Abstract
Structural genomics is a new approach in functional assignment of proteins identified via whole-genome sequencing programs. Its rationale is that nonhomologous proteins performing similar or related biological functions might have similar tertiary structure. We used dye pseudoaffinity chromatography, two-dimensional gel electrophoresis, and mass spectrometry to identify two novel Escherichia coli nucleotide-binding proteins, YnaF and YajQ. YnaF exhibited significant sequence identity with MJ0577, an ATP-binding protein from a hyperthermophile (Methanococcus jannaschii), and with UspA, a protein from Haemophilus influenzae that belongs to the Universal Stress Protein family. YnaF conserves the ATP-binding site and the dimeric structure observed in the crystal of MJ0577. The protein YajQ, present in many bacterial genomes, is missing in eukaryotes. In the absence of significant similarities of YajQ to any solved structure, we determined its structural and ligand-binding properties by NMR and isothermal titration calorimetry. We demonstrate that YajQ is composed of two domains, each centered on a β-sheet, that are connected by two helical segments. NMR studies, corroborated with local sequence conservation among YajQ homologs in various bacteria, indicate that one of the β-sheets is mostly involved in biological activity.
Keywords: Structural genomics, YajQ protein, YnaF protein, E. coli, nucleotide binding
One challenging issue of the whole-genome sequencing programs is related to genes encoding proteins of unknown function that represent ∼40% of the bacterial genomes (Riley and Serres 2000). An approach similar to that which transformed gene sequencing into genomics is underway to transform structural biology into structural genomics. The rationale is simple and rewarding from a long-term perspective: the function of an unknown protein might be inferred after comparison with a related known structure for which the function has already been determined. Cost reduction in structure analysis by automation of basic techniques from gene cloning and protein overproduction/purification to structure determination by NMR spectroscopy or X-ray crystallography is a prerequisite of such an approach (Burley et al. 1999; Orengo et al. 1999). Complementary strategies to decipher the functional role of newly discovered genes have been proposed. Among them, the two-hybrid method (Fields and Song 1989) and the affinity purification of protein complexes (Rigaut et al. 1999) are filling the gap toward function assignment using the interaction of unknown proteins with known targets.
Affinity chromatography, and its variant affinity elution chromatography, is another way of deducing the functional properties of proteins from their interaction with defined ligands. In this study, we combined dye pseudoaffinity chromatography, two-dimensional gel electrophoresis, and mass spectrometry to select proteins interacting with nucleotides. Soluble Escherichia coli extract adsorbed on Blue Sepharose at neutral pH was eluted with ATP and NAD+. Among ∼40 eluted proteins, representing ∼2% of the soluble extract, we identified two species of unknown function. One, YnaF, showed significant sequence similarities to UspA, which belongs to the Universal Stress Protein (USP) family (Sousa and McKay 2001), and MJ0577, an ATP-binding protein from the hyperthermophile Methanococcus jannaschii. The second, YajQ, common to several bacterial species but absent in eukaryotes, binds as predicted, ATP but also GTP and tRNA. The physiological roles of YajQ and YnaF are discussed here on the basis of sequence and structural analysis.
Results and Discussion
Identification of YajQ and YnaF in the bacterial proteome
Blue Sepharose is widely used in protein purification and particularly in the isolation of nucleotide-binding proteins (Thompson et al. 1975; Dean and Watson 1979). Free or matrix-bound Cibacron blue interacts with ATP- or NAD+-binding sites of various enzymes. On the other hand, the presence of ionizable groups in the dye molecule confers ion-exchange properties to the matrix. At pH 7.4, ∼20% of an E. coli soluble extract was adsorbed to Blue Sepharose, and ∼10% of this (i.e., 2% of the initial amount of proteins) was eluted with 2 mM ATP, a nucleotide concentration in which the "salt effect" is negligible. As shown in Figure 1 ▶, among 25 spots identified in two-dimensional (2D) gel electrophoresis, five spots represent ∼60% of the total ATP-eluted proteins as estimated from the intensity of Coomassie blue staining: phosphoglycerate kinase, 22%; malate dehydrogenase, 15%; elongation factor, 9.4%; nucleoside diphosphate kinase, 7.9%; and 6-phosphogluconate dehydrogenase, 6.3%. Other clearly represented spots correspond to ADP-glyceromannoheptose-6-epimerase, phosphoglycerate mutase, acetate kinase, glycerol kinase, pantothenate synthetase, YajQ (SWISS-PROT P77482), and YnaF (SWISS-PROT P37903), the second comigrating with nucleoside diphosphate kinase. Except for phosphoglycerate mutase, which uses bisphosphoglycerate as a cofactor, all other identified spots correspond as expected to nucleotide-binding proteins.
Fig. 1.

Identification by 2D gel electrophoresis and mass spectrometry of proteins eluted from a Blue Sepharose column with 2 mM ATP. The identity of the spots is the following: Pgk, phosphoglycerate kinase; Mdh, malate dehydrogenase; TufA, elongation factor; Ndk, nucleoside diphosphate kinase; Gnd, 6-phosphogluconate dehydrogenase; RfaD, ADP-glycero mannoheptose-6-epimerase; GpmA, phosphoglycerate mutase; AckA, acetate kinase; GlpK, glycerol kinase; PanC, panthotenate synthase; GlnB, the regulatory subunit of glutamine synthetase; AtpD, the β subunit of ATP synthase; Eda, 2-keto-3-deoxygluconate 6 phosphate aldolase.
