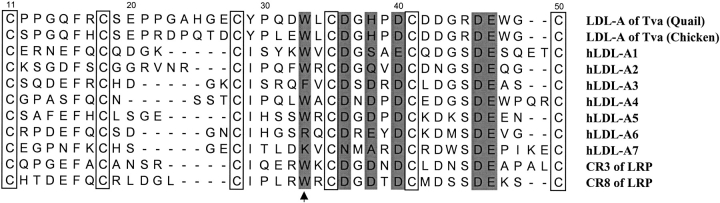Abstract
Tva is the cellular receptor for subgroup A Rous sarcoma virus (RSV-A), and the viral receptor function is solely determined by a 40-residue motif called the LDL-A module of Tva. In this report, an integral approach of molecular, biochemical, and biophysical techniques was used to examine the role of a well-conserved tryptophan of the LDL-A module of Tva in protein folding and ligand binding. We show that substitution of tryptophan by glycine adversely affected the correct folding of the LDL-A module of Tva, with only a portion giving a calcium-binding conformation. Furthermore, we show that the misfolded LDL-A conformations of Tva could not efficiently bind to its ligand. These results indicate that this conserved tryptophan in the LDL-A module of Tva plays an important role in correct protein folding and ligand recognition. Furthermore, these results suggest that the familial hypercholesterolemia (FH) French Canadian-4 mutation is likely caused by protein misfolding of low-density lipoprotein receptor, thus explaining the defect for this class of FH.
Keywords: Tva, LDL-A module, protein folding, familial hypercholesterolemia (FH)
Tva is the cellular receptor for subgroup A Rous sarcoma virus (RSV-A) (Bates et al. 1993; Young et al. 1993). One important feature of this receptor is that it contains a 40-residue motif, called the LDL-A module, which is highly homologous to the human low-density lipoprotein receptor (LDLR) ligand binding repeats (LDL-A modules). In human LDLR, seven adjacent LDL-A modules at the amino terminus of the protein are responsible for binding to its ligands, apoB and apoE (Esser et al. 1988; Russell et al. 1989). Interactions between Tva and EnvA, the RSV-A envelope protein, mediate viral entry (Bates et al. 1993; Young et al. 1993). Remarkably, the viral receptor function is solely determined by the LDL-A module of Tva (Rong and Bates 1995). Furthermore, the LDL-A module of Tva can be functionally replaced by human LDLR repeat 4 (LDL-A4) with only a few amino-acid substitutions to mediate efficient viral infection (Rong et al. 1998b). Thus, analysis of the Tva/EnvA interaction may provide a simple system to elucidate the mechanism of LDLR/ligand interactions in general.
LDLR is the primary receptor for the uptake of plasma cholesterol into cells. Normally, cholesterol-containing low-density lipoprotein (LDL) particles bind to the LDLR and are imported into the cells through receptor-mediated endocytosis (Brown and Goldstein 1986). Thus, in individuals who inherit defective LDLR (familial hypercholesterolemia [FH]) the cells become deficient in their capacity to take up LDL particles from the blood, resulting in pathologically high levels of blood cholesterol and development of premature heart disease in these individuals (Hobbs et al. 1992). One class of FH, called the FH French Canadian-4 mutation, has a single missense mutation in the LDLR (Trp66 to Gly), and is particularly prevalent in the French Canadian population (Leitersdorf et al. 1990).
LDLR is the prototype for an expanding family of cell-surface receptors, such as the LDLR-related protein (LRP), gp330, and the very low-density lipoprotein (VLDL) receptors (for review, see Hussain et al. 1999). Each of these proteins contains multiple cysteine-rich motifs, each ∼40 residues in length (referred to as the LDL-A module), and these repeats form the interaction domains with their ligands. In contrast, some members of the LDLR superfamily such as Tva only have a single LDL-A module. Although the structure of LDLR has not yet been reported, structures of a number of individual LDL-A modules have been determined by X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy (Daly et al. 1995a,b; Fass et al. 1997; Huang et al. 1999; Dolmer et al. 2000; North and Blacklow 2000b; Simonovic et al. 2001). These LDL-A modules share two important structural features. First, the overall structure consists of two short loops that are stabilized by three pairs of disulfide bonds formed by the six invariable cysteines. Second, six residues near the carboxyl terminus form a calcium "cage" (Brown et al. 1997), contributing to the stability of the LDL-A structure (Blacklow and Kim 1996; Atkins et al. 1998). We have previously characterized the role of calcium in protein folding of the Tva LDL-A module and demonstrated that like human LDL-A modules, calcium is not only required for the correct folding of the Tva LDL-A module but also for the maintenance of structural integrity (Wang et al. 2001). Furthermore, structural analysis demonstrated that like other LDL-A modules, the calcium cage of the Tva LDL-A module is coordinated by six amino-acid residues including four highly conserved acidic residues via their side chains and two nonacidic residues via their carbonyl oxygens (Tonelli et al. 2001; Wang et al. 2002). To date, the roles of the calcium-coordinating acidic residues of LDL-A modules of Tva in protein folding and viral receptor function have been well characterized by biochemical and functional analysis (Zingler et al. 1995; Zingler and Young 1996; Rong et al. 1998a; Wang et al. 2001). However, the roles of the nonacidic, calcium-coordinating residues in protein folding of the Tva or other LDL-A modules have not been characterized.
Sequence alignment indicates that one of the nonacidic, calcium-coordinating residues in Tva, tryptophan 33, is well conserved among hundreds of LDL-A modules. Substitution of this conserved tryptophan by a glycine in the human LDLR repeat 2 (Trp66 to Gly) is referred to as the FH French Canadian-4 mutation (Hobbs et al. 1992). In this report, we used an integral approach of molecular, biochemical, and biophysical techniques to examine the role of tryptophan 33 in protein folding and ligand binding of Tva. We found that substitution of this residue by glycine caused protein misfolding, indicating that this conserved residue plays an important role in protein packing in the Tva LDL-A module. The large fraction of misfolded molecules was unable to bind to the ligand EnvA efficiently. These results suggest that the FH French Canadian-4 mutation may cause protein misfolding of LDLR, thus explaining the defect for this class of FH.
