Abstract
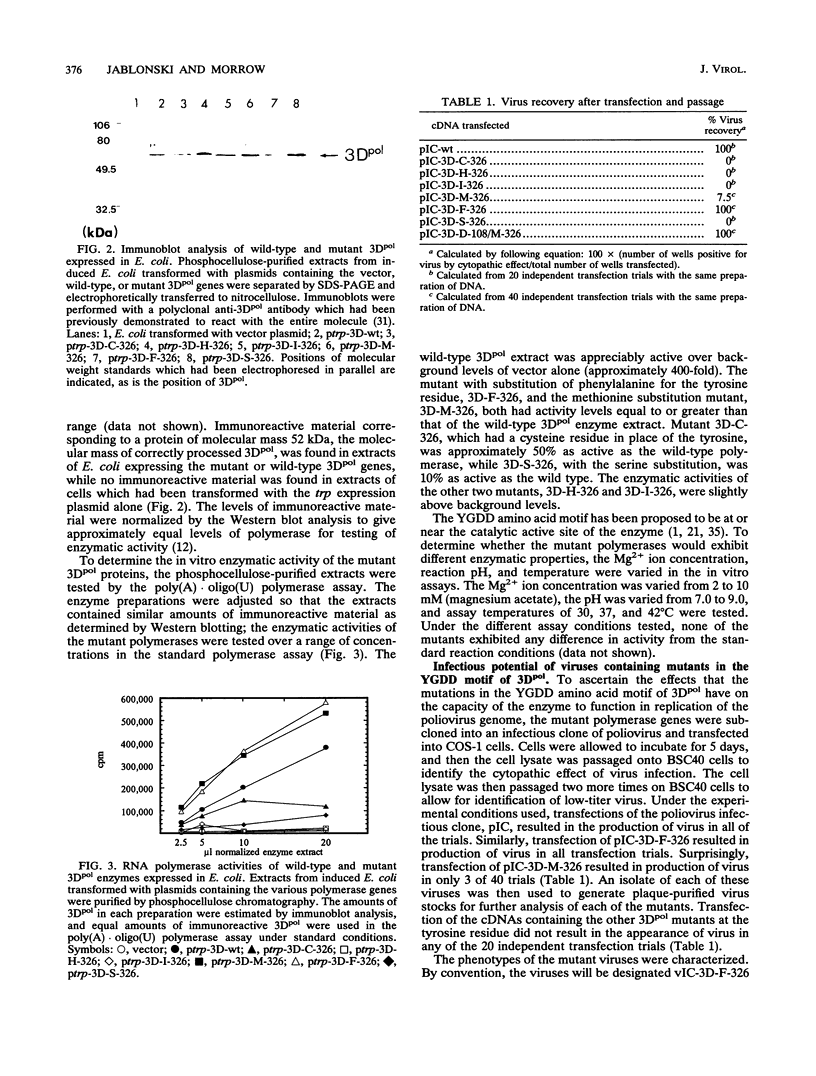
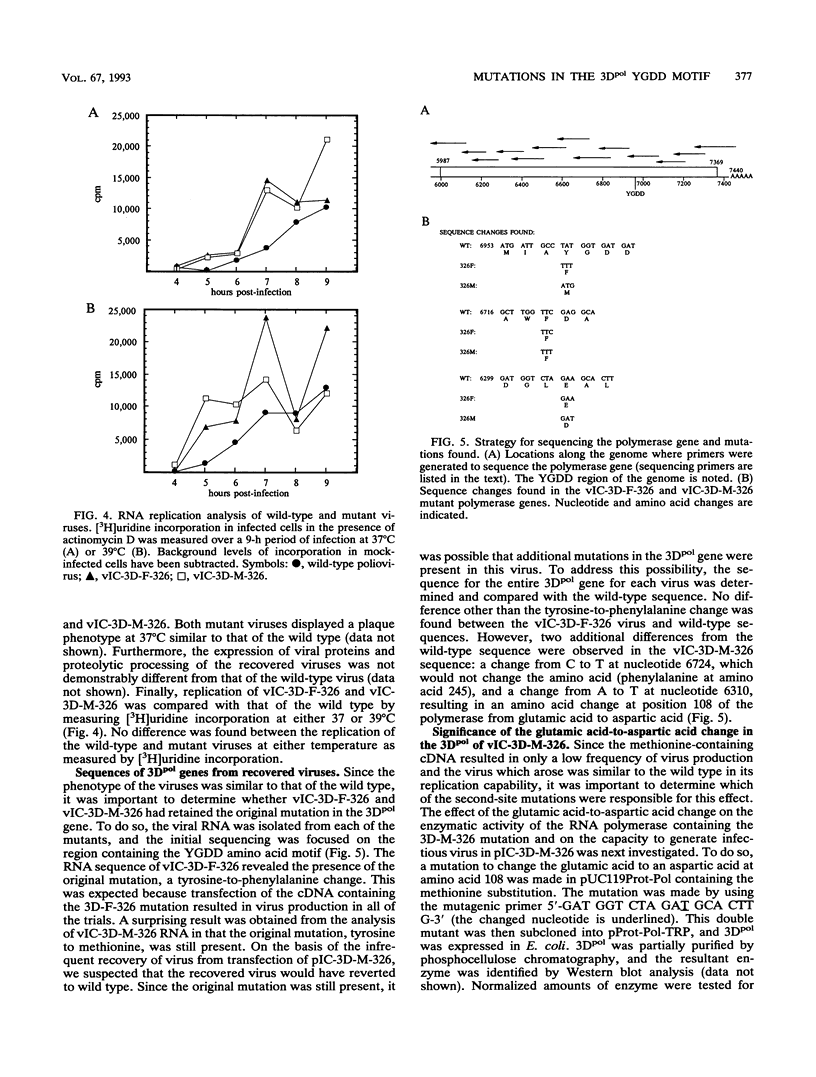
The poliovirus RNA-dependent RNA polymerase (3Dpol) shares a region of homology with all RNA polymerases, centered around the amino acid motif YGDD, which has been postulated to be involved in the catalytic activity of the enzyme. Using oligonucleotide site-directed mutagenesis, we substituted the tyrosine at this motif of the poliovirus RNA-dependent RNA polymerase with cysteine, histidine, isoleucine, methionine, phenylalanine, or serine. The enzymes were expressed in Escherichia coli, and in vitro enzyme activity was tested. The phenylalanine and methionine substitutions resulted in enzymes with activity equal to that of the wild-type enzyme. The cysteine substitution resulted in an enzyme with approximately 50% of the wild-type activity, while the serine substitution resulted in an enzyme with approximately 10% of the wild-type activity; the isoleucine and histidine substitutions resulted in background levels of enzyme activity. To assess the effects of the mutants in viral replication, the mutant polymerase genes were subcloned into the infectious cDNA clone of poliovirus. Transfection of poliovirus cDNA containing the phenylalanine mutation in 3Dpol gave rise to virus in all of the transfection trials, while cDNA containing the methionine mutation resulted in virus in only 3 of 40 transfections. Transfection of cDNAs containing the other substitutions at the tyrosine residue did not result in infectious virus. The recovered viruses demonstrated kinetics of replication similar to those of the wild-type virus, as measured by [3H]uridine incorporation at either 37 or 39 degrees C. RNA sequence analysis of the 3Dpol