The 2 mM NAD+ eluted several other nucleotide-binding proteins, for example, thioredoxin reductase, isocitrate dehydrogenase, and lysyl-tRNA synthetase, to which the remaining YnaF, malate dehydrogenase, and elongation factor Tu should be added (data not shown). The two proteins with unknown function identified by this procedure as nucleotide-binding species were therefore YajQ and YnaF. It is worth mentioning that both YnaF and YajQ were previously visualized on 2D gels (Pasquali et al. 1996; Fountoulakis et al. 1999), the second only after enrichment by hydroxylapatite chromatography. Because YajQ was separated from the other ATP-eluted species even after one-dimensional (1D) SDS-PAGE, its band was also microsequenced after transfer onto a Problott membrane filter (Applied Biosystems). The first four N-terminal amino acids (Pro Ser Ala Phe) corresponded to those deduced from the gene sequence, except that the N-terminal Met was missing.
YnaF homologs and structure modeling
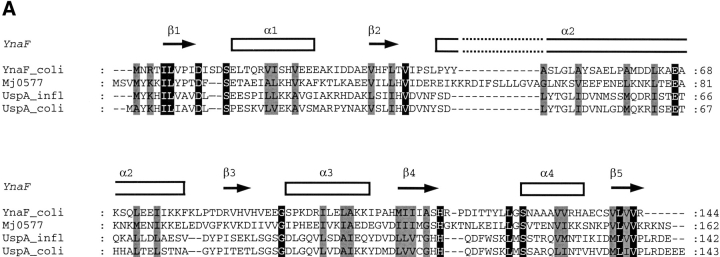
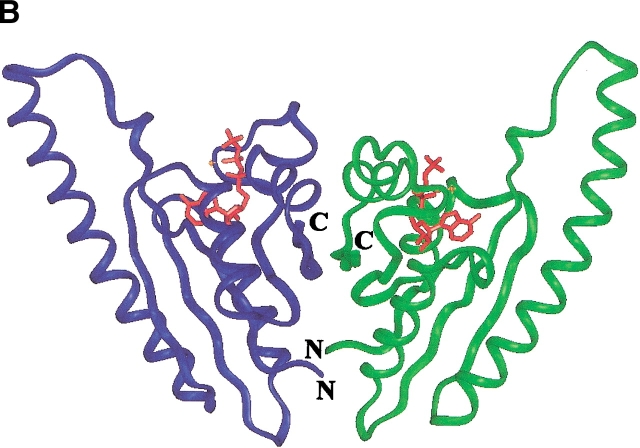
BLAST sequence searches using YnaF as a query in the nonredundant protein sequence database and in the microbial genome database (NCBI of the USA NIH, http:// www.ncbi.nlm.nih.gov/) revealed a cluster of proteins present in many bacterial genomes, notably those of γ-Proteobacteria and the Bacillus/Clostridium group, Staphylococci, and Actinobacteria. No homologs of YnaF were detected in Chlamydiales or Neisseriaceae. An alignment of the top four sequences is shown in Figure 2A ▶. Searches for compatible folds revealed 27% sequence identity with MJ0577, an ATP-binding protein from M. jannaschii of unknown function (Zarembinski et al. 1998). A second hit was the more distantly related protein UspA (16% sequence identity; PDB1JMV) induced in response to bacterial stress (Sousa and McKay 2001). Structure comparison of UspA and MJ0577 showed that they belong to the same structure family (UPF0022) comprising various proteins with distinct ligand specificities (Sousa and McKay 2001). Based on the sequence alignment derived from threading using the program mGenTHREADER (Jones 1999), a complete model was built using MODELLER (Sali and Blundell 1993). The ATP molecule present in the template (PDB1MJH) was included and a dimer was modeled. The final structure was validated using the programs PROSA (Sippl and Weitckus 1992) and Verify3D (Eisenberg et al. 1997; scores: −1.0 and +0.35, respectively, versus −1.6 and +0.45, for the original template PDB1MJH). The model indicates the conservation of the ATP-binding site and the dimeric structure observed in the crystal of MJ0577 (Fig. 2B ▶). A similar dimeric form was observed in UspA, indicating that this represents a common structure motif in the USP family.
Fig. 2.
(A) Sequence alignment of YnaF-like proteins. The identical residues over all sequences are shown on a black background, and the conservative substitutions on gray. On the top of the sequences, the secondary structure elements as assigned on the modeled structure of YnaF are indicated. MJ0577 is the Methanococcus jannaschii ATP-binding protein (Y577-METJA; SWISS-PROT Q57997); UspA Haemophilus influenzae (SWISS-PROT P44880) has a similar fold to the MJ0577 protein but does not bind ATP. (B) Modeled 3D structure of the YnaF protein. The dimeric model of YnaF is shown with bound Mg-ATP in pink as in MJ0577. The blue and green ribbons represent individual monomers of the dimeric structure.
YajQ homologs
Protein sequences similar to YajQ were found in several bacterial genomes, notably those of γ-Proteobacteria (Haemophilus influenzae, Pseudomonas aeruginosa), Vibrionaceae, and the Bacillus/Clostridium genus (Bacillus subtilis, Enterococcus faecalis) and Actinobacteria (Mycobacterium tuberculosis). An alignment of selected sequences is shown in Figure 3A ▶. No homologs of YajQ were detectable in Mollicutes, Chlamydia, Spirochaetales, Neisseriaceae, or Ricketsiales. The genomic context may be useful in assigning functions to uncharacterized genes that show no obvious homology to known genes (Huynen et al. 2000). In the simplest case, an operon-like organization of the neighbors is predictive for the function of the encoded protein in a metabolic pathway. However, the analysis of the local neighborhood of yajQ-like genes in different bacterial genomes showed a high degree of diversity. In E. coli and Salmonella typhimurium, the yajQ open reading frame has an opposite orientation compared with that of its neighbors: yajR, encoding a protein of unknown function, and apbA/panE, encoding ketopantoate reductase and involved in coenzyme A metabolism. abpA and the next gene, yajL (thiJ), are the only genes found close to yajQ in E. coli, S. typhi, S. typhimurium, and Salmonella dublin. The Haemophilus influenzae yajQ gene (HI1034) is flanked by serB, encoding a phosphoserine phosphatase, and by corA, encoding a metal ion-transport protein. Therefore, no operon-like organization could be found for yajQ and its neighbors.
Fig. 3.

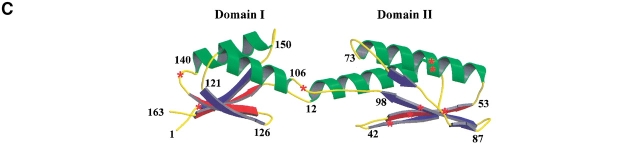
(A) Sequence alignment of proteins from the YajQ family. Identical residues over all sequences are shown on a black background, and the conservative substitutions are shown on gray. On the top of the sequences are indicated the secondary structure elements as determined by NMR spectroscopy, and the residues affected by the interaction with tRNA (labeled with +). (B) Topology of the two β-sheets established by NMR spectroscopy. The NOE contacts between protons of adjacent strands are indicated by arrows. Residues found to be most affected by the interaction with tRNA are indicated by a continuous box, and the interrupted-line boxes indicate the residues showing smaller changes. (C) Modeled 3D structure of the YajQ protein. Helices are in green and β-sheets in blue. Some amino acid positions are labeled to facilitate the recognition of secondary structure elements. The β-strands β1 and β7, which are the most affected by interaction with tRNA, are in red, whereas other, less affected residues are indicated by red stars.
HCA analysis and automatic secondary structure prediction using Jpred2 (Callebault et al. 1997) indicated that the βαββαβ sequence is a common motif in all the orthologous bacterial sequences, permitting us to propose a structural model and a functional hypothesis. The best candidates as template structure were the two-domain RNA-binding proteins (Draper 1999), like the two-RRM domain of hnRNP A1 (PDB2UP1) and the Poly(A)-Binding Protein (PDB1CVJ). Despite a low sequence similarity (10% sequence identity over 120 amino acids), the secondary structures match well, indicating that YajQ might adopt a similar fold. This structural prediction is in agreement with the nucleotide-binding capacity of YajQ indicated by the dye pseudoaffinity chromatography and confirmed by isothermal titration calorimetry (see below).
Purification and characterization of recombinant YajQ
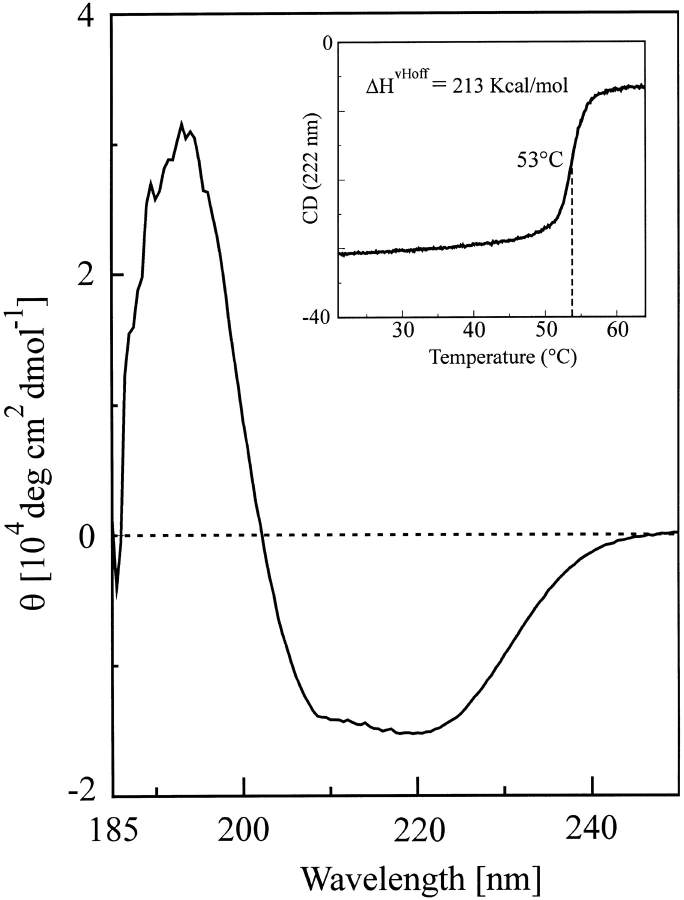
The above observations prompted us to undertake a structural analysis of YajQ. The protein overproduced in strain BL21 (DE3) was purified by Blue Sepharose and Ultrogel AcA54 chromatography (Fig. 4 ▶). The molecular mass determined by ESI-MS (18,212 ± 0.72 D) was in agreement with that calculated from the sequence, assuming that the N-terminal methionine residue is missing. Gel permeation chromatography indicated that the recombinant protein is monomeric. The circular dichroism (CD) spectrum (Fig. 5 ▶) showed that YajQ is well structured. Based on the CDsstr program (Johnson 1999), we estimated that 42% of the secondary structure is α-helix, 10% 3d10-helix, 13% extended β-strand, 11% turn, 5% polyproline-like, and 19% others. Although the CD analysis agreed fairly well in predicting the α-helix content, it underestimated the amount of β-structure by a factor of 2 (see below). The denaturation curve in the inset of Figure 5 ▶ shows a highly cooperative thermal transition with a midpoint at 53°C, which is common to many mesophilic α/β proteins. The curve, recorded at 222 nm, indicates that the observed process reflects the loss of almost 80% of the α-helix secondary structure, strongly indicating that the whole structure unfolds cooperatively at this temperature. The experimental data were well fitted to a simple, one-step model assuming a transition between two thermodynamic states. The calculated high unfolding enthalpy (ΔHvHoff = 213 kcal/mole) indicates that the denaturation reveals a large hydrophobic surface to the solvent.
