Abstract
Basic region leucine zipper (bZip) proteins contain a bipartite DNA-binding motif consisting of a coiled-coil leucine zipper dimerization domain and a highly charged basic region that directly contacts DNA. The basic region is largely unfolded in the absence of DNA, but adopts a helical conformation upon DNA binding. Although a coil → helix transition is entropically unfavorable, this conformational change positions the DNA-binding residues appropriately for sequence-specific interactions with DNA. The N-terminal residues of the GCN4 DNA-binding domain, DPAAL, make no DNA contacts and are not part of the conserved basic region, but are nonetheless important for DNA binding. Asp and Pro are often found at the N-termini of α-helices, and such N-capping motifs can stabilize α-helical structure. In the present study, we investigate whether these two residues serve to stabilize a helical conformation in the GCN4 basic region, lowering the energetic cost for DNA binding. Our results suggest that the presence of these residues contributes significantly to helical structure and to the DNA-binding ability of the basic region in the absence of the leucine zipper. Similar helix-capping motifs are found in approximately half of all bZip domains, and the implications of these findings for in vivo protein function are discussed.
Keywords: Basic region, leucine zipper, N-cap, DNA binding, conformational change
Basic region leucine zipper (bZip) proteins contain a bipartite DNA-binding motif consisting of a coiled-coil leucine zipper dimerization domain and a highly charged basic region that directly contacts DNA (for reviews, see Hu and Saver 1992; Hurst 1995). In the presence of DNA, the bZip domain forms a dimer of uninterrupted α-helices (Ellenberger et al. 1992; König and Richmond 1993; Keller et al. 1995). The leucine zipper region is both necessary and sufficient for dimerization (Gentz et al. 1989; Landschulz et al. 1989; Ransone et al. 1990), forming a stable, two-stranded, parallel coiled coil (O’Shea et al. 1989a, 1989b, 1991). The basic region, on the other hand, is largely unstructured in the absence of DNA, but adopts a helical structure upon DNA binding (O’Neil et al. 1990; Patel et al. 1990; Talanian et al. 1990; Weiss 1990; Weiss et al. 1990; O’Neil et al. 1991; Saudek et al. 1991; Talanian et al. 1992; Cuenoud and Schepartz 1993; Richards et al. 1996).
Although the coil → helix transition observed in bZip proteins upon DNA binding is entropically unfavorable, this conformational change positions the DNA-binding residues appropriately for sequence-specific interactions with DNA. However, the requirement for a structural change can lead to a reduction in specificity if there are nonspecific modes of binding that do not require the same folding transition (Johnson et al. 1994). Indeed, Record et al. have observed that extended, 17-residue tetracationic peptides bind nonspecifically to DNA with lower affinity than do compact, five-residue peptides with the same overall charge. This result suggests an energetic penalty for helix formation or other conformational changes required for the longer peptides to bind to DNA (Padmanabhan et al. 1997). Nonetheless, a number of proteins in a variety of structural classes undergo folding transitions upon protein•DNA complexation (Spolar and Record 1994).
Recent studies of bZip variants suggest that a coil → helix conformational change is not necessary for high-affinity, specific binding (Takemoto and Fisher 1995; Lajmi et al. 2000). Shin and coworkers have shown that an Ala-rich variant of the GCN4 basic region binds to DNA with only a sevenfold decrease in affinity relative to wild-type (Lajmi et al. 2000). Importantly, high-affinity specific DNA binding is maintained despite the loss of favorable Coulombic interactions. As expected, this variant is substantially more helical than wild-type in the absence of DNA. Similarly, multiple alanine substitutions in the basic domain of b-HLH-ZIP proteins enhance DNA binding affinity (Takemoto et al. 1995). Taken together, these results suggest that the energetic penalty associated with DNA binding by a flexible domain can be avoided, at least in part, by stabilizing nascent or partial helix formation in the absence of DNA.
The C-terminal 56 residues of the yeast transcriptional activator GCN4 (residues 226–281) are both necessary and sufficient for DNA binding (Hope and Struhl 1986; Oakley and Dervan 1990). This domain contains both the leucine zipper dimerization domain and the basic region. The N-terminal residues, DPAAL, make no DNA contacts and are not part of the conserved basic region, but are nonetheless important for DNA binding (Oakley and Dewan 1990). Asp and Pro are often found at the N-termini of α-helices, and they are followed by the excellent helix-forming residues Ala and Leu (Richardson and Richardson 1988). We therefore considered the possibility that residues 226–230 of GCN4 play a structural role, facilitating helix formation and reducing the entropic cost of the coil → helix transition that takes place upon DNA binding.
Statistical analysis of peptides and proteins has shown that there is a strong preference for certain amino acid residues at the N- and C-termini of α-helices (Richardson and Richardson 1988). Because the first four amide groups and the last four carbonyl groups necessarily lack the intrahelical i → i + 4 hydrogen bonds that are characteristic of α-helices, there is a preference at these positions for residues that can accommodate the unique hydrogen bonding requirements of the backbone amide and carbonyl groups. Amide hydrogens at the helix N-terminus often pair with side-chain H-bond acceptors, and carbonyl oxygens at the C-terminus are satisfied primarily by hydrogen bonds with backbone amide groups from the adjacent loop or turn (Presta and Rose 1988).
Helix-capping motifs have been shown to stabilize α-helices in peptide models (Bruch et al. 1991; Chakrabartty et al. 1993; Forood et al. 1993; Harper and Rose 1993; Lyu et al. 1993; Yumoto et al. 1993; Seale et al. 1994; Zhou et al. 1994; Petukhov et al. 1996; Esposito et al. 1997; Reymond et al. 1997) and in intact proteins (Serrano and Fersht 1989; Bell et al. 1992; Thapar et al. 1996). In the present study, we investigate whether the Asp and Pro residues at the N-terminus of the GCN4 bZip domain serve to stabilize a helical conformation for the GCN4 basic region, lowering the energetic cost for DNA binding. In addition, we investigate the role of these residues on the affinity and specificity of GCN4•DNA interactions. The implications of our findings for in vivo protein function are discussed.