Results
We have demonstrated previously that correct folding of the Tva LDL-A module, like other LDL-A modules, is Ca2+-dependent, and calcium is coordinated by four highly conserved acidic residues (D36, D40, D46, and E47) and two nonacidic residues (W33 and H38). Sequence alignment of the wild-type Tva LDL-A module with other LDL-A modules including all seven repeats of human LDLR indicates that W33 of Tva is also conserved, while H38 of Tva is not conserved in other LDL-A modules (Fig. 1 ▶). The conservation of tryptophan at position 33 implies that W33 may have another role in protein folding of the LDL-A module in addition to calcium binding, because both W33 and H38 are involved in calcium binding via their carbonyl oxygens (Tonelli et al. 2001; Wang et al. 2002). To directly test the role of W33 of Tva in protein folding, five amino-acid substitutions of W33 were generated: two nonpolar (A and G), one aromatic (F), one basic (K), and one acidic (E) residue, and the mutant proteins were expressed in and purified from Escherichia coli strain BL21. The in vitro folding properties of these mutant proteins were then characterized by high-performance liquid chromatography (HPLC) and two-dimensional NMR. We predicted that the nonconserved substitutions of W33 of Tva would adversely affect correct folding and structural integrity of the Tva LDL-A module if W33 plays another role in addition to calcium binding, as suggested by the residue conservation in other LDL-A modules. Alternatively, if W33 is only involved in calcium binding via its carbonyl oxygen, the nonconserved substitutions at this position should not impair the correct protein folding and function of Tva.
Fig. 1.
Amino-acid sequence alignment of the LDL-A modules of the wild-type Tva, human low-density lipoprotein receptor (LDLR), and LDLR-related protein. The six conserved cysteines are boxed, and the six residues involved in calcium coordination are shaded. The arrow indicates the residue targeted for mutational analysis in this study. The residues of the Tva LDL-A module from both quail and chicken are numbered from 11 to 50 according to the mature Tva.
Examination of effects of W33 mutations on protein folding by reverse-phase HPLC
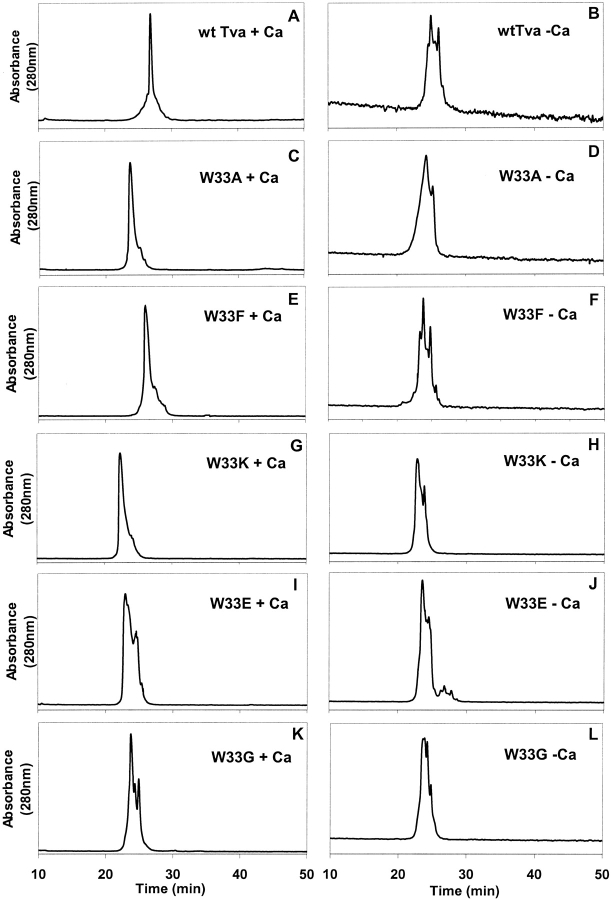
The purified W33 mutant proteins, after cleavage of the glutathione S-transferase (GST) portion by thrombin were first examined by reverse-phase HPLC following the in vitro refolding either in the presence or absence of 10 mM CaCl2. Wild-type Tva and all five W33 mutant proteins were eluted as multiple peaks in HPLC profiles when the proteins were refolded in the absence of calcium (Figs. 2B, 2D, 2F, 2H, 2J, 2L ▶). Interestingly, in the presence of calcium, W33A, W33F, and W33K mutant proteins were eluted predominantly as a sharp peak with several detectable "shoulders" (Figs. 2C, 2E, 2G ▶); while W33E and W33G mutant proteins were eluted as several peaks (Figs. 2I, 2K ▶). In contrast, under the same conditions, the wild-type Tva LDL-A module protein was eluted as a single sharp peak (Fig. 2A ▶). These results suggest that correct folding of the W33 mutants, like that of wild-type Tva, is Ca2+-dependent. In addition, mutations of W33 adversely affected protein folding. Different substitutions exerted different levels of effect on Tva folding, with W33E and W33G mutations having a greater influence than W33A, W33F, and W33K mutations. Because W33 was substituted by residues with different polarity and charges, each of these mutant proteins has a different retention time on a hydrophobic Vydac C18 column. Thus as expected, W33K, W33E, and W33G mutant proteins were eluted earlier than W33A mutant protein, while wild-type Tva and W33F mutant protein were eluted later in HPLC profiles (Fig. 2 ▶).
Fig. 2.
Elution profiles of the wild-type Tva LDL-A module and its W33 mutants by reverse-phase high-performance liquid chromatography (HPLC). The purified proteins were folded in vitro either in the presence (A, C, E, G, I, and K ) or absence (B, D, F, H, J, and L) of calcium, and then eluted by reverse-phase HPLC. X-axis, elution time (min), Y-axis, absorbance (280 nm).
Examination of effects of W33 mutations on protein folding by NMR spectroscopy
The effect of W33 mutations on protein folding and structure was further characterized by NMR spectroscopy. The 15N-labeled wild-type Tva LDL-A module and W33 mutant proteins were prepared and used for acquiring the [1H-15N] HSQC spectra. This two-dimensional NMR technique allows differentiation of correctly folded protein from misfolded ones, as only well-defined protein conformations give good chemical shift dispersion away from the random-coil values in both proton (1H) and amide 15N dimensions.
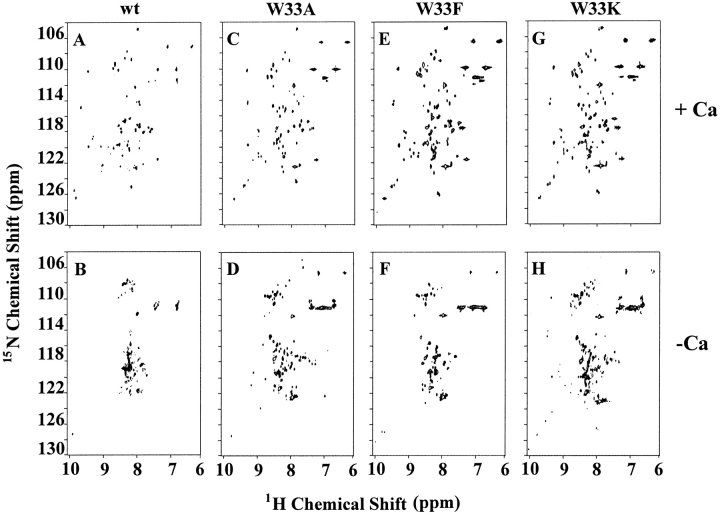
As shown above, W33A, W33F, and W33K mutant proteins were eluted as one predominant peak in HPLC profiles in the presence of calcium (see Fig. 2 ▶). These peak fractions were individually collected and used for acquisition of HSQC spectra. Like the wild-type Tva LDL-A module, W33A, W33F, and W33K mutant proteins gave good spectral dispersions in both proton and amide 15N dimensions (Figs. 3A, 3C, 3E, 3G ▶) when calcium was added before collecting the NMR data. In contrast, the same peak fractions gave poorly dispersed spectra when calcium was omitted during the NMR spectral data collection (Figs. 3B, 3D, 3F, 3H ▶). These results further demonstrated that the majority of these W33 mutant proteins (A, F, and K) can be well refolded as a single isomer in vitro in the presence of calcium, and that calcium is required to maintain the structural integrity, as previously demonstrated for the wild-type Tva module (Wang et al. 2001).
Fig. 3.
2D [1H-15N]-HSQC spectra of the LDL-A module of Tva and its W33 mutants. The two-dimensional [1H-15N]-HSQC spectra were acquired either in the presence (A, C, E, and G) or absence (B, D, F, and H) of calcium. X-axis, 1H chemical shift (ppm), Y-axis, 15N chemical shift (ppm).
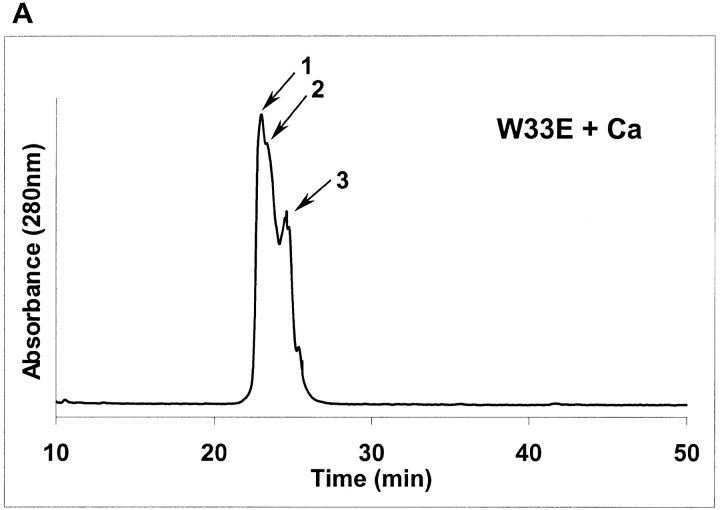
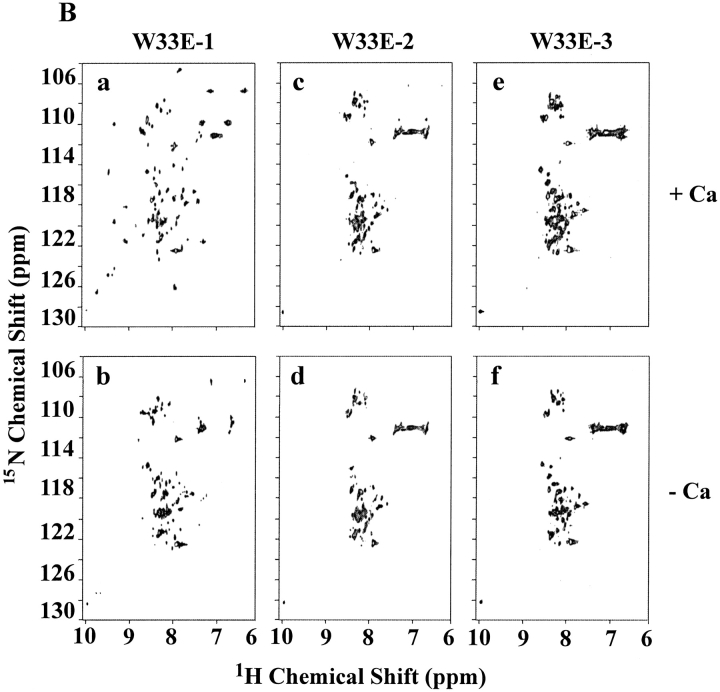
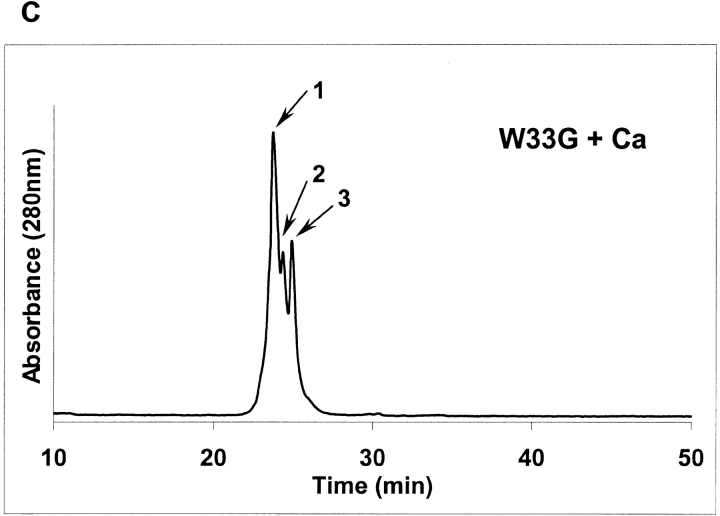
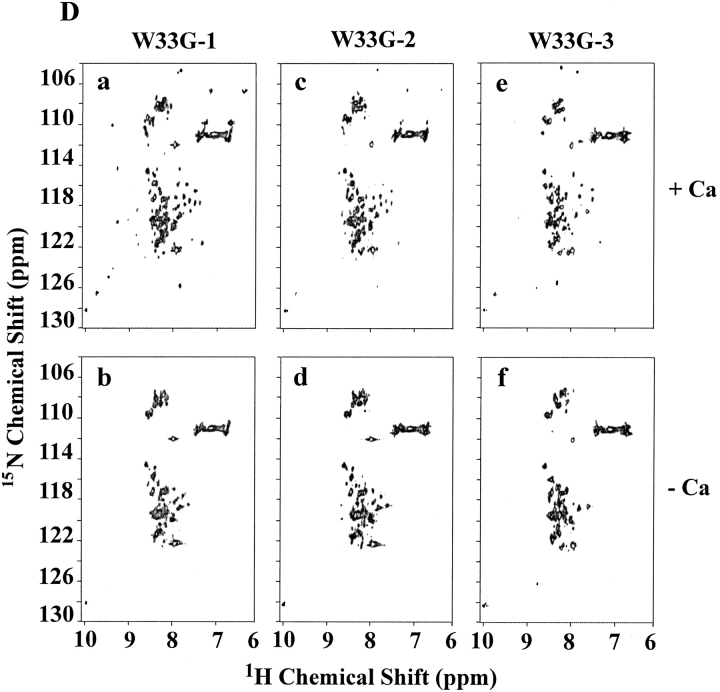
As shown above, W33E and W33G mutant proteins were eluted as multiple peaks by HPLC, even in the presence of calcium (see Figs. 2I, 2K ▶). Each of these peaks (labeled as peaks 1, 2, and 3 in Figs. 4A, 4C ▶) was collected separately, and these samples were then subjected to NMR analysis as described in Figure 3 ▶. It was found that peak fraction 1 of W33E mutant protein (about 30% of the total protein collected in three peak fractions) was able to bind calcium and displayed good chemical shift dispersion when calcium was added (Fig. 4B ▶, panel a). However, both peaks 2 and 3 of W33E mutant protein had poorly dispersed NMR spectra in the absence of calcium and showed no change upon addition of calcium even at high concentration (10 mM, Fig. 4B ▶, panels c–f). These results strongly suggest that only peak fraction 1 in Figure 4A ▶ represents the correctly folded protein for W33E, while both fractions 2 and 3 were not correctly folded or capable of binding calcium. In contrast, substitution of W33 by glycine has more dramatic effect on protein folding. There was a relatively good chemical shift dispersion of the W33G peak 1 fraction (about 47% of total collected protein) when 10 mM calcium was used in the spectrum acquisition, indicating that it responds to calcium binding. These results suggest that this fraction of the protein has the correct disulfide-bonding pattern. However, the spectral dispersions of this fraction were greatly reduced compared to those of the other correctly folded W33 mutants (Fig. 4D ▶, panel a). This was not because of partial saturation of the calcium-binding site as even with higher calcium concentration (50 mM), this fraction did not give a better spectral dispersion (data not shown). Peak fractions 2 and 3 of W33G mutant protein gave poorly resolved spectra both in the absence and presence of calcium (Fig. 4D ▶, panels c–f), indicating that these fractions were not capable of binding calcium, presumably as a result of misfolding. The W33G mutant protein therefore displayed the most dramatic folding defect.
Fig. 4.
2D [1H-15N]-HSQC spectra of W33E and W33G mutants. The purified 15N-labeled proteins were folded in vitro in the presence of calcium and were further purified by reverse-phase high-performance liquid chromatography. Three fractions were collected for W33E mutant protein (A), and W33G mutant protein (C), respectively. The two-dimensional [1H-15N]-HSQC spectra of W33E mutant protein (B), and that of W33G mutant protein (D) were acquired either in the presence (a, c, and e) or absence (b, d, and f) of calcium.
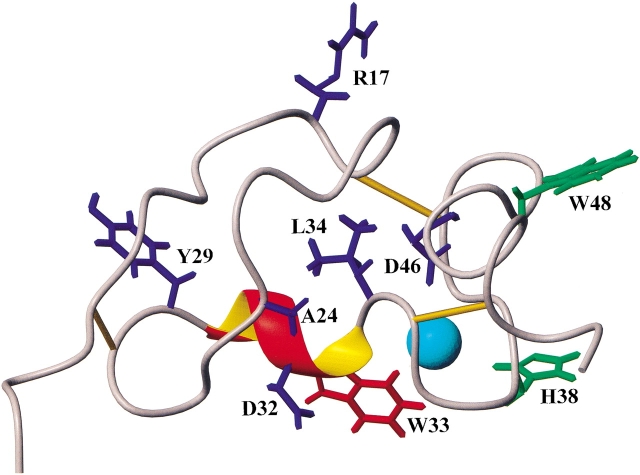
To better evaluate the effects of substitution mutations of W33 on protein folding, we compared the [1H-15N]-HSQC spectra of the W33 mutant proteins to that of the wild-type Tva LDL-A module based on the NMR resonance assignment of all of the backbone amides of the wild-type module that we have previously made. W33F and W33K mutant proteins did not show many chemical shift changes compared to the wild-type Tva module. For W33F and W33K, the cross peaks corresponding to residues Y29 and D46 were changed in the spectra. In addition, the position of residue L34 of W33F was altered. More chemical shift changes were observed for W33A mutant protein, including residues R17, A24, Y29, D32, L34, and D46. These results suggest that substitution of W33 by alanine, a smaller residue, has more influence on protein folding. Furthermore, most of the structural changes upon substitution mutation occur at the vicinity of the mutational site for these mutants (Fig. 5 ▶). The absence of significant changes in chemical shift outside the mutated residue suggests that these mutations of W33 do not alter the overall three-dimensional conformation of the Tva LDL-A module.
Fig. 5.
Residues of the Tva LDL-A module affected by amino-acid substitutions of W33. The residues within the Tva LDL-A module whose chemical shifts have been changed as a result of the W33 substitutions in the [1H-15N]-HSQC spectra are labeled in blue. W33F: Y29, L34 and D46; W33K: Y29 and D46; W33E (peak fraction 1): Y29 and D46; and W33A: R17, A24, Y29, D32, L34, and D46. W33G mutation caused changes in chemical shifts of numerous residues (not shown). The likely ligand contact residues, H38 and W48, are shown in green. The Tva LDL-A module structure is from Wang et al. (2002).
Spectral comparison between peak fraction 1 of W33E and the wild-type Tva module showed that only the cross peaks corresponding to residues Y29 and D46 were shifted, indicating that peak fraction 1 of W33E mutant protein was correctly folded. In contrast, many more cross peak changes were observed for peak fraction 1 of the W33G mutant protein. In fact, we could only unambiguously identify 15 residues (out of 40 total) that did not display changed chemical shifts for peak fraction 1 of this mutant protein. Interestingly, among the 15 residues are H38 and W48, two residues that have been implicated in ligand recognition. This finding may help explain why peak fraction 1 of W33G mutant protein, although apparently having a different conformation from wild-type Tva, was biologically active (see below). Together, these results indicate that substitution of W33 with glycine or glutamate resulted in Ca2+-dependent folding defects as revealed by both HPLC profiles and NMR HSQC spectra.
Envelope binding of the wild-type Tva LDL-A module, W33E and W33G mutant proteins
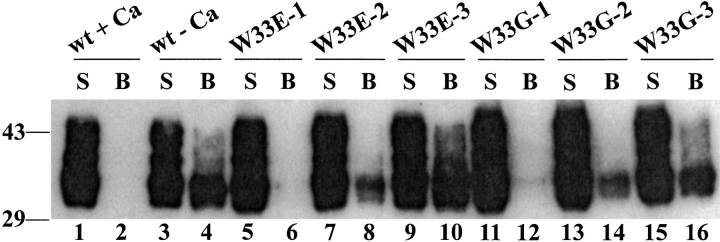
As shown above, the folding property of W33E and W33G mutant proteins is different from that of wild-type Tva and the three other W33 mutants (A, F, and K). The W33E and W33G proteins after in vitro folding in the presence of calcium were eluted as several distinct peaks in the HPLC profiles (see Figs. 2I, 2K ▶). To further test which fractions of W33E and W33G proteins had biological activity, the peak fractions of these mutant proteins in the HPLC profiles (Figs. 4A, 4C ▶) were collected (labeled as peaks 1, 2, and 3, respectively) and used to block binding of a myc-tagged Tva to the RSV-A SU-immunoadhesin (SUA-rIgG) in a ligand-binding assay as described in details in Materials and Methods. Briefly the peak fractions of W33E (W33E-1, W33E-2, and W33E-3) and W33G (W33G-1, W33G-2, and W33G-3) mutant proteins were first incubated with the purified SUA-rIgG preimmobilized on protein A Sepharose beads. After washings, the 293T lysate containing the myc-Tva protein was added and incubated with the immobilized protein A Sepharose beads. The myc-Tva was determined as bound (beads associated, B) or not bound (in supernatant, S) after SDS-PAGE and Western blot analysis using an anti-myc Mab, 9E10. If the W33E and W33G mutant proteins could bind EnvA and form stable complexes, incubation of SUA-rIgG with oversaturating amounts of W33E or W33G mutant proteins should completely block myc-Tva binding to the bead-associated SUA-rIgG.
When the wild-type Tva module protein was refolded in the presence of calcium and used in this binding assay, the myc-Tva protein was detected only in the supernatant (S), not on the beads (B), indicating that the wild-type Tva protein can completely block myc-Tva/SUA-rIgG binding (Fig. 6 ▶, lanes 1–2). In contrast, if the wild-type Tva module protein was refolded in the absence of calcium, it could not block myc-Tva binding (Fig. 6 ▶, lanes 3–4). These results are consistent with the notion that calcium is required for the correct folding of Tva protein and thus for its biological function. For W33E mutant protein, peak fraction 1 (W33E-1) was able to completely block myc-Tva binding to its ligand (Fig. 6 ▶, lanes 5–6). In contrast, neither peak fractions 2 nor 3 could efficiently block myc-Tva binding (Fig. 6 ▶, lanes 7–10). Similarly, for W33G mutant protein, only peak fraction 1 (W33G-1) could efficiently block myc-Tva binding, while peak fractions 2 and 3 could not efficiently block it (Fig. 6 ▶, lanes 11–16). Although this assay is not very quantitative in nature, it seems that for both W33E and W33G mutant proteins, peak fraction 1 was more active in blocking myc-Tva binding than fraction peak 2, while the fraction peak 3 had the least activity among the three peaks. These results are consistent with our assumption that peak fraction 1 of W33G has correct disulfide-bonding and hence calcium-binding site, and can thus efficiently compete with the wild-type Tva for ligand recognition and binding, even if the packing in the core of the module has been adversely affected by the loss of the indole side chain.
Fig. 6.
Binding of wild-type Tva and its W33 mutants to ASLV-A SU-immunoadhesin. The purified W33 mutant proteins were folded in vitro in the presence of calcium. Their ability to block binding of myc-Tva expressed in 293T cells to ASLV-A SU-immunoadhesin was examined by a ligand-binding assay (as described in Materials and Methods). The purified wild-type Tva LDL-A module proteins refolded either in the presence (+) or absence (−) of calcium were used as the positive and the negative control, respectively. The Western blot was probed with a monoclonal anti-myc antibody.
Discussion
All the reported LDL-A modules share two important structural features: three pairs of disulfide bonds formed by six invariable cysteines, and an octahedral calcium cage coordinated by six residues near the carboxyl terminus of the motif. The calcium cage is coordinated by the side chains of four highly conserved acidic residues and the carbonyl oxygens of two nonacidic residues. In the absence of calcium, an LDL-A module that has the correct disulfides formed is nevertheless unstructured, but adopts a unique, tightly folded state upon calcium binding. Furthermore, because a large fraction of the residues (12 aa) is involved in the critical disulfides and calcium cage for each LDL-A module (about 40 aa in size), and because disulfides are present both within the calcium-binding C-terminal lobe and connecting the calcium-binding lobe to the N-terminal lobe, it seems likely that only LDL-A modules with the correct disulfide-bonding pattern could bind calcium. Conversely, modules that do not bind calcium, despite having the necessary primary structure, are likely to have a nonnative pattern of disulfides. In considering the effects of single-site mutations on the conformation of the LDL-A modules, there are three distinct effects possible. One is a direct change on a calcium-coordinating ligand that might weaken or abolish calcium binding. The second is an effect on formation of the correct disulfides. The third is an effect on the overall conformation of the module, which might still have correct disulfides and an intact calcium-binding site. A given mutation may have more than one of these effects.
In an attempt to understand the structural basis for the linkage between the French Canadian-4 mutation (Trp66 to Gly) of LDL-A2 and FH, we have used the single LDL-A module of Tva as a model, and characterized variants containing a series of substitutions at this position. Of the two nonacidic residues that coordinate calcium through their carbonyls, one is W33. Sequence alignment among the 132 LDL-A modules from a wide range of LDLR members such as LDLR, LRP, and membrane serine proteases indicates that approximately 90% of the residues at this position are aromatic or bulky residues (66W, 11F, 9Y, 15R or K, and 16L), while the other position (H38 in Tva) is not well conserved, suggesting an additional role of the conserved residue (W33 of Tva) besides calcium coordination. Our results suggest that mutations at this position collectively had three, possibly linked effects, with the relative importance depending on the nature of the substitution. One effect was to reduce the efficiency of formation of the correct disulfides. A second was to alter the overall packing of the module and the third was to alter the ability of the Tva module to bind to its ligand EnvA.
The smallest effects were seen for the most conservative mutations, phenylalanine and lysine (the lysine mutation is conservative in that the four methylene groups that represent the buried portion of the side chain at this position are hydrophobic). In addition, the alanine mutation had less adverse effect than the glutamate or glycine mutation. These mutations (F, K, and A) caused only a small amount of misfolding of the disulfides, evidenced by small shoulders in the HPLC profiles. The predominant fraction of correctly folded protein also gave HSQC NMR spectra in the presence of calcium that were all well dispersed and with perturbations restricted mostly to residues immediately adjacent to the 33 position. In fact, phenylalanine, arginine, and lysine are the residues corresponding in position to W33 of Tva in human LDL-A3, A6, and A7, respectively (see Fig. 1 ▶), consistent with the results in this study. Very reasonably therefore, the largest effects among these three mutations were seen for the smallest side-chain, alanine.
For the nonconservative change to glutamate, the major effect was on the efficiency of formation of the correct disulfides. For the W33E variant only about 30% of the protein formed correct disulfides, with 70% giving two other major isomeric forms. Whereas the misfolded forms could neither bind calcium or bind to EnvA, the minor 30% correctly folded species still gave a well-dispersed NMR spectrum in the presence of calcium, with only a few changes from the other three variants, and was able to bind EnvA well. The most perturbing mutation was that to glycine, which also represents the largest loss of side-chain volume from the small module interior. For this variant, the efficiency of formation of the correct disulfides reduced to about 50%. In addition, the HSQC NMR spectrum, while showing spectral dispersion consistent with a unique conformation, had chemical shift perturbations to a majority of the residues. These results suggest that the loss of the indole side chain had resulted in an alteration in the packing of the module interior, with effects that were propagated to most residues. Nevertheless, while the misfolded fractions of this variant could bind only very poorly to EnvA, the fraction with presumed correctly folded disulfides, W33G-1, bound well, though not as well as the W33E-1 fraction or wild-type Tva.