Fig. 4.
SDS-PAGE (12.5%) of fractions obtained during the purification of recombinant YajQ from Escherichia coli. (Lane 1) Bacterial extract (25 μg of protein); (lane 2) Blue Sepharose chromatography (20 μg of protein); (lane 3) pure protein after Ultrogel AcA54 chromatography (16 μg of protein); (lane 4) standard proteins from top to bottom: phosphorylase a (94 kD); bovine serum albumin (68 kD); ovalbumin (43 kD); carbonic anhydrase (30 kD); soybean trypsin inhibitor (20.1 kD); and lysozyme (14.4 kD).
Fig. 5.
The far-UV CD spectrum of Escherichia coli YajQ at pH 7.4 and 25°C. The inset shows the thermal denaturation curve recorded at 222 nm.
As a first step in the detailed structural characterization of YajQ, we performed the 1H, 13C, and 15N resonance assignement of the backbone and side chains. This was done by the analysis of a pair of complementary triple-resonance experiments, recorded at 500 MHz (HNCA/HN(CO)CA), and of a high-sensitivity three-dimensional (3D) 15N-NOESY-HSQC spectrum at 800 MHz (Miron et al. 2001). The elements of secondary structure were delineated from several NMR parameters including the short- and medium-range NOEs between backbone protons and the 13C and 1H secondary chemical shifts. Overall, four α-helices and eight β-strands, were identified with the following limits: β1: 3–8, α1: 12–29, β2: 37–42, β3: 47–53, α2: 55–73, β4: 79–81 and 85–87, β5: 91–98, α3: 106–117, β6: 121–126, β7: 129–134, α4: 139–150, and β8: 156–161 (represented in Fig. 3A ▶). Characteristic NOE interactions between Hα protons belonging to different strands indicate an antiparallel arrangement. Analysis of the ensemble of long-range NOE cross-peaks between β-strand protons enabled us to define two four-stranded antiparallel β-sheets (I and II), whose topology is shown in Figure 3B ▶. β-Sheet II includes the contiguous strands β2 to β5, whereas β-sheet I integrates the N-terminal β-strand and the last three β-strands, thus bringing the two ends of the protein chain close in space. Preliminary analysis of NOESY spectra revealed no long-range interproton NOEs between the two sheets, indicating that there is no close contact between them. Therefore, it can be reasonably assumed that YajQ is composed of two domains, each centered on a β-sheet, connected by helices 1 and 3. Delineation of secondary structure elements and the topology of the β-structure allows a more comprehensive analysis of the sequence superimposition in the YajQ family (Fig. 3A ▶). The most striking observation is the absence of any sequence conservation within the strands constituting β-sheet II, in contrast with β-sheet I, in which almost 80% of the residues are identical or conservatively substituted. This is a strong indication that the two domains play a different role in the structural stability/dynamics and/or the functional mechanism. In addition, the loops connecting α1/β2, α3/β6, and β7/α4 show an unusual proportion of sequence similarity, which may reflect their participation to the protein function.
3D structure modeling
The putative global fold of YajQ was further assessed through comparative molecular modeling using the softwares TITO (Labesse and Mornon 1998) and MODELLER (Sali and Blundell 1993) with NMR-derived restraints for the β-sheet topology and secondary structure length. For domain I, comprising β1, β6–β8, α3, and α4, a better structure compatibility was observed with the small copper chaperonin Hah1, but the residues critical for metal binding are not conserved. No good template could be detected for the second domain (α1, α2, β2–β5). The topology of the two β-sheets in YajQ implies a duplication and a rearrangement by exchange of two strands (β1 and β5) from one sheet to another compared with that of Hah1. Starting from hnRNP or poly(A)-binding protein, a β-strand swapping is also required, indicating that YajQ adopts a unique (α + β) fold distantly related to RRMs (Draper 1999). A hypothetical template was made of two Hah1 domains facing each other as in the crystal structure PDB1FE0. The relative position of the two domains was changed to place the two β-sheets in the same plane and at a distance ∼7 Å apart, allowing "correct" loop modeling in between the two domains (loops β1/α1 and β5/α3). The topology of the β-sheets was modified according to the NMR assignment, simply by renumbering the residues for β1 and β5. NMR-derived secondary structures and β-sheet topology were also included as additional constraints in MODELLER (Sali and Blundell 1993). The sequence-to-structure alignment was refined using TITO to optimize the insertion/deletion positions. The model with the lowest objective energy produced by MODELLER was kept (Fig. 3C ▶). The pseudoenergy derived from TITO (Labesse and Mornon 1998), PROSA (Sippl and Weitckus 1992), and the score from Verify3D (Eisenberg et al. 1997) validate the model (−41.9, −1.0, and +0.35, respectively). Keeping in mind the absence of significant sequence identity, the values of the different validation scores compare favorably with those of the crystal structure (for the original template PDB1FE0, TITO: −100.6; PROSA: −2.4, and Verify3D: +0.45).