Results
To investigate the importance of putative helix-capping residues for the structure and function of the GCN4 basic region, the bZip peptides GCN4–56 and GCN4–54 were overexpressed and purified from Escherichia coli. GCN4–56 contains the 56 C-terminal residues (226–281) of GCN4, including both the leucine zipper dimerization domain and the basic region (Fig. 1a ▶). GCN4–56 also contains a Met-Lys dipeptide at the N-terminus for efficient translation initiation of the recombinant protein fragment in E. coli (Looman et al. 1987). The resulting 58-residue peptide is the same GCN4 construct that was used for crystallization of the peptide complex with AP-1 DNA (Ellenberger et al. 1992) and for NMR studies of basic region dynamics in the absence of DNA (Bracken et al. 1999), with the exception of a single Met-to-Val substitution to avoid problems with Met oxidation. GCN4–54 differs only in the absence of two amino acid residues, Asp226 and Pro227, at the N-terminus of the bZip domain (Fig. 1a ▶).
Fig. 1.
Sequences from N- to C-termini of (A) GCN4–56 and GCN4–54 and (B) BR31 and BR29. The sequence of GCN4–56 corresponds to residues 226–281 of GCN4, preceded by an N-terminal Met-Lys dipeptide (Hollenbeck 2000). The sequence of BR31 corresponds to residues 226–252 of GCN4, followed by a Gly-Gly-Cys-Trp tetrapeptide at the C-terminus. In addition, because oxidation of the Met residue in the linker region (Met 250) leads to a reduction in helical content, this residue has been changed to Val in each peptide to allow a more accurate comparison of helical content. (C) Sequences of the DNA oligonucleotides used in the mobility shift assays and CD studies. The arrows indicate the relative orientations of each half-site within the full binding site. The center of the pseuosymmetric AP-1 site is indicated with a circle.
The effect of helix-capping residues on basic region structure
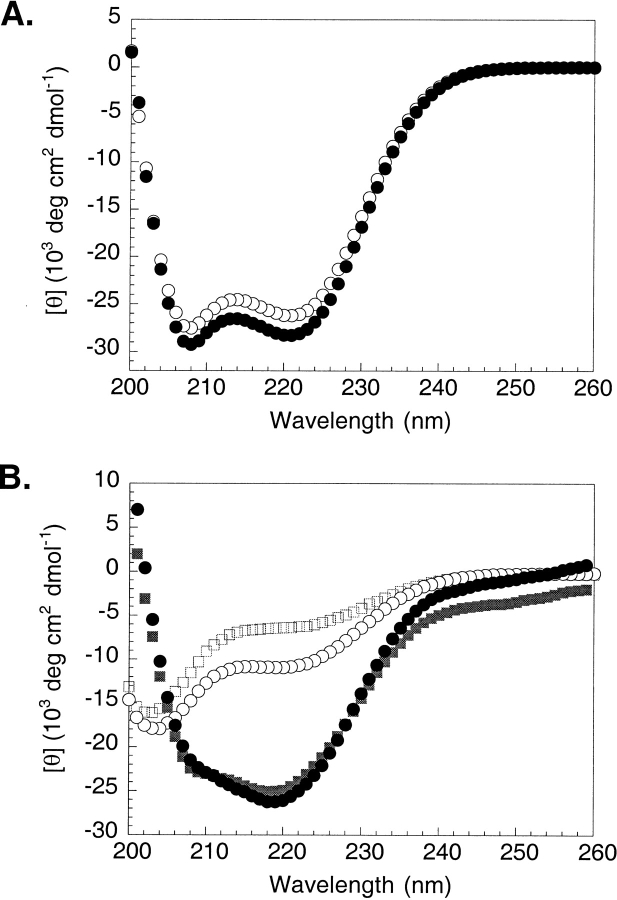
We used CD spectroscopy to determine whether the presence of the Asp-Pro dipeptide affects the degree of helicity in the GCN4 bZip domain in the absence of DNA. At 25°C, GCN4–56 is ∼74% α-helical based on a mean residue ellipticity of −28,100 deg cm2 dmol−1 at 222 nm (Fig. 2a ▶). This observation is consistent with a helical leucine zipper domain and basic region that is not well structured, as others have observed (O’Neil et al. 1990; Talanian et al. 1990; Weiss, 1990; Weiss et al. 1990; Saudek et al. 1991; Cuenoud and Schepartz 1993). GCN4–54 is slightly less helical (∼69%) under the same conditions. This small difference in θ222 is reproducible. However, the CD signal of both proteins is dominated by the helical leucine zipper domain, making differences in the structure of the basic region difficult to detect within the error of the experiment (see Materials and Methods).
Fig. 2.
(A) CD spectra of GCN4–54 (open circles) and GCN4–56 (filled circles) in 50 mM KCl and 12.5 mM potassium phosphate at pH 7.0 and 25°C. Values of [θ]222 (deg cm2 dmol−1) are −26,100 for GCN4–54 and −28,100 for GCN4–56. These values correspond to folded peptides that are 69 and 74% helical, respectively (Chen et al. 1974). (B) CD spectra of BR29ox (open squares) and BR31ox (open circles) in the absence (open symbols) and presence (filled symbols) of CRE DNA under the same conditions. Values of [θ]222 (deg cm2 dmol−1) are −6270 for BR29ox alone, −10,700 for BR31ox alone, −23,900 for BR29ox bound to CRE DNA, and −25,000 for BR31ox bound to CRE DNA. These values correspond to folded peptides that are 17, 29, 66, and 69% helical, respectively (Chen et al. 1974).