gene of both viruses demonstrated that the tyrosine-to-phenylalanine or tyrosine-to-methionine mutation was still present. No other differences in the 3Dpol gene between the wild-type and phenylalanine-containing virus were found. The virus containing the methionine mutation also contained two other nucleotide changes from the wild-type 3Dpol sequence; one resulted in a glutamic acid-to-aspartic acid change at amino acid 108 of the polymerase, and the other resulted in a C-to-T base change at nucleotide 6724, which did not result in an amino acid change. To confirm that the second amino acid mutation found in the 3Dpol gene of the methionine-substituted virus allowed for replication ability, a mutation corresponding to the glutamic acid-to-aspartic acid change was made in the polymerase containing the methionine substitution, and this double-mutant polymerase was expressed in E. coli. The double-mutant enzyme was as active as the wild-type enzyme under in vitro assay conditions.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988 Nov 11;16(21):9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Salas M. Site-directed mutagenesis of the YCDTDS amino acid motif of the phi 29 DNA polymerase. Gene. 1990 Sep 28;94(1):45–51. doi: 10.1016/0378-1119(90)90466-5. [DOI] [PubMed] [Google Scholar]
- Bernad A., Lázaro J. M., Salas M., Blanco L. The highly conserved amino acid sequence motif Tyr-Gly-Asp-Thr-Asp-Ser in alpha-like DNA polymerases is required by phage phi 29 DNA polymerase for protein-primed initiation and polymerization. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. S., Pal-Ghosh R., Morrow C. D. Expression of human immunodeficiency virus type 1 (HIV-1) gag, pol, and env proteins from chimeric HIