to the proposed 7/2 (or 7s) helix of Okuyama et al. (1977). Conversely, a Δθ of 107° from chain to chain is 3.36 residues per 360° turn, and Δθ of 40° from triplet to triplet is 9 triplets per turn—this is somewhat compacted compared with the ideal Rich-Crick 10/3 (or 107) helix (Rich and Crick 1961).
c Note that a negative chain-to-chain Δθ corresponds to a positive Δz displacement.
d Angle or distance from Cα to Cα.
e N indicates number of residues in data set; μ, mean; and σ, standard deviation (see Materials and Methods for details).
f Value >1% away from mean calculated by four iterations of removal of outliers at μ ± 2σ (see Materials and Methods). Magnitude of difference is 0.3°.
g Mean distance rounded down (by >0.001 Å) to provide a triplet-to-triplet stagger of 8.53 Å.
h Actual mean is 8.656 Å—rounded down to allow use of directly measured means of to chain-to-chain Δz and Cα to Cα Δz values.
Table 3.
Statistics required for addition of backbone and Cβ to a Cα trace of triple helix. Derived from indicated regions in all available high-resolution triple-helical model peptide crystal structures (Table 1)
| N | C′ | O | Cβ | |||||
| Atom Statistic | Nb | μ (σ)b | Nb | μ (σ)b | Nb | μ (σ)b | Nb | μ (σ)b |
| Iminorich region | ||||||||
| rh (Å) | ||||||||
| G | 182 | 1.54 (0.25)c | 184 | 2.63 (0.27) | 184 | 3.01 (0.27) | — | — |
| X | 187 | 3.35 (0.24) | 187 | 3.21 (0.25) | 195 | 2.10 (0.31) | 185 | 5.16 (0.24) |
| Y | 188 | 3.82 (0.26) | 185 | 2.61 (0.24) | 189 | 3.52 (0.25) | 190 | 4.29 (0.28) |
| Δθ (°)a to Cα | ||||||||
| G | 175 | −42 (6.1) | 175 | −12 (3.2) | 174 | −35 (5.1) | — | — |
| X | 188 | 6.8 (1.6) | 183 | −3.7 (1.2) | 182 | 6.5 (3.7) | 197 | 13 (1.5) |
| Y | 192 | 6.8 (1.7) | 191 | 26 (3.2) | 195 | 41 (4.2) | 189 | −5.3 (1.7) |
| Δz (Å)a to Cα | ||||||||
| G | 185 | −0.69 (0.08) | 189 | 1.21 (0.06) | 184 | 1.55 (0.09) | — | — |
| X | 198 | −1.18 (0.06) | 180 | 1.22 (0.05) | 186 | 1.24 (0.08) | 197 | 0.20 (0.10)c |
| Y | 197 | −1.23 (0.05) | 188 | 0.63 (0.08) | 190 | 0.47 (0.16) | 193 | 0.89 (0.08)c |
| Aminorich region in 1BKV | ||||||||
| rh (Å) | ||||||||
| G | 12 | 1.53 (0.22) | 12 | 2.54 (0.28) | 12 | 2.93 (0.22) | — | — |
| X | 12 | 3.30 (0.29) | 12 | 3.16 (0.24) | 12 | 2.05 (0.26) | 12 | 5.16 (0.28) |
| Y | 12 | 3.77 (0.24) | 12 | 2.64 (0.23) | 12 | 3.59 (0.27) | 12 | 4.30 (0.23) |
| Δθ (°)a to Cα | ||||||||
| G | 12 | −44 (8.2) | 12 | −12 (2.9) | 12 | −36 (4.3) | — | — |
| X | 12 | 6.8 (1.3) | 12 | −4.8 (1.5) | 12 | 6.0 (3.8) | 12 | 13 (1.4) |
| Y | 12 | 9.2 (1.3) | 12 | 24 (2.4) | 12 | 39 (3.3) | 12 | −8.1 (2.1) |
| Δz (Å)a to Cα | ||||||||
| G | 12 | −0.79 (0.08) | 12 | 1.19 (0.04) | 12 | 1.52 (0.08) | — | — |
| X | 12 | −1.16 (0.03) | 12 | 1.19 (0.05) | 12 | 1.25 (0.09) | 12 | 0.33 (0.05) |
| Y | 12 | −1.19 (0.03) | 12 | 0.80 (0.10) | 12 | 0.79 (0.16) | 12 | 0.83 (0.07) |
a A positive Δθ is a counter-clockwise rotation. A positive Δz is a translation along the helical long-axis from N-terminal to C-terminal.
b N indicates number of residues in data set; μ, mean; and σ, standard deviation (see Materials and Methods for details).
c These values >1% away from mean calculated by four iterations of removal of outliers at μ ± 2σ (see Materials and Methods). Magnitudes of difference are 0.01 to 0.02 Å.
Table 4.
Comparison of model triple-helix bond lengths to expected values from Engh and Huber (EH) and dihedral angles to the data set for iminorich model region (I) versus aminorich model region (A)
| Gly | X | Y | ||||
| Bond | Length in model (Å) | EH—μ (σ) (Å)b | Length in model (Å) | EH—μ (σ) (Å)b | Length in model (Å) | EH—μ (σ) (Å)b |
| I | ||||||
| N → Cα | 1.42 | 1.451 (0.016) | 1.47 | 1.466 (0.015) | 1.46 | 1.466 (0.015) |
| Cα → C′ | 1.52 | 1.516 (0.018) | 1.53 | 1.525 (0.021) | 1.53 | 1.525 (0.021) |
| C′ → O | 1.23 | 1.231 (0.020) | 1.20 | 1.231 (0.020) | 1.22 | 1.231 (0.020) |
| C′ → Na | 1.31 | 1.329 (0.014) | 1.32 | 1.341 (0.016) | 1.34 | 1.341 (0.016) |
| Cα →Cβ | — | — | 1.50 | 1.530 (0.020) | 1.49 | 1.530 (0.020) |
| A | ||||||
| N → Cα | 1.46 | 1.451 (0.016) | 1.45 | 1.458 (0.019) | 1.45 | 1.458 (0.019) |
| Cα → C′ | 1.51 | 1.516 (0.018) | 1.52 | 1.525 (0.021) | 1.53 | 1.525 (0.021) |
| C′ → O | 1.24 | 1.231 (0.020) | 1.21 | 1.231 (0.020) | 1.24 | 1.231 (0.020) |
| C′ → Na | 1.31 | 1.329 (0.014) | 1.35 | 1.329 (0.014) | 1.28 | 1.329 (0.014) |
| Cα → Cβ | — | — | 1.54 | 1.530 (0.020) | 1.50 | 1.530 (0.020) |
| Dihedral angle | Model (°) | Data set—μ (σ) (°)c | Model (°) | Data set—μ (σ) (°)c | Model (°) | Data set—μ (σ) (°)c |
| a C′ (n − 1) to N(n) of given residue number n. | ||||||
| b Means (μ) and standard deviations (σ) from Engh and Huber (1991) chosen as appropriate to G-P-P triplet (I) or G-X-Y triplet (A), where X and Y are not G, P, or A. | ||||||
| c Mean indicates μ; standard deviation, σ. | ||||||
| I | ||||||
| φ | −71 | −72 (5.5) | −74 | −74 (3.6) | −59 | −60 (3.2) |
| ψ | 174 | 176 (4.7) | 162 | 163 (4.3) | 152 | 152 (6.2) |
| ω | 180 | 180 (0.84) | 177 | 178 (2.3) | 178 | 178 (2.3) |
| A | ||||||
| φ | −73 | −68 (3.6) | −71 | −71 (5.1) | −69 | −66 (3.4) |
| ψ | 169 | 167 (4.0) | 161 | 160 (5.4) | 152 | 148 (2.2) |
| ω | 180 | 179 (0.4) | 180 | 179 (0.2) | 179 | 180 (0.6) |
The most apparent differences between the iminorich regions and the iminodeficient region are in the residue-to-residue Δz and the triplet-to-triplet Δθ. The triplet-to-triplet Δθ of 53° and chain-to-chain Δθ of −102° for the iminorich region generate ∼6.8 triplets and ∼3.5 residues per 360° revolution, corresponding quite closely to the 7/2 (or 75) helix proposed initially by Okuyama et al. (1977) Those of 40° and −107° in the iminodeficient region give nine triplets and 3.36 residues per 360°, which is somewhat compressed compared with the Rich-Crick 10/3 (or 107) helix (Rich and Crick 1961). For the iminodeficient region, the statistics are entirely based on a single region of one model peptide structure (1BKV)—these parameters will definitely improve as further (G-X-Y)n structures are solved. We have chosen to keep the statistically derived values in both cases, rather than values based on the ideal Okuyama or Rich-Crick models.
In comparison to existing homology modeling methods, our parameterization has the following differences. First, the peptide backbone used for prediction of the triple-helix is a statistical compilation of all existing high-resolution structures. Second, the parameter set allows for easy prediction of a triple-helix of any length—extrapolation of an arbitrarily long structure from a short peptide using homology modeling tools is by no means trivial. Finally, consideration of the primary sequence of a triple-helical molecule would allow the prediction of regions of high verus low triple-helical propensity, as extensively studied by Persikov et al. (2000, 2002). The parameters given herein very readily allow the production of a triple-helix, with various regions predicted to be of differing helical stability.
A program such as SCWRL (Bower et al. 1997) takes one existing peptide backbone from a PDB file and adds side-chain atoms onto it. This is a highly valuable methodology for homology modeling; however, unlike this approach, the attempt here is to produce a statistically representative generalization of the triple-helix. In a strict homology model, one would need either to arbitrarily choose only a single triple-helical structure to use as the homologous scaffold with SCWRL, or to generate a parameter set equivalent to that given herein. Our statistically produced triple-helix provides a structure with an overall average root mean square difference (RMSD) of 3.70 Å in comparison to model peptides generated by SCWRL using each of the iminorich sequences used herein (Table 1). Notably, the statistical parameters produce a triple-helix with only a 0.263 Å RMSD from the SCWRL-modeled chains A–C of PDB 1K6F and 0.269 Å from chains D–F (for atoms N, Cα, C‘, O, and Cβ), showing an uncanny closeness to the extremely high-resolution structure 1K6F solved by Berisio et al. (2002). This could be interpreted to imply overrepresentation of 1K6F in the overall parameter set, because it comprises just <30% of the values included. The equivalent parameter set calculated without the inclusion of 1K6F, however, provides a predicted structure with an RMSD of only 0.053 Å compared to that with the parameter set given herein. Therefore, 1K6F improves the statistical values rather than skewing the overall parameter set. As a result, homology models constructed with 1K6F would likely be highly similar to structures generated with the iminorich parameter set given in Tables 2 and 3. However, to generate a triple-helix of arbitrary length, statistical parameters such as those herein would still need to be produced from 1K6F and would be based on only this single structure, which is subtly different from the statistically derived data set. Software such as Gencollagen has been used extremely successfully for molecular dynamic predictions of short model peptides (Klein and Huang 1999) but is based on idealized bond parameters. Our parameter set does not rely in any way on an idealized triple-helix; instead, we present a statistically derived framework, independent of any single crystal structure, from which triple-helical collagen structures may be accurately predicted.
Given the agreement in bond lengths and backbone dihedral angles (as in Table 4), as well as with the interchain H-bonding patterns, we believe that the statistical parameters given herein are very reasonable for predicting a triple-helix—especially in iminorich regions. Source code is freely available on request for the generation of triple-helical structures of any primary sequence and length, and a Web-based interface will be available at http://www.chem.utoronto.ca/staff/MCG/. We also plan to maintain an updated parameter set at this web site as further structures become available.
Materials and methods
The atomic coordinates for the high-resolution model peptide structures listed in Table 1 were obtained directly from the PDB (http://www.rcsb.org/pdb; Bernstein et al. 1977; Berman et al. 2000) for the cases in which PDB entry codes were given. In the remaining two cases, K. Okuyama, of Tokyo University of Agriculture and Technology, Japan, was kind enough to provide atomic coordinates. Data analysis was performed using Interactive Data Language (IDL 5.5; Research Systems, Inc.) on a Silicon Graphics Inc. Octane workstation. Additional statistical elaboration was performed in MATLAB 6.1 (The MathWorks) on an Octane. Source code is freely available on request.
Before statistical analysis, each triple-helical structure was converted to cylindrical polar coordinates. This process, briefly, involved the following steps. Assuming net directionality running N-terminal to C-terminal for a triple-helix, a vector composed of the sum of atom-to-atom vectors for a given model peptide structure will be primarily aligned with the helical long-axis. Depending on the structure in question, the optimal composition of this aggregate vector sum varied. For example, Σ[Cαn-1 → Cαn + Nn-1 → Nn + C‘n-1 → C‘n] seemed to provide the best alignment for 1QSU. Other structures were better aligned by an aggregate vector summing each bond along the backbone. See the Supplementary Material for the details of the compositions of aggregate vectors used.
The atomic coordinates for the entire structure were then rotated such that this aggregate vector was aligned along an arbitrary axis which was chosen as the Z-axis in an (rh, θ, z) style cylindrical polar-coordinate system, where rh is the distance from the center-line of the cylinder, θ is the counterclockwise angle from an arbitrary line in the plane perpendicular to z (i.e., Cα of G1 lies at θ = 0 in Fig. 1 ▶), and z is the translational distance along the cylinder. The means and standard deviations given in Tables 2 and 3 could then be readily calculated.
For structure 1BKV, residues 10–21 of each chain were analyzed separately owing to the more extended conformation observed in this amino acid–rich X and Y region (note that the last triplet in this region is the imino triplet Gly-Pro-Hyp—it was included because of its better agreement with the amino portion) compared with the other nine structures that are iminorich. Rather than picking specific triplets to exclude from the overall data set (such as terminal triplets or those containing nonstandard residues), any values lying outside of ±1.645 SD from the mean of the original data set were excluded from the values included in Figure 1 ▶ and in Tables 2 and 3. Because there is no single accepted method for rejection of outliers (for extensive discussion, see Barnett and Lewis 1994), this is necessarily an arbitrary trimming of each mean. Were the initial data sets normally distributed, 10% of the data points would be excluded by this filter; in almost all cases, <10% of the data was excluded, and all resulting normal distributions had a better qualitative fit to the central desired portion of the data, implying that the trimmed means and standard deviations are representative. Also, in all cases but four that are indicated in the table, trimmed means calculated in this manner were <1% different from those calculated by the often used method with four iterations of trimming outliers at μ ± 2σ, where a new μ and σ are calculated after each trim. Normally distributed values were not assumed; comparison to normal distributions was simply used as a qualitative aid during analysis. Note that no such filtering was applied in the separate data set for the middle section of 1BKV for any statistics reported with an N of <85. The proportions excluded by this statistical filtering are available in the Supplementary Material. In general, such filtering will become less and less necessary as more model peptide structures become incorporated into the parameter set.
SCWRL 2.95 (Bower et al. 1997) freely available from F.E. Cohen (University of California at San Francisco, USA) and R.L. Dunbrack (Fox Chase Cancer Center, Philadelphia, PA, USA) at http://www.fccc.edu/research/labs/dunbrack/scwrl/) was used for comparison with homology models. Each PDB file listed in Table 1 was used as a homology model backbone template for comparison to PDB files produced using the parameters in Tables 2 and 3. Note that for the iminorich regions, SCWRL was used to produce a homology model with pure (GPP) triplet repeats to ensure prediction of a similar Cβ orientation to the statistical average. RMSD calculations were performed with ProFit 2.2, freely available from A.C.R. Martin, University of Reading, UK, (http://www.bioinf.org.uk/software/profit/), which uses the McLachlan algorithm (McLachlan 1982).
Finally, it should be noted that the parameters given herein are amenable to the production of triple-helical structures lying along the long axis of a cylinder described in cylindrical polar coordinates of the form (rh, θ, z). The sign convention used is as follows: A positive Δθ is a counterclockwise rotation, and a positive Δz corresponds to a translation along the helical long-axis from N-terminal to C-terminal. Conversion of such a structure to cartesian coordinates in form (x, y, z) is straightforward: x = rh(cosθ), y = rh(sinθ), and z remains unchanged.
Electronic supplemental material
Two PDB format files containing coordinates for triple-helical [(GAA)10]3 sequences generated using the iminorich (GAA_PVTHim.pdb) and iminodeficient (GAA_PVTHam.pdb) parameter sets given above. Additional details about the Materials and Methods are given in Sup_PVMeth.pdf, an Adobe Acrobat format file.
Acknowledgments
This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC). J.K.R. is grateful to NSERC for a Postgraduate Scholarship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0218502.
References
- Barnett, V. and Lewis, T. 1994. Outliers in statistical data, 3rd ed. John Wiley & Sons, Chichester, United Kingdom.
- Beck, K. and Brodsky, B. 1998. Supercoiled protein motifs: The collagen triple-helix and the α-helical coiled coil. J. Struct. Biol. 122 17–29. [DOI] [PubMed] [Google Scholar]
- Bella, J., Eaton, M., Brodsky, B., and Berman, H.M. 1994. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science 266 75–81. [DOI] [PubMed] [Google Scholar]
- Bella, J., Brodsky, B., and Berman, H.M. 1995. Hydration structure of a collagen peptide. Structure 3 893–906. [DOI] [PubMed] [Google Scholar]
- Berisio, R., Vitagliano, L., Mazzarella, L., and Zagari, A. 2002. Crystal structure of the collagen triple-helix model [(Pro-Pro-Gly)10]3. Protein Sci. 11 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, F.C., Koetzle, T.F., Williams, G.J., Meyer, Jr., E.E., Brice, M.D., Rodgers, J.R., Kennard, O., Shimanouchi, T., and Tasumi, M. 1977. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 112 535–542. [DOI] [PubMed] [Google Scholar]
- Bower, M.J., Cohen, F.E., and Dunbrack, Jr., R.L. 1997. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: A new homology modeling tool. J. Mol. Biol. 267 1268–1282. [DOI] [PubMed] [Google Scholar]
- Brodsky, B. and Shah, N.K. 1995. Protein motifs, 8: The triple-helix motif in proteins. FASEB J. 9 1537–1546. [DOI] [PubMed] [Google Scholar]
- Engh, R.A. and Huber, R. 1991. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. A 47 392–400. [Google Scholar]
- Harbury, P.B., Tidor, B., and Kim, P.S. 1995. Repacking protein cores with backbone freedom: Structure prediction for coiled coils. Proc. Natl. Acad. Sci. 92 8408–8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kajava, A.V. 2001. Proteins with repeated sequence: Structural prediction and modeling. J. Struct. Biol. 134 132–144. [DOI] [PubMed] [Google Scholar]
- Klein, T.E. and Huang, C.C. 1999. Computational investigations of structural changes resulting from point mutations in a collagen-like peptide. Biopolymers 49 167–183. [DOI] [PubMed] [Google Scholar]
- Kramer, R.Z., Vitagliano, L., Bella, J., Berisio, R., Mazzarella, L., Brodsky, B., Zagari, A., and Berman, H.M. 1998. X-ray crystallographic determination of a collagen-like peptide with the repeating sequence (Pro-Pro-Gly). J. Mol. Biol. 280 623–638. [DOI] [PubMed] [Google Scholar]
- Kramer, R.Z., Bella, J., Mayville, P., Brodsky, B., and Berman, H.M. 1999. Sequence-dependent conformational variations of collagen triple-helical structure. Nat. Struct. Biol. 6 454–457. [DOI] [PubMed] [Google Scholar]
- Kramer, R.Z., Venugopal, M.G., Bella, J., Mayville, P., Brodsky, B., and Berman, H.M. 2000. Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair. J. Mol. Biol. 301 1191–1205. [DOI] [PubMed] [Google Scholar]
- Mayo, K.H. 1996. NMR and X-ray studies of collagen model peptides. Biopolymers 40 359–370. [DOI] [PubMed] [Google Scholar]
- McLachlan, A.D. 1982. Rapid comparison of protein structures. Acta Crystallogr. A 38 871–873. [Google Scholar]
- Nagarajan, V., Kamitori, S., and Okuyama, K. 1998. Crystal structure analysis of collagen model peptide (Pro-Pro-Gly)10. J. Biochem. (Tokyo) 124 1117–1123. [DOI] [PubMed] [Google Scholar]
- Nagarajan, V., Kamitori, S., and Okuyama, K. 1999. Structure analysis of a collagen-model peptide with a (Pro-Hyp-Gly) sequence repeat. J. Biochem. (Tokyo) 125 310–318. [DOI] [PubMed] [Google Scholar]
- Okuyama, K., Takayanagi, M., Ashida, T., and Kakudo, M. 1977. New structural model for collagen. Polym. J. 9 341–343. [Google Scholar]
- Paige, M.F. and Goh, M.C. 2001. Ultrastructure and assembly of segmental long spacing (SLS) collagen studied by atomic force microscopy. Micron 32 355–361. [DOI] [PubMed] [Google Scholar]
- Paige, M.F., Rainey, J.K., and Goh, M.C. 2001. A study of fibrous long spacing collagen ultrastructure and assembly by atomic force microscopy. Micron 32 341–353. [DOI] [PubMed] [Google Scholar]
- Persikov, A.V., Ramshaw, J.A., Kirkpatrick, A., and Brodsky, B. 2000. Amino acid propensities for the collagen triple-helix. Biochemistry 39 14960–14967. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Peptide investigations of pairwise interactions in the collagen triple-helix. J. Mol. Biol. 316 385–394. [DOI] [PubMed] [Google Scholar]
- Rich, A. and Crick, F.H.C. 1961. Molecular structure of collagen. J. Mol. Biol. 3 483–506. [DOI] [PubMed] [Google Scholar]
- Schiffer, M. and Edmundson, A.B. 1967. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys. J. 7 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick, J., Kolinski, A., and Mohanty, D. 1999. De novo predictions of the quaternary structure of leucine zippers and other coiled coils. Int. J. Quantum Chem. 75 165–176. [Google Scholar]
- Vitagliano, L., Berisio, R., Mazzarella, L., and Zagari, A. 2001. Structural bases of collagen stabilization induced by proline hydroxylation. Biopolymers 58 459–464. [DOI] [PubMed] [Google Scholar]
- Yang, P.K., Tzou, W.S., and Hwang, M.J. 1999. Restraint-driven formation of α-helical coiled coils in molecular dynamics simulations. Biopolymers 50 667–677. [DOI] [PubMed] [Google Scholar]
Web references
- http://www.bioinf.org.uk/software/profit; ProFit 2.2.
- http://www.rcsb.org/pdb; Protein Data Bank.
- http://www.fccc.edu/research/labs/dunbrack/scwrl; SCWRL 2.95.
- http://www.chem.utoronto.ca/staff/MCG/; Web-based interface for the generation of triple-helical structures of any primary sequence and length.