We therefore synthesized two basic region peptides, BR31 and BR29 (Fig. 1b ▶), corresponding to residues 226–252 and 228–252 of GCN4, respectively, with the Met-to-Val substitution discussed above. A Gly-Gly-Cys-Trp tetrapeptide was appended to the C-terminus of each peptide for disulfide-bond formation and concentration determination. The oxidized peptide dimers, BR31ox and BR29ox, functionally mimic the full bZip domain, as the essential contribution of the leucine zipper to DNA binding is dimerization (Talanian et al. 1990, 1992). Similar peptides have been shown previously to bind specifically to DNA (Talanian et al. 1990).
As expected, the basic region peptides are largely unfolded in the absence of DNA at room temperature (Fig. 2b ▶). However, BR31ox, which contains the helix-capping residues, Asp and Pro, at the N-terminus, is substantially more helical than BR29ox (∼29% and ∼17%, respectively), suggesting that the presence of the Asp-Pro dipeptide at the N-terminus of the bZip domain stabilizes helix formation in the GCN4 basic region. Upon the addition of CRE DNA, the α-helical content of both peptides increases substantially (Fig. 2b ▶), to approximately 69% for BR31ox and approximately 65% for BR29ox. Importantly, there is little difference in the helical content of either peptide when bound to DNA, suggesting a higher entropic cost for DNA binding in the absence of the Asp-Pro dipeptide. Helical structure has also been observed for the GCN4 basic region at low temperatures in the absence of DNA (Weiss 1990; Weiss et al. 1990; Saudek et al. 1991), and the Asp-Pro dipeptide also affects this temperature-dependent transition. Although helix formation for BR31ox is stabilized by approximately the same extent at 0°C as in the presence of DNA (∼65% helical; data not shown), only partial folding is observed for BR29ox at the same temperature (∼41% helical; data not shown). Thus, our data suggest that the entropic penalty for helix formation in the presence or the absence of DNA is reduced by the putative helix-capping residues, Asp and Pro.
The CD difference spectra shown in Figure 2b ▶ were obtained by subtracting the CD spectrum of the CRE DNA solution from the CD spectrum of each peptide•DNA complex. The CD spectrum of the DNA duplex (data not shown) is consistent with B-form DNA, with a minimum at ∼247 nm and a maximum at ∼221 nm. We note that upon complexation, there is a small change in the difference spectra at ∼247 nm, where the protein contributes little, if any, to the observed signal. It therefore appears that a small conformational change in the DNA accompanies protein binding, consistent with the observations of previous workers (Cuenoud et al. 1993; Podust et al. 2001). This DNA conformational change is not surprising, as the DNA fragment in the crystal structure of the GCN4•CRE complex is distorted from B-form by a 20° bend toward the leucine zipper (König and Richmond 1993).
The effect of helix-capping residues on DNA binding
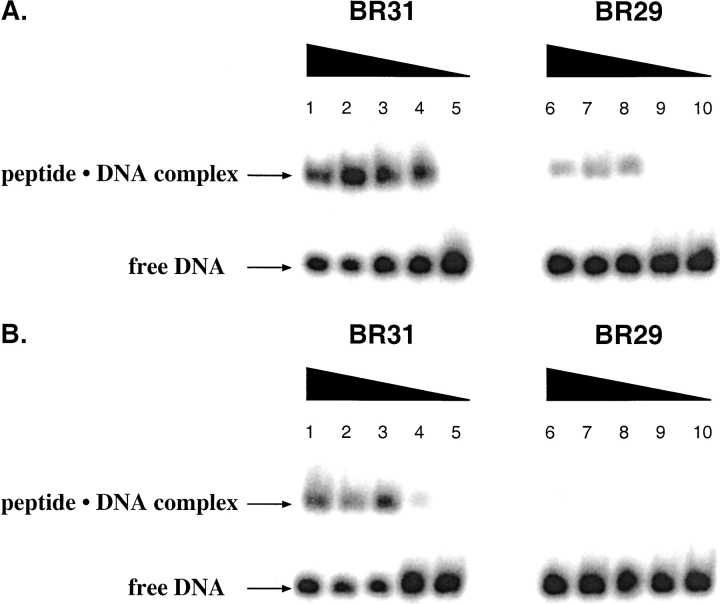
To determine if the presence of helix-capping residues contributes to the DNA-binding affinity and specificity of the GCN4 bZip domain, we probed the DNA binding properties of both BR31ox and BR29ox using an electrophoretic mobility-shift assay. We monitored peptide•DNA complex formation in the absence (Fig. 3a ▶) and presence (Fig. 3b ▶) of excess nonspecific DNA (calf thymus, 100 μM in bp). Although the pseudosymmetric AP-1 sequence, 5′-TGA(C/G)TCA-3′, is the optimal recognition site for GCN4 (Hill et al. 1986; Oliphant et al. 1989; Mavrothalassitis et al. 1990), high-affinity binding to the perfectly symmetric CRE site, 5′-ATGACGTCAT-3′ has also been observed (Sellers et al. 1990). Interestingly, we were not able to detect DNA binding in the presence of AP-1 DNA with either BR31ox or BR29ox (data not shown). This observation is consistent with those of Talanian et al. (1992).
Fig. 3.