Ligand-binding properties of YajQ
Binding of nucleotides to YajQ was assayed using isothermal titration microcalorimetry (ITC). The binding isotherm observed at 25°C with ATP as the ligand (Fig. 6A ▶) indicated 1:1 stoichiometry, a binding constant of (1.0 ± 0.3) × 104 M−1, a low unfavorable binding enthalpy (ΔH° = 0.37 ± 0.06 kcal/mole) and a large favorable binding entropy (TΔS° = 5.84 ± 0.24 kcal/mole). The binding enthalpy remained positive in the 20°–35°C interval, with a linear temperature dependence corresponding to a small negative heat capacity change during complexation (ΔCp = −9.7 ± 1.5 cal mole−1 K−1; Fig. 6B ▶). By increasing the temperature, the binding entropy varied almost in parallel with the binding enthalpy so that enthalpy and entropy changes nearly canceled out in the binding free energy, which varied little in the temperature range studied. Taken together, the negative heat capacity change on binding and the entropy-driven character of the binding reaction (−TΔS°/ΔG° = 106.3% at 25°C) are characteristic for an association reaction dominated by hydrophobic interactions (Spolar et al. 1989; Makhatadze and Privalov 1990; Luque and Freire 1998). GTP was found to bind to YajQ with similar parameters, within experimental errors, to those measured with ATP. The interaction of YajQ with ATP was further explored using 1D and 2D (1H–15N)-HSQC NMR spectra. No spectral changes were noted up to a threefold molar excess of ATP over the protein, indicating that the nucleotide induces very small structural perturbations.
Fig. 6.
Binding of ATP to YajQ as determined by high-sensitivity isothermal titration calorimetry. (A) The binding isotherm at 25°C. The shape of the binding isotherm corresponds to that obtained with C = 2 (Wiseman et al. 1989). (B) The enthalpy (ΔH, circles), entropy (TΔS, squares), and free energy (ΔG, triangles) changes for binding of ATP to YajQ versus temperature.
Because the structural homology search indicated that YajQ might bind RNA, we investigated the possible intermolecular interaction using ITC and NMR spectroscopy. As a first step, we titrated total E. coli tRNA, which may contain ∼100 different molecular species (Lowe and Eddy 1997), with YajQ. The measured peaks, whose area represents the mixing enthalpy for a titration step, are positive, indicating that the protein and tRNA molecules do interact and the process is endothermic, as for the case of YajQ/ATP. The corrected peak area exhibits a slight decrease along the titration pathway, without reaching the transition phase and a final plateau, which indicates that the total injected protein did not saturate the existing binding tRNA sites. Nevertheless, according to the average area of the first titration peaks, the reaction enthalpy is of the order of 2 kcal/mole, larger than for the nucleotides reported here, but significantly smaller than those generally encountered in protein/nucleic acid interactions (Lopez et al. 1999; Sieber and Allemann 2000). Some cases of endothermic protein/DNA complex formation were reported in the literature (Hyre and Spicer 1995; Privalov et al. 1999; Shi et al. 1999), and were usually explained by a preponderant hydrophobic contribution. A more detailed thermodynamic analysis of the tRNA binding reaction to YajQ, depending on the type and concentration of RNAs, and the environmental factors, is presently underway in our laboratory.
NMR (1H–15N)-HSQC spectra of the uniformly 15N-labeled YajQ (1 mM) in the presence of a mixture of E. coli tRNAs were recorded at 35°C. The concentrations were calculated to have an approximate equimolar ratio. Overall, the HSQC spectrum of the protein conserves its global appearance, with a general broadening of all the cross-peaks (Fig. 7 ▶), which may be caused by the increase of the rotational correlation time owing to complex formation. It appeared, however, that the increase in linewidth is unequally distributed over the spectrum, indicating that some particular residues or molecular fragments, more closely involved in the binding process, undergo a conformational exchange with an intermediate-to-slow kinetics on the NMR time scale. The most affected cross-peaks (F4, D5, I6, V7, S8, I130, R131, V132, N160) belong to the strands β1 and β7 (shown in red in Fig. 3C ▶), constituting the core of β-sheet I (Fig. 3B ▶). Other signals, exhibiting an intermediate broadening, correspond to residues from β-sheet II (L41, K49, V50, S52, W92, V94), α-helix 2 (I64, L65), and loops of domain I (S104, R137), and are indicated by red stars in Figure 3C ▶. The amplitude of the spectral perturbations and their structural distribution (Fig. 3C ▶) strongly indicate that domain I is mostly involved in the interaction with tRNA, and β-sheet II plays a secondary role. A similar NMR titration experiment using yeast total tRNA showed a much smaller spectral broadening, affecting roughly all the cross-peaks (data not shown). The difference may be due to the existence of a certain specificity of the interaction between E. coli YajQ and its own tRNA. These NMR observations support the previous sequence conservation analysis, conferring a critical functional role to a limited structural region of the protein. Although they indicate the existence of a molecular interaction between YajQ and tRNA, the present ITC and NMR experiments are still far from elucidating the biological role of the protein and its mechanism. More specific investigations of binding properties (e.g., with homogenous tRNA molecules or shorter RNA motifs) are necessary for finding the specific target, the interaction site, and thermodynamic characteristics.
Fig. 7.

Comparison of HSQC spectra of YajQ alone (A) and in the presence of Escherichia coli tRNA (B). The spectra were recorded in potassium phosphate buffer (pH 6.5) in 95% H20/5% D20, at 35°C on a Varian NMR spectrometer Unity 500. The cross-peaks affected by the interaction with tRNA are boxed and labeled with the type and number of the corresponding amino acid residue.
Concluding remarks
Structural comparison of YnaF, UspA, and MJ0577 showed that they belong to the same structure family (UPF0022), comprising various proteins with distinct ligand specificities (Sousa and McKay 2001). YnaF would represent an ATP-dependent USP in E. coli, whereas UspA and the other related USPs in E. coli (YecG, YiiT, YdaA, and YbdQ) would bind yet uncharacterized ligands. This result indicates that oriented proteomics revealing the ligands of a set of proteins can help structure-based functional assignment.