The ability of Tva modules with perturbed internal packing, but otherwise intact calcium binding sites and correct disulfides, such as W33E-1 and W33G-1, to bind well to the ligand EnvA is readily understood in terms of two of the most important residues for ligand binding being side chains of H38 and W48, which earlier structural studies on Tva have shown to project out from the body of the module (Tonelli et al. 2001; Wang et al. 2002). As long as the relative positions of their backbones have not been changed significantly by the mutations to glutamate or glycine, which is what the present results would suggest, it would be expected that binding to EnvA would not be greatly affected.
Although it must be recognized that Tva is only a model for the LDL-A2 module of LDLR, the findings for the W33G mutation in Tva suggest the following as the likely consequences of this mutation in LDLR itself. The bulky, and largely conserved, indole side chain at position 33 plays an important role in packing of LDL-A modules. Its complete loss, through mutation to glycine, causes a large drop in the efficiency of formation of the correct disulfides. Because the modules with misformed disulfides do not bind to target ligands, this would result in a reduction in functional LDLR molecules. The final effect is that the altered packing of the modules that form correct disulfides correlates with a reduced, but not abolished, affinity of binding to ligand. Therefore, even the fraction of functional LDLR molecules with this mutation might be expected to have reduced efficacy as binders of LDL particles.
These effects together may have important implications in explaining French Canadian-4 mutations of LDLR in FH. This class of FH was first discovered in the French Canadian population, which makes up ∼7% (or 17.8% by another group) of the mutant LDL receptor alleles causing FH in this population (Leitersdorf et al. 1990). Another study showed that FH French Canadian-4 mutation accounted for roughly 15% of the diagnosed FH families in Denmark (Jensen et al. 1996). Thus this missense mutation (Trp66 to Gly) is responsible for a significant number of FH cases in some ethnic groups. Because previous work showed that the W→G mutation does not interfere with the processing of the LDLR protein (Leitersdorf et al. 1990) and because it is shown that each of the modules fold independently and different modules or combinations interact with various ligands to determine the receptor specificity (Russell et al. 1989; Bieri et al. 1998; North and Blacklow 1999, 2000a), it is possible that structural alteration by this mutation is not as global as shown by other mutations such as the calcium-binding, acidic residues in an LDL-A module, which renders LDLR degraded during the biosynthesis and transport. Instead, this mutation may alter the local structure around the second repeat of LDLR that makes it inefficient to bind to its ligands, causing the FH phenotype in patients with this type of mutation.
Materials and methods
Cloning, expression, purification, and in vitro refolding of the Tva LDL-A modules
The W33 mutant constructs of quail Tva were cloned into a GST fusion vector, pGEX-4T-1, and expressed in E. coli, BL21, as GST fusion proteins. Expression of the fusion proteins was induced with 1 mM IPTG (Sigma) at OD600 = 0.6 as previously described (Wang et al. 2001). After purification by GSH-Sepharose affinity chromatography and thrombin cleavage, the LDL-A module proteins were further purified by reverse-phase HPLC on a Vydac C-18 column operated at a flow rate of 3.00 mL/min using a linear gradient of 0.1% trifluoroacetic acid (TFA) (A) and 90% acetonitrile (B) (10–60% B) at room temperature.
Characterization of in vitro folding of the Tva LDL-A modules
The folding properties of the Tva LDL-A modules were examined by reverse-phase HPLC and by two-dimensional NMR spectroscopy ([1H-15N] HSQC spectra) as described in Wang et al. (2001).
Reverse-phase HPLC
The purified and lyophilized proteins were first dissolved in 6 M guanidinium chloride, 50 mM Tris-HCl, 1 mM dithiothreitol, pH 8.5, then diluted to ∼100 μg/mL with refolding buffer (50 mM Tris, pH 8.5, 1 mM reduced glutathione, and 0.5 mM oxidized glutathione) and incubated in the refolding buffer containing either 10 mM CaCl2 or 1 mM EDTA. Following the refolding reactions, the proteins were analyzed by reverse-phase HPLC on a Vydac C18 column under the same conditions as for protein purification described above.
NMR spectroscopy
To acquire the [1H-15N]-HSQC spectra, we used the 15N labeled Tva LDL-A module proteins following a previous protocol (Wang et al. 2001), using ∼200 μM of the HPLC purified wild type or W33 mutant proteins in each of these experiments. The NMR data were collected on a Bruker DRX600 spectrometer equipped with a pulse-field–gradient accessory and operating at 600.13 MHz for 1H, and were processed and analyzed using Triad 6.3. The central frequencies were 4.70 and 118 ppm for 1H and 15N, respectively.
Ligand-binding assay
The ability of wild-type Tva LDL-A module and its W33 mutant proteins to block binding of myc-Tva to SUA-rIgG (RSV-A SU-immunoadhesin; Zingler and Young 1996) was examined by a ligand-binding assay developed by us. In this assay, 1 μg of purified SUA-rIgG protein was incubated with protein A Sepharose beads (Pharmacia) overnight at 4°C. After washing, 300 ng of the HPLC-purified Tva LDL-A proteins (either wild type or W33 mutants) after in vitro refolding reactions were added, and the mixture was further incubated at 4°C for 1 h. After washing, 50 μL of the cell lysate from 293T cells expressing myc-Tva were added and incubated for an additional hour at 4°C. After centrifugation, both the supernatant (S) and the beads (B) were subjected to SDS-PAGE and Western blot analysis using a monoclonal antibody, 9E10, to detect the myc-tagged Tva proteins.
Acknowledgments
We thank John Young (University of Wisconsin, Madison) for providing the SUA-rIgG plasmid. This work was supported by National Institutes of Health grants CA092459, AI48056 (L.R.), and GM54414 (P.G.W.G.). The Bruker DRX600 was purchased with a grant from the National Science Foundation DIR9601705. L.R. was a recipient of the Schweppe Foundation Career Development Award. Q.-Y.W. was a recipient of the American Heart Association Midwest Affiliate Predoctoral fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0219802.