Equilibrium binding of BR29 and BR31 to CRE DNA in PBS buffer in the (A) absence and (B) presence of calf thymus DNA (100 μM in base pairs). Peptide concentrations are 6 μM (lanes 1 and 6), 1 μM (lanes 2 and 7), 600 nM (lanes 3 and 8), and 100 nM (lanes 4 and 9). Lanes 5 and 10 contained 32P-end-labeled DNA only.
Both BR31ox and BR29ox form a stable complex with CRE DNA in the absence of excess nonspecific DNA. However, BR29ox appears to bind to DNA with somewhat lower affinity than does BR31ox (cf. lanes 4 and 9 of Fig. 3a ▶). Unfortunately, the degree of DNA binding site saturation is low for both peptides, precluding precise measurements of apparent dissociation constants. It appears, however, that the binding site saturation is lower for BR29ox than for BR31ox, perhaps due to faster dissociation rates for the less stable protein•DNA complex.
Upon the addition of nonspecific competitor DNA (calf thymus, 100 μM in bp) to the binding reactions, no complex formation is observed with BR29ox, even at the highest peptide concentration (6 μM) (Fig. 3b ▶). Significantly, although BR31ox appears to have slightly lower affinity for CRE DNA in the presence of competitor DNA than in its absence, the helix-cap-containing peptide still forms a detectable protein•DNA complex at a protein concentration of 100 nM. Thus, removal of the Asp-Pro dipeptide from the N-terminus of the basic region appears to reduce both the affinity and specificity of DNA binding.
Because helix-capping residues have a clear impact on DNA binding by disulfide-linked basic region peptides, we examined the effect of these residues in the context of the intact bZip domain. We measured the apparent Kd of each GCN4 peptide for AP-1 DNA in the presence of increasing amounts of calf thymus DNA. Calculated values for the apparent monomeric equilibrium dissociation constants are shown in Table 1. In the absence of nonspecific competitor DNA, GCN4–56 binds AP-1 DNA with an apparent dissociation constant of (4 ± 2) × 10−8 M, in agreement with previously published values (Hope and Smith 1985; Weiss et al. 1990; Metallo and Schepartz 1994; Berger et al. 1996; Hollenbeck and Oakley 2000). GCN4–54 has comparable affinity for the AP-1 site (Kd (apparent) = (3 ± 1) × 10−8 M). Importantly, at concentrations of calf thymus DNA between 0 and 300 μM bp, there was no detectable difference in the affinity of either protein for AP-1 DNA, suggesting that the helix cap does not affect DNA-binding affinity or specificity in the context of the dimeric bZip domain.
Table 1.
Binding constants for GCN4-56 and GCN4-54 to AP-1 DNA in the presence of increasing amounts of competitor DNA as measured by an electrophoretic mobility-shift assay
| [CT DNA] (μM bp) | Kd (M)a | |
| GCN4-56 | GCN4-54 | |
| 0 | 4 ± 2 × 10−8 | 3 ± 1 × 10−8 |
| 100 | 1.1 ± 0.2 × 10−7 | 1.2 ± 0.3 × 10−7 |
| 300 | 2.8 ± 0.7 × 10−7 | 1.8 ± 0.4 × 10−7 |
a Values reported are the mean values ± the standard deviation in the values measured from three individual data sets.
Discussion
In addition to defining the beginnings and ends of α-helices, helix-capping motifs play a role in stabilizing protein structure. The importance of N-terminal capping has been demonstrated using synthetic model peptides (Chakrabartty et al. 1993; Harper and Rose 1993; Yumoto et al. 1993; Seale et al. 1994; Petukhov et al. 1996) and site-directed mutagenesis of helix termini in native proteins (Serrano and Fersht 1989; Bell et al. 1992; Thapar et al. 1996). Indeed, N-capping motifs can stabilize monomeric helices by up to 2 kcal mol−1 relative to the unfolded random coil state (Lyu et al. 1992; Doig and Baldwin 1995; Petukhov et al. 1996).
In a statistical survey of α-helices in globular protein structures, Richardson and Richardson observed a preference for Asn, Asp, Ser, Thr, and Gly in the N-cap position and a strong preference for Pro in the N-cap + 1 (N1) position (Richardson and Richardson 1988). As an N-cap, Asp can interact favorably with the helix dipole (Wada 1976; Hol 1985) as well as form a hydrogen bond to one of the exposed backbone NH groups in the first turn of helix. In the N + 1 position, Pro is ideal because it lacks an exposed backbone NH group that would need to be satisfied by a side-chain H-bond acceptor (Presta and Rose 1988; Richardson and Richardson 1988). In the crystal structure of the GCN4•AP-1 DNA complex, the side chain of Asp226 in one monomer is indeed folded in a conformation suitable for H-bond formation to the exposed backbone amide of Ala228. However, the distance between the side chain carbonyl oxygen and the nitrogen of Ala228 is 4.31 Å, outside of H-bonding distance. In the crystal structure of the GCN4•CRE DNA complex, no electron density is visible for Asp226 suggesting multiple conformations for its side chain. Thus, it is not clear whether Asp226 forms an energetically significant H-bond with an unpaired amide nitrogen at the N-terminal end of the basic region helix.
Nonetheless, we have found that the residues Asp and Pro at the N-terminus of the GCN4 basic region do indeed increase the extent of helix formation in the absence of DNA (Fig. 2b ▶). Importantly, upon DNA binding, there is little difference in the helical content of either peptide, suggesting that the two DNA-bound complexes are similar in structure. Thus, the presence of helix-capping residues at the N-terminus reduces the amount of free energy required to complete the coil → helix transition observed upon binding DNA.