In the absence of significant similarities with any solved structure, the NMR characterization of YajQ was undertaken. In conjunction with sequence analysis, a putative function—RNA binding—has been hypothesized and addressed by means of NMR experiments. As the crystal structure of the equivalent protein in H. influenzae has been deposited in the PDB but is not accessible, it will be of interest to compare the predicted structure of the E. coli and H. influenzae proteins, as they exhibit 61% sequence identity.
Materials and methods
Chemicals
Nucleotides, E. coli tRNA, restriction enzymes, T4 DNA ligase, and T7 DNA polymerase were from Roche Diagnostics or from Sigma. Cibacron Blue 3G-A Sepharose CL-6B (Blue Sepharose) was from Bio-Rad Laboratories.
Bacterial strains, plasmids, growth conditions and DNA manipulations
General DNA manipulations were performed as described by Sambrook et al. (1989). The open reading frame corresponding to the yajQ gene from E. coli was amplified by polymerase chain reaction using genomic DNA as template. The 491-bp DNA fragment was amplified using the following primers: forward, 5′-GGGTCTCATATGCCATCTTTCGATATTGTCTC-3′, with an NdeI restriction site (underlined); and reverse, 5′-CCGGAATTCTTAATCGCGGAACTT TTTGAACTG-3′ with an EcoRI restriction site (underlined). The amplified fragment was inserted into the corresponding NdeI and EcoRI sites of the pET24a expression plasmid (Novagen Inc.), producing the plasmid pAOT7. The protein was expressed in the E. coli BL21(DE3)/pDIA17 strain (Munier et al. 1991), grown at 37°C in LB medium containing 50 mg/L kanamycin and 34 mg/L chloramphenicol. IPTG was added to the final concentration of 1 mM at an OD600 of 0.8–1.0, and cells were grown for another 4 h. 15N/13C uniform labeling was performed by growing cells in the minimal medium M9, supplemented with 2 mM MgSO4, 0.1 mM CaCl2, 3.6 μM FeSO4, 3 μM vitamin B1, 3 g/L U-13C-glucose (Cambridge Isotope Laboratories), and 1.5 g/L 15N-ammonium sulfate (Eurisotop). Cells were grown at 37°C in the presence of antibiotics, and protein expression was induced by IPTG, as indicated above.
Dye pseudoaffinity chromatography, 2D gel electrophoresis, and mass spectrometry
E. coli K-12 cells grown in LB medium until the late exponential phase were harvested, then broken by sonication in 50 mM Tris-HCl (pH 7.4). The soluble extract clarified by centrifugation was loaded onto a Blue Sepharose column equilibrated with the same buffer. The column was washed with 10 volumes of Tris-HCl buffer until the optical density at 280 nm was <0.02, when proteins were eluted with 2 volumes of 2 mM ATP in 50 mM Tris-HCl (pH 7.4). A second elution step was then initiated with 2 volumes of 2 mM NAD+ in the same buffer. The peak fractions corresponding to ATP and NAD+ elutions were concentrated to ∼5 mg of protein/mL, then were prepared for 2D electrophoresis as described in Hommais et al. (2001). For electrophoresis, 50 μg of protein from each pool was loaded onto the IEF gels (pH gradient 3–10, Genomic Solutions); the second dimension was performed on 12.5% slab gels. Proteins were visualized by Coomassie blue staining. In-gel digestion of proteins was performed by the protocol of Shevchenko et al. (1996), using bovine trypsin (Roche Molecular Biochemicals). The generated peptides were cleaned on a reversed-phase support using Zip-Tip C18 Millipore or Poros R2 (Perseptive Biosystems). The mixture of peptides was analyzed by tandem mass spectrometry on a triple quadrupole API365 mass spectrometer (Applied Biosystems-MDS-Sciex) equipped with a nanoelectrospray source (Protana). Nucleotide and protein databases were searched with PeptideSearch, using the peptide sequence tag algorithm (Mann and Wilm 1994).
Purification of YajQ expressed in E. coli
E. coli cells overproducing YajQ protein were disrupted by sonication in 50 mM Tris-HCl (pH 7.4). The bacterial extract was centrifuged at 10,000g for 30 min, and YajQ, representing >20% of soluble protein, was purified by Blue Sepharose and gel permeation chromatography. The protein adsorbed on the Blue Sepharose column was eluted with 0.5 M NaCl in 50 mM Tris-HCl (pH 7.4). The fractions containing YajQ were pooled and concentrated, and then loaded onto an Ultrogel AcA54 column (120 × 1 cm) equilibrated with 50 mM Tris-HCl (pH 7.4). The peak fraction corresponding to YajQ was concentrated to 10–20 mg of protein/mL and conserved under the frozen state at −20°C. Uniformly 15N- or/and 13C-labeled YajQ was purified using the same protocol.
NMR analysis
NMR samples were at a concentration of 1.0–1.3 mM in 50 mM potassium phosphate buffer (pH 6.5) in 95% H2O/5% D2O or in 99.98% D2O. Assignment was mainly performed by the analysis of 2D homonuclear and double and triple resonance (HSQC, NOESY-HSQC, TOCSY-HSQC, HNCA, and HN(CO)CA) NMR experiments (Wüthrich 1986; Cavanagh et al. 1996) at 500 MHz (Varian Unity-500). Additional 2D 15N HSQC and 3D 15N NOESY-HSQC spectra were also acquired on a Bruker 800 MHz spectrometer (ICSN, Gif-sur-Yvette). The NMR data were processed and analyzed using Felix98 software (Accelrys), running on a Silicon Graphics Indigo Workstation.
Circular dichroism measurements
The CD spectra were acquired on a Jasco715 spectropolarimeter equipped with a device for automatic temperature control. The denaturation curve was recorded on a 15 μM protein solution in 10 mM potassium phosphate buffer at pH 7.4 by monitoring the ellipticity at 222 nm as a function of temperature (1°C/min).