References
- Atkins, A.R., Brereton, I.M., Kroon, P.A., Lee, H.T., and Smith, R. 1998. Calcium is essential for the structural integrity of the cysteine-rich, ligand-binding repeat of the low-density lipoprotein receptor. Biochemistry 37 1662–1670. [DOI] [PubMed] [Google Scholar]
- Bates, P., Young, J.A.T., and Varmus, H.E.. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74 1043–1051. [DOI] [PubMed] [Google Scholar]
- Bieri, S., Atkins, A.R., Lee, H.T., Winzor, D.J., Smith, R., and Kroon, P.A. 1998. Folding, calcium binding, and structural characterization of a concatemer of the first and second ligand-binding modules of the low-density lipoprotein receptor. Biochemistry 37 10994–11002. [DOI] [PubMed] [Google Scholar]
- Blacklow, S.C. and Kim, P.S. 1996. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat. Struct. Biol. 3 758–762. [DOI] [PubMed] [Google Scholar]
- Brown M.S. and Goldstein, J.L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science 232 34–47. [DOI] [PubMed] [Google Scholar]
- Brown, M.S., Herz, J., and Goldstein, J.L. 1997. Calcium cage, acid baths and recycling receptors. Nature 388 629–630. [DOI] [PubMed] [Google Scholar]
- Daly, N.L., Scanlon, M.J., Tjordjevic, J.T., Kroon, P.A., and Smith, R. 1995a. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. 92 6334–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, N.L., Djordjevic, J.T., Kroon, P.A., and Smith, R. 1995b. Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry 34 14474–14481. [DOI] [PubMed] [Google Scholar]
- Dolmer, K., Huang, W., and Gettins, P.G. 2000. NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human α(2)-macroglobulin. J. Biol. Chem. 275 3264–3269. [DOI] [PubMed] [Google Scholar]
- Esser, V., Limbird, L.E., Brown, M.S., Goldstein, J.L., and Russell, D.W. 1988. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 263 13282–13290. [PubMed] [Google Scholar]
- Fass, D., Blacklow, S., Kim, P.S., and Berger, J.M. 1997. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388 691–693. [DOI] [PubMed] [Google Scholar]
- Hobbs, H.H., Brown, M.S., and Goldstein, J.L. 1992. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Human Mutation 1 445–466. [DOI] [PubMed] [Google Scholar]
- Huang, W., Dolmer, K., and Gettins, P.G. 1999. NMR solution structure of complement-like repeat CR8 from the low density lipoprotein receptor-related protein. J. Biol. Chem. 274 14130–14136. [DOI] [PubMed] [Google Scholar]
- Hussain, M.M., Strickland, D.K., and Bakillah, A. 1999. The mammalian low-density lipoprotein receptor family. Annu. Rev. Nutr. 19 141–172. [DOI] [PubMed] [Google Scholar]
- Jensen, H.K., Jensen, L.G., Hansen, P.S., Faergeman, O., and Gregersen, N. 1996. The Trp23-Stop and Trp66-Gly mutations in the LDL receptor gene: Common causes of familial hypercholesterolemia in Denmark. Atherosclerosis 120 57–65. [DOI] [PubMed] [Google Scholar]
- Leitersdorf, E., Tobin, E.J., Davignon, J., and Hobbs, H.H. 1990. Common low-density lipoprotein receptor mutations in the French Canadian population. J. Clin. Invest. 85 1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North, C.L. and Blacklow, S.C. 1999. Structural independence of ligand-binding modules five and six of the LDL receptor. Biochemistry 38 3926–3935. [DOI] [PubMed] [Google Scholar]
- North, C.L. and Blacklow, S.C. 2000a. Evidence that familial hypercholesterolemia mutations of the LDL receptor cause limited local misfolding in an LDL-A module pair. Biochemistry 39 13127–13135. [DOI] [PubMed] [Google Scholar]
- North, C.L. and Blacklow, S.C. 2000b. Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry 39 2564–2571. [DOI] [PubMed] [Google Scholar]
- Rong, L. and Bates, P. 1995. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: The 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J. Virol. 69 4847–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, L., Gendron, K., Strohl, B., Shenoy, R., Wool-Lewis, R.J., and Bates, P. 1998a. Characterization of determinants for envelope binding and infection in Tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 72 4552–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, L., Gendron, K., and Bates, P. 1998b. Conversion of a human low-density lipoprotein receptor ligand-binding repeat to a virus receptor: Identification of residues important for ligand specificity. Proc. Natl. Acad. Sci. 95 8467–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, D.W., Brown, M.S., and Goldstein, J.L. 1989. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 264 21682–21688. [PubMed] [Google Scholar]
- Simonovic, M., Dolmer, K., Huang, W., Strickland, D.K., Volz, K., and Gettins, P.G. 2001. Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry 40 15127–15134. [DOI] [PubMed] [Google Scholar]
- Tonelli, M., Peters, R.J., James, T.L., and Agard, D.A. 2001. The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus. FEBS Lett. 509 161–168. [DOI] [PubMed] [Google Scholar]
- Wang, Q.Y., Dolmer, K., Huang, W., Gettins, P.G., and Rong, L. 2001. Role of calcium in protein folding and function of Tva, the receptor of subgroup A avian sarcoma and leukosis virus. J. Virol. 75 2051–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q.Y., Huang, W., Dolmer, K., Gettins, P.G., and Rong, L. 2002. Solution structure of the viral receptor domain of Tva and its implications in viral entry. J. Virol. 76 2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.A.T., Bates, P., and Varmus, H.E. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avain leukosis and sarcoma viruses. J. Virol. 67 1811–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler, K. and Young, J.A.T. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70 7510–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler, K., Belanger, C., Peters, R., Agard, D., and Young, J.A.T. 1995. Identification and characterization of the viral interaction determinants of the subgroup A avian leukosis virus receptor. J. Virol. 69 4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]