Because these residues are important for GCN4 basic region structure, we searched for helix-capping motifs in other naturally occurring bZip proteins (see Materials and Methods). A search of the Swiss-Prot database identified 104 unique bZip sequences. Of these, 50 (∼50%) have putative helix-capping residues near the N-terminus of the bZip domain. One representative sequence of each of the 13 families of cloned bZip proteins identified by Hurst (1995) is shown in Figure 4 ▶. Importantly, the presence or absence of helix-capping residues is strongly conserved within each family. Taken together, these data suggest that helix-stabilizing residues occur commonly at the N-terminus of the basic region and that these sequences are functionally significant in the bZip classes in which they are found.
Fig. 4.
Amino acid sequences of naturally occurring bZip basic regions. Putative helix-capping residues are shown in bold and underlined.
Intriguingly, although the Asp-Pro dipeptide has a clear impact on DNA binding by disulfide-linked basic region peptides, the presence of these residues at the N-terminus of the basic region does not appear to affect DNA-binding affinity or specificity in the context of the full bZip domain (Table 1). One explanation for this observation is that a helical leucine zipper domain itself stabilizes nascent or partial helix formation in the basic region. Both GCN4–56 and GCN4–54 are considerably more helical in the absence of DNA than would be expected if only the leucine zipper region were folded. Because approximately 30 residues within the coiled coil are folded (O’Shea et al. 1991; Ellenberger et al. 1992), we would expect GCN4–56 to be 52% helical in the absence of DNA. The observed higher degree of helicity (∼74%, Fig. 2a ▶) can be explained by partial helix formation in the basic region. Indeed, NMR studies have demonstrated that the GCN4 basic region adopts fluctuating helical structure in the absence of DNA (Saudek et al. 1991; Bracken et al. 1999). Similarly, the GCN4 leucine zipper region appears to stabilize DNA-dependent α-helix formation in a hybrid protein in which the GCN4 basic region is grafted into a small helical domain from the cytoskeletal protein villin (Morii et al. 2002). If the leucine zipper stabilizes helix formation more effectively than does the Asp-Pro dipeptide, or if the effects of the N-cap and the leucine zipper are not additive, little or no change in the DNA binding properties of the dimeric bZip domain would be expected.
Significantly, the relatively high (μM) dimerization constants of bZip proteins (O’Shea et al. 1989a; O’Neil et al. 1991; Kohler eta l. 1999) and the short lifetime of the GCN4 dimer (Weiss et al. 1990) suggest that GCN4 exists in vivo primarily as unfolded monomers. Indeed, several groups have observed that bZip peptides can bind to DNA sequentially as monomers that subsequently dimerize on the DNA (Metallo and Schepartz 1997; Berger et al. 1998; Kohler et al. 1999). In this case, the coiled-coil domain will not affect helix formation in the basic region as GCN4 leucine zipper monomers are primarily unfolded in solution (Zitzewitz et al. 1995). The impact of helix-stabilizing residues at the N-terminus of the basic region is therefore expected to be maximal in the case of monomer•DNA binding. Schepartz et al. have demonstrated that excess nonspecific DNA slows the dimer binding pathway, but not the sequential monomer binding pathway, suggesting that the monomer pathway may allow bZip proteins to find their target sites more quickly (Kohler et al. 1999). Thus, in vivo, in the presence of a large excess of nonspecific DNA, the basic region helix-capping motif is likely to contribute significantly to DNA-binding affinity and specificity.
Materials and methods
Peptide production and purification
Construction of the plasmid, pGCN4–56, encoding the basic region and leucine zipper of GCN4 (residues 226–281), was described previously (Hollenbeck and Oakley 2000). pGCN4–54, encoding residues 228–281 of GCN4, was constructed by mutagenesis using QuikChange Site-Directed Mutagenesis (Strategene). The primary sequence of each construct was confirmed by DNA sequencing (Sanger and Coulson 1977). GCN4–54 and GCN4–56 were overexpressed in E. coli strain BL21(DE3) pLysS using the T7 expression system (Studier et al. 1990) and purified from cell lysates as described previously (Hollenbeck and Oakley 2000).
The peptides BR29 and BR31 were synthesized by solid-phase methods and purified by reverse-phase HPLC with a Vydac C18 column and a linear water/acetonitrile gradient containing 0.1% (v/v) trifluoroacetic acid as described previously (McClain et al. 2001). All peptides were acetylated at the N-terminus and amidated at the C-terminus. Peptide concentrations were determined by UV absorbance in 6 M guanidinium hydrochloride and 0.02 M phosphate (pH 6.5) assuming an extinction coefficient of 5690 M−1 cm−1 for Trp at 280 nm (Edelhoch 1967). The molecular mass of each purified peptide was confirmed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Covalently constrained dimers were produced through air oxidation (phosphate buffer, pH 8–9), and purified by reverse-phase HPLC as described previously (O’Shea et al. 1989a).
Electrophoretic mobility shift assays
Oligonucleotides (Genosys) containing the DNA binding sites AP-1 and CRE (Fig. 1c ▶) were purified by preparative denaturing (19% polyacrylamide, 19:1 acrylamide:bisacrylamide) gel electrophoresis and 5′-end-labeled by standard procedures (Sambrook et al. 1989). Radiolabeled oligonucleotides were annealed to their complement by heating an equimolar mixture of the two single-strand DNAs to 90°C for 2 min and then cooling slowly to room temperature.
Protein•DNA complex formation was monitored in PBS binding buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM sodium phosphate, 1.4 mM potassium phosphate, 1 mM EDTA, 1 mM DTT, 0.1% Igepal CA-630, and 0.4 mg/mL BSA [pH 7.4]) (Metallo and Schepartz 1994; Hollenbeck and Oakley 2000) supplemented with calf thymus DNA (Hollenbeck and Oakley 2000). Apparent equilibrium dissociation constants were determined for the GCN4–56•AP-1 DNA and GCN4–54•AP-1 DNA complexes as described previously (Hollenbeck and Oakley 2000).