Calorimetric measurements
The interaction between nucleotides and YajQ was measured by isothermal titration calorimetry (ITC) using a MCS ultrasensitive system (MicroCal Inc.) as previously described (Goldberg et al. 1999; Schaeffer et al. 2002). Protein at 0.14–0.20 mM in 50 mM Tris-HCl (pH 7.4) was titrated in the calorimeter cell (1.4 mL) by 10-μL successive injections of 4.0 mM ATP or GTP in the same buffer, until a final nucleotide/YajQ molar ratio of 4:1. Raw calorimetric data, that is, the heats evolved after each aliquot injection, were corrected for the heats of dilution of the protein and the nucleotide, normalized to the concentration of nucleotide added, and analyzed using the software package ORIGIN (Wiseman et al. 1989) provided by the manufacturer. Calorimetric binding isotherms were fitted by an iterative nonlinear least squares algorithm to a binding model including a single set of independent sites. The association constant (Ka), the molar binding stoichiometry (N), and the molar binding enthalpy (ΔH°) were determined directly from the fitted curve. The Gibbs free energy and molar entropy of binding were calculated using the equations ΔG° = −RT lnKa and ΔS° = (ΔH° − ΔG°)/T, respectively, where R is the gas constant and T the absolute temperature in kelvins. The molar change in heat capacity (ΔCp) accompanying binding was determined using a linear regression analysis of binding enthalpies versus temperature. Experimental variations of the free energy and entropy of binding with temperature were compared with the calculated variations of these parameters using the equations ΔS(T) = ΔS(TR) + ΔCp ln(T/TR), and ΔG(T) = ΔH(T) − TΔS(T), with ΔH(T) = ΔH(TR) + ΔCp(T − TR), and TR, a reference temperature taken as 25°C (Fig. 6 ▶). Binding of tRNA to YajQ was measured as follows: The tRNA solution in the cell (12.5 mg/mL) in 50 mM Tris-HCl (pH 7.4) was titrated with 260 μM YajQ in the same buffer by 26 injections of 10 μL.
Sequence comparison and molecular modeling
Protein sequence database searches were performed with the PSI-BLAST version 2.0.5 program (Altschul et al. 1997) with default parameters and the recently developed meta-server for fold recognition and structure modeling (Douguet and Labesse 2001). For YnaF, the fully automatic procedure available on the meta-server led to satisfactory models. For YajQ, pairwise and multiple alignments were confirmed by Hydrophobic Cluster Analysis as previously described (Callebault et al. 1997), in order to delineate structurally conserved regions along the amino acid sequences. Alignment refinement was subsequently performed using the program TITO (Labesse and Mornon 1998) with putative templates (PDB2UP1, PDB1CVJ, PDB1FE0). The NMR-derived secondary structures were used as additional restraints in the modeling steps. Three-dimensional models were built using two rearranged Hah1 domains as template in MODELLER 4.0 (Sali and Blundell 1993). Models of YnaF and YajQ were assessed using Verify3D (Eisenberg et al. 1997) and PROSA (Sippl and Weitckus 1992). These 3D structures were visualized on a UNIX workstation using XmMol (Tuffery 1995).
Acknowledgments
This work was supported by grants from Institut Pasteur, the Centre National de la Recherche Scientifique (URA 2185), Institut Curie, and Institut National de la Santé et de la Recherche Médicale. We thank Régine Lambrecht for excellent secretarial help.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0217502.
References
- Altschul, S.F., Madden, T.L., Schaeffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S.K., Almo, S.C., Bonanno, J.B., Capel, M., Chance, M.R., Gaasterland, T., Lin, D., Sali, A., Studier, F.W., and Swaminathan, S. 1999. Structural genomics: Beyond the human genome project. Nat. Genet. 23 151–157. [DOI] [PubMed] [Google Scholar]
- Callebault, I., Labesse, G., Durand, P., Poupon, A., Canard, L., Chomilier, J., Henrissat, B., and Mornon, J.-P. 1997. Deciphering protein sequence information through Hydrophobic Cluster Analysis (HCA): Current status and perspectives. Cell. Mol. Life Sci. 53 621–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh, J., Fairbrother, W.J., Palmer III, A.G., and Skelton, N.J. 1996. Protein NMR spectroscopy. Principles and practice. Academic Press, San Diego.