CD spectroscopy
CD spectra were acquired with a Jasco J-715 spectropolarimeter. Samples were prepared in 50 mM KCl and 12.5 mM potassium phosphate (pH 7.0). The wavelength dependence of [θ] was monitored at 25°C by five scans acquired in 1-nm increments with a sampling time of 4 sec. The helix content was determined by the method of Chen et al. (1974). We find from repeated measurements that the error in [θ]222 for protein samples prepared from the same stock solution is approximately 1%, and we estimate that the error in the concentration determination of the protein stock solutions is approximately 5%. The CD spectrum shown for each peptide•DNA complex was obtained by subtracting the CD spectrum of the DNA solution from the CD spectrum of the complex. The spectrum of the CRE DNA duplex is consistent with B-form DNA, with a minimum at ca. 247 nm and a maximum at ca. 221 nm.
Statistical survey of bZip protein sequences
A search of the Swiss-Prot database using the identifier "bZip" yielded 176 sequences. These sequences were aligned with the GCN4 basic region using ClustalW (European Bioinformatics Institute, http://www.ebi.ac.uk/clustalw/). Three sequence fragments were eliminated as were 18 sequences bearing little or no homology to the consensus sequence NXX(A)(A)XX(C/S)R (Hurst 1995). The unique sequences (104/155) were surveyed for the presence of helix-capping residues at the N-terminus of the basic region.
Acknowledgments
We thank A. Hansen for assistance with mass spectrometry. This work was supported by Grant GM57571 from the National Institutes of Health and Grant 32029-G4 from the American Chemical Society–Petroleum Research Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
bZip, basic region leucine zipper
b-HLH-ZIP, basic/helix-loop-helix/leucine zipper
CD, circular dichroism
[θ], molar ellipticity
[θ]222, molar ellipticity at 222 nm
CRE, cAMP-responsive element
bp, base pairs
Kd, apparent monomeric equilibrium dissociation constant
HPLC, high-performance liquid chromatography
MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight
PBS, phosphate-buffered saline
EDTA, ethylenediaminetetraacetic acid
DTT, dl-dithiothreitol
BSA, bovine serum albumin
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0211102.
References
- Bell, J.A., Becktel, W.J., Sauer, U., Baase, W.A., and Matthews, B.W. 1992. Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr59. Biochemistry 31 3590–3596. [DOI] [PubMed] [Google Scholar]
- Berger, C., Jelesarov, I., and Bosshard, H.R. 1996. Coupled folding and site-specific binding of the GCN4-bZip transcription factor to the AP-1 and ATF/CREB DNA sites studied by microcalorimetry. Biochemistry 35 14984–14991. [DOI] [PubMed] [Google Scholar]
- Berger, C., Piubelli, L., Haditsch, U., and Bosshard, H.R. 1998. Diffusion-controlled DNA recognition by an unfolded, monomeric bZip transcription factor. FEBS Lett. 425 14–18. [DOI] [PubMed] [Google Scholar]
- Bracken, C., Carr, P.A., Cavanagh, J., and Palmer III, A.G. 1999. Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: Implications for the entropy of association with DNA. J. Mol. Biol. 285 2133–2146. [DOI] [PubMed] [Google Scholar]
- Bruch, M.D., Dhingra, M.M., and Gierasch, L.M. 1991. Sidechain-backbone hydrogen bonding contributes to helix stability in peptides derived from an alpha-helical region of carboxypeptidase A. Proteins Struct. Funct. Genet. 10 130–139. [DOI] [PubMed] [Google Scholar]
- Chakrabartty, A., Doig, A.J., and Baldwin, R.L. 1993. Helix capping propensities in peptides parallel those in proteins. Proc. Natl. Acad. Sci. 90 11332–11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-H., Yang, J.T., and Chau, K.H. 1974. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13 3350–3359. [DOI] [PubMed] [Google Scholar]
- Cuenoud, B. and Schepartz, A. 1993. Altered specificity of DNA-binding proteins with transition metal dimerization domains. Science 259 510–513. [DOI] [PubMed] [Google Scholar]
- Doig, A.J. and Baldwin, R.L. 1995. N- and C-capping preferences for all 20 amino acids in α-helical peptides. Protein Sci. 4 1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6 1948–1954. [DOI] [PubMed] [Google Scholar]
- Ellenberger, T.E., Brandl, C.J., Struhl, K., and Harrison, S.C. 1992. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: Crystal structure of the protein–DNA complex. Cell 71 1223–1237. [DOI] [PubMed] [Google Scholar]
- Esposito, G., Dhanapal, B., Dumy, P., Varma, V., Mutter, M., and Bodenhausen, G. 1997. Lysine as helix C-capping residue in a synthetic peptide. Biopolymers 41 27–35. [DOI] [PubMed] [Google Scholar]
- Forood, B., Feliciano, E.J., and Nambiar, K.P. 1993. Stabilization of α-helical structures in short peptides via end capping. Proc. Natl. Acad. Sci. 90 838–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz, R., Rauscher III, F.J., Abate, C., and Curran, T. 1989. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science 243 1695–1699. [DOI] [PubMed] [Google Scholar]
- Harper, E.T. and Rose, G.D. 1993. Helix stop signals in proteins and peptides: The capping box. Biochemistry 32 7605–7609. [DOI] [PubMed] [Google Scholar]
- Hill, D.E., Hope, I.A., Macke, J.P., and Struhl, K. 1986. Saturation mutagenesis of the yeast his3 regulatory site: Requirements for transcriptional induction and for binding by GCN4 activator protein. Science 234 451–457. [DOI] [PubMed] [Google Scholar]
- Hol, W.G.J. 1985. The role of the α-helix dipole in protein function and structure. Prog. Biophys. Mol. Biol. 45 149–195. [DOI] [PubMed] [Google Scholar]
- Hollenbeck, J.J. and Oakley, M.G. 2000. GCN4 binds with high affinity to DNA sequences containing a single consensus half-site. Biochemistry 39 6380–6389. [DOI] [PubMed] [Google Scholar]
- Hope, I.A. and Struhl, K. 1985. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: Implications for general control of amino acid biosynthetic genes in yeast. Cell 43 177–188. [DOI] [PubMed] [Google Scholar]
- ———. 1986. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46 885–894. [DOI] [PubMed] [Google Scholar]
- Hu, J.C. and Sauer, R.T. 1992. The basic region leucine zipper family of DNA binding proteins. Nucleic Acids Mol. Biol. 6 82–101. [Google Scholar]
- Hurst, H.C. 1995. bZip Proteins. Protein Profile 2 105–168. [PubMed] [Google Scholar]
- Johnson, N.P., Lindstrom, J., Baase, W.A., and von Hippel, P.H. 1994. Double-stranded DNA templates can induce α-helical conformation in peptides containing lysine and alanine: Functional implications for leucine zipper and helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. 91 4840–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, W., König, P, and Richmond, T.J. 1995. Crystal structure of a bZIP/DNA complex at 2.2 Å: Determinants of DNA specific recognition. J. Mol. Biol. 254 657–667. [DOI] [PubMed] [Google Scholar]
- Kohler, J.J., Metallo, S.J., Schneider, T.L., and Schepartz, A. 1999. DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl. Acad. Sci. 96 11735–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König, P. and Richmond, T.J. 1993. The x-ray structure of the GCN4-bZip bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol. 233 139–154. [DOI] [PubMed] [Google Scholar]
- Lajmi, A.R., Lovrencic, M.E., Wallace, T.R., Thomlinson, R.R., and Shin, J.A. 2000. Minimalist, alanine-based, helical protein dimers bind to specific DNA sites. J. Am. Chem. Soc. 122 5638–5639. [Google Scholar]
- Landschulz, W.H., Johnson, P.F., and McKnight, S.L. 1989. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 243 1681–1688. [DOI] [PubMed] [Google Scholar]
- Looman, A.C., Bodlaender, J., Comstock, L.J., Eaton, D., Jhurani, P., deBoer, H.A., and van Knippenberg, P.H. 1987. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 6 2489–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, P.C., Zhou, H.X., Jelveh, N., Wemmer, D.E., and Kallenbach, N.R. 1992. Position-dependent stabilizing effects in α-helices: N-terminal capping in synthetic model peptides. J. Am. Chem. Soc. 114 6560–6562. [Google Scholar]
- Lyu, P.C., Wemmer, D.E., Zhou, H.X., Pinker, R.J., and Kallenbach, N.R. 1993. Capping interactions in isolated α helices: Position-dependent substitution effects and structure of a serine-capped peptide helix. Biochemistry 32 421–425. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis, G., Beal, G., and Papas, T.S. 1990. Defining target sequences of DNA-binding proteins by random selection and PCR: Determination of the GCN4 binding sequence repertoire. DNA Cell Biol. 9 783–788. [DOI] [PubMed] [Google Scholar]
- McClain, D.L., Binfet, J.P., and Oakley, M.G. 2001. Evaluation of the energetic contribution of interhelical Coulombic interactions for coiled coil helix orientation specificity. J. Mol. Biol. 313 371–383. [DOI] [PubMed] [Google Scholar]
- Metallo, S.J. and Schepartz, A. 1994. Distribution of labor among bZip segments in the control of DNA affinity and specificity. Chem. Biol. 1 143–151. [DOI] [PubMed] [Google Scholar]
- ———. 1997. Certain bZip peptides bind DNA sequentially as monomers and dimerize on the DNA. Nat. Struct. Biol. 4 115–117. [DOI] [PubMed] [Google Scholar]
- Morii, T., Sato, S.-i., Hagihara, M., Mori, Y., Imoto, K., and Makino, K. 2002. Structure-based design of a leucine zipper protein with new DNA contacting region. Biochemistry 41 2177–2183. [DOI] [PubMed] [Google Scholar]
- O’Neil, K.T., Hoess, R.H., and DeGrado, W.F. 1990. Design of DNA-binding peptides based on the leucine zipper motif. Science 249 774–778. [DOI] [PubMed] [Google Scholar]
- O’Neil, K.T., Shuman, J.D., Ampe, C., and DeGrado, W.F. 1991. DNA-induced increase in the α-helical content of C/EBP and GCN4. Biochemistry 30 9030–9034. [DOI] [PubMed] [Google Scholar]
- O’Shea, E.K., Rutkowski, R., and Kim, P.S. 1989a. Evidence that the leucine zipper is a coiled coil. Science 243 538–542. [DOI] [PubMed] [Google Scholar]
- O’Shea, E.K., Rutkowski, R., Stafford III, W.F., and Kim, P.S. 1989b. Preferential heterodimer formation by isolated leucine zippers from Fos and Jun. Science 245 646–648. [DOI] [PubMed] [Google Scholar]
- O’Shea, E.K., Klemm, J.D., Kim, P.S., and Alber, T. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254 539–544. [DOI] [PubMed] [Google Scholar]
- Oakley, M.G. and Dervan, P.B. 1990. Structural motif of the GCN4 DNA binding domain characterized by affinity cleaving. Science 248 847–850. [DOI] [PubMed] [Google Scholar]
- Oliphant, A.R., Brandl, C.J., and Struhl, K. 1989. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: Analysis of yeast GCN4 protein. Mol. Cell. Biol. 9 2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, S., Zhang, W., Capp, M.W., Anderson, C.F., and Record, Jr., M.T. 1997. Binding of cationic (+4) alanine- and glycine-containing oligopeptides to double-stranded DNA: Thermodynamic analysis of effects of Coulombic interactions and α-helix induction. Biochemistry 36 5193–5206. [DOI] [PubMed] [Google Scholar]
- Patel, L., Abate, C., and Curran, T. 1990. Altered protein conformation on DNA binding by Fos and Jun. Nature 347 572–575. [DOI] [PubMed] [Google Scholar]
- Petukhov, M., Yumoto, N., Murase, S., Onmura, R., and Yoshikawa, S. 1996. Factors that affect the stabilization of α-helices in short peptides by a capping box. Biochemistry 35 387–397. [DOI] [PubMed] [Google Scholar]
- Podust, L.M., Krezel, A.M., and Kim, Y. 2001. Crystal structure of the CCAAT box/Enhancer-Binding Protein β Activation Transcription Factor-4 basic leucine zipper heterodimer in the absence of DNA. J. Biol. Chem. 276 505–513. [DOI] [PubMed] [Google Scholar]
- Presta, L.G. and Rose, G.D. 1988. Helix signals in proteins. Science 240 1632–1641. [DOI] [PubMed] [Google Scholar]
- Ransone, L.J., Visvader, J., Wamsley, P., and Verma, I. M. 1990. Trans-dominant negative mutants of Fos and Jun. Proc. Natl. Acad. Sci. 87 3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, M.T., Huo, S., Duggan, B., Wright, P.E., and Dyson, H.J. 1997. Contribution of increased length and intact capping sequences to the conformational preference for helix in a 31-residue peptide from the C terminus of myohemerythrin. Biochemistry 36 5234–5244. [DOI] [PubMed] [Google Scholar]
- Richards, J.P., Bächinger, H.P., Goodman, R.H., and Brennan, R.G. 1996. Analysis of the structural properties of cAMP-responsive element-binding protein (CREB) and phosphorylated CREB. J. Biol. Chem. 271 13716–13723. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. and Richardson, D.C. 1988. Amino acid preferences for specific locations at the ends of α-helices. Science 240 1648–1652. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, R. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Sanger, F.S.N. and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudek, V., Pasley, H.S., Gibson, T., Gausepohl, H., Frank, R., and Pastore, A. 1991. Solution structure of the basic region from the transcriptional activator GCN4. Biochemistry 30 1310–1317. [DOI] [PubMed] [Google Scholar]
- Seale, J.W., Srinivasan, R., and Rose, G.D. 1994. Sequence determinants of the capping box, a stabilizing motif at the N-termini of α-helices. Protein Sci. 3 1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers, J.W., Vincent, A.C., and Struhl, K. 1990. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol. Cell. Biol. 10 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, L. and Fersht, A.R. 1989. Capping and α-helix stability. Nature 342 296–299. [DOI] [PubMed] [Google Scholar]
- Spolar, R.S. and Record, Jr., M.T. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263 777–784. [DOI] [PubMed] [Google Scholar]
- Studier, F.W., Rosenberg, A.H., Dunn, J.J., and Dubendorff, J.W. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185 60–89. [DOI] [PubMed] [Google Scholar]
- Takemoto, C. and Fisher, D.E. 1995. Structural constraints for DNA recognition by Myc and other b-HLH-Zip proteins: Design of oncoprotein analogs. Gene Expr. 4 311–317. [PMC free article] [PubMed] [Google Scholar]
- Talanian, R.V., McKnight, C.J., and Kim, P.S. 1990. Sequence-specific DNA binding by a short peptide dimer. Science 249 769–771. [DOI] [PubMed] [Google Scholar]
- Talanian, R.V., McKnight, C.J., Rutkowski, R., and Kim, P.S. 1992. Minimum length of a sequence-specific DNA-binding peptide. Biochemistry 31 6871–6875. [DOI] [PubMed] [Google Scholar]
- Thapar, R., Nicholson, E.M., Rajagopal, P., Waygood, E.B., Scholtz, J.M., and Klevit, R.E. 1996. Influence of N-cap mutations on the structure and stability of Escherichia coli HPr. Biochemistry 35 11268–11277. [DOI] [PubMed] [Google Scholar]
- Wada, A. 1976. The α-helix as an electric macro-dipole. Adv. Biophys. 9 1–63. [PubMed] [Google Scholar]
- Weiss, M.A. 1990. Thermal unfolding studies of a leucine zipper domain and its specific DNA complex: Implications for scissor’s grip recognition. Biochemistry 29 8020–8024. [DOI] [PubMed] [Google Scholar]
- Weiss, M.A., Ellenberger, T., Wobbe, C.R., Lee, J.P., Harrison, S.C., and Struhl, K. 1990. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature 347 575–578. [DOI] [PubMed] [Google Scholar]
- Yumoto, N., Murase, S., Hattori, T., Yumamoto, H., Tatsu, Y., and Yoshikawa, S. 1993. Stabilization of α-helix in C-terminal fragments of neuropeptide Y. Biochem. Biophys. Res. Commun. 196 1490–1495. [DOI] [PubMed] [Google Scholar]
- Zhou, H.X., Lyu, P.C., Wemmer, D.E., and Kallenbach, N.R. 1994. Structure of a C-terminal α-helix cap in a synthetic peptide. J. Am. Chem. Soc. 116 1139–1140. [Google Scholar]
- Zitzewitz, J.A., Bilsel, O., Luo, J., Jones, B.E., and Matthews, C.R. 1995. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry 34 12812–12819. [DOI] [PubMed] [Google Scholar]