- Dean, P. and Watson, D. 1979. Protein purification using immobilized triazine dyes. J. Chromatogr. 165 301–319. [DOI] [PubMed] [Google Scholar]
- Douguet, D. and Labesse, G. 2001. Easier threading efficiency through Web-based comparisons and cross-validations. Bioinformatics 17 752–753. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. 1999. Themes in RNA–protein recognition. J. Mol. Biol. 293 255–270. [DOI] [PubMed] [Google Scholar]
- Eisenberg, D., Luthy, R., and Bowie, J.U. 1997. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 277 396–404. [DOI] [PubMed] [Google Scholar]
- Fields, S. and Song, O. 1989. A novel genetic system to detect protein–protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Fountoulakis, M., Takacs, M.-F., Berndt, P., langen, H., and Takacs, B. 1999. Enrichment of low abundance proteins of Escherichia coli by hydroxyapatite chromatography. Electrophoresis 20 2181–2195. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.E., Schaeffer, F., Guillou, Y., and Djavadi-Ohaniance, L. 1999. Pseudo-native motifs in the noncovalent heme-apocytochrome c complex. Evidence from antibody binding studies by enzyme-linked immunosorbent assay and microcalorimetry. J. Biol. Chem. 274 16052–16061. [DOI] [PubMed] [Google Scholar]
- Hommais, F., Krin, E., Laurent-Winter, C., Soutourina, O., Malpertuy, A., Le Caer, J.P., Danchin, A., and Bertin, P. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein H-NS. Mol. Microbiol. 40 20–36. [DOI] [PubMed] [Google Scholar]
- Huynen, M., Snel, B., Lathe III, W., and Bork, P. 2000. Predicting protein function by genomic context: Quantitative evaluation and qualitative inferences. Genome Res. 10 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyre, D.E. and Spicer, L.D. 1995. Thermodynamic evaluation of binding interactions in the methionine repressor system of Escherichia coli using isothermal titration calorimetry. Biochemistry 34 3212–3221. [DOI] [PubMed] [Google Scholar]
- Johnson, W.C. 1999. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35 307–312. [PubMed] [Google Scholar]
- Jones, D.T. 1999. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287 797–815. [DOI] [PubMed] [Google Scholar]
- Labesse, G. and Mornon, J.-P. 1998. A tool for incremental threading optimization (TITO) to help alignment and modelling of remote homologs. Bioinformatics 14 206–211. [DOI] [PubMed] [Google Scholar]
- Lopez, M.M., Yutani, K., and Makhatadze, G.I. 1999. Interactions of the major cold shock protein of Bacillus subtilis CspB with single-stranded DNA templates of different base composition. J. Biol. Chem. 274 33601–33618. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque, I. and Freire, E. 1998. Structure-based prediction of binding affinities and molecular design of peptide ligands. Methods Enzymol. 295 100–127. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I. and Privalov, P.L. 1990. Heat capacity of proteins. I. Partial molar heat capacity of individual amino acid residues in aqueous solution: Hydration effect. J. Mol. Biol. 213 375–384. [DOI] [PubMed] [Google Scholar]
- Mann, M. and Wilm, M. 1994. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66 4390–4399. [DOI] [PubMed] [Google Scholar]
- Miron, S., Borza, T., Saveanu, C., Gilles, A.-M., Bârzu, O., and Craescu, C.T. 2001. 1H, 13C and 15N resonance assignment of YajQ, a protein of unknown structure and function from Escherichia coli. J. Biomol. NMR 20 287–288. [DOI] [PubMed] [Google Scholar]
- Munier, H., Gilles, A.-M., Glaser, P., Krin, E., Danchin, A., Sarfati, R.S., and Bârzu, O. 1991. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 196 469–474. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Todd, A.E., and Thornton, J.M. 1999. From protein structure to function. Curr. Opin. Struct. Biol. 9 374–382. [DOI] [PubMed] [Google Scholar]
- Pasquali, C., Frutiger, S., Wilkins, M.R., Hughes, G.J., Appel, R.D., Bairoch, A., Schaller, D., Sanchez, J.C., and Hochstrasser, D.F. 1996. Two-dimensional gel electrophoresis of Escherichia coli homogenates: The Escherichia coli SWISS-2DPAGE database. Electrophoresis 17 547–555. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L., Jelesarov, I., Read, C.M., Dragan, A.I., and Crane-Robinson, C. 1999. The energetics of HMG box interactions with DNA: Thermodynamics of the DNA binding of the HMG box from mouse sox-5. J. Mol. Biol. 294 997–1013. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Riley, M. and Serres, M.H. 2000. Interim report on genomics of Escherichia coli. Ann. Rev. Microbiol. 54 341–411. [DOI] [PubMed] [Google Scholar]
- Sali, A. and Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaeffer, F., Matuschek, M., Guglielmi, G., Miras, I., Alzari, P.M., and Béguin, P. 2002. Duplicated dockerin subdomains of Clostridium thermocellum endoglucanase CelD bind to a cohesin domain of the scaffolding protein CipA with distinct thermodynamic parameters and a negative cooperativity. Biochemistry 41 2106–2114. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68 850–858. [DOI] [PubMed] [Google Scholar]
- Shi, Y., Wang, S., Krueger, S., and Schwarz, F.P. 1999. Effect of mutations at the monomer–monomer interface of cAMP receptor protein on specific DNA binding. J. Biol. Chem. 274 6946–6956. [DOI] [PubMed] [Google Scholar]
- Sieber, M. and Allemann, R.K. 2000. Thermodynamics of DNA binding of MM17, a ‘single chain dimer’ of transcription factor MASH-1. Nucleic Acids Res. 28 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl, M.J. and Weitckus, S. 1992. Detection of native-like models for amino acid sequences of unknown three-dimensional structure in a data base of known protein conformations. Proteins 13 258–271. [DOI] [PubMed] [Google Scholar]
- Sousa, M.C. and McKay, D.B. 2001. Structure of the universal stress protein of Haemophilus influenzae. Structure 9 1135–1141. [DOI] [PubMed] [Google Scholar]
- Spolar, R.S., Ha, J.H., and Record, Jr., M.T., 1989. Hydrophobic effect in protein folding and other noncovalent processes involving proteins. Proc. Natl. Acad. Sci. 86 8382–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, S.T., Cass, K.H., and Stellwagen, E. 1975. Blue dextran-sepharose: An affinity column for the dinucleotide fold in proteins. Proc. Natl. Acad. Sci. 72 669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffery, P. 1995. XmMol: An X11 and motif program for macromolecular visualization and modeling. J. Mol. Graph. 13 67–72. [DOI] [PubMed] [Google Scholar]
- Wiseman, T., Williston, S., Brandts, J.F., and Lin, L.N. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179 131–137. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Zarembinski, T.I., Hung, L.W., Mueller-Dieckmann, H.J., Kim, K.K., Yokota, H., Kim, R., and Kim, S.H. 1998. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural genomics. Proc. Natl. Acad. Sci. 95 15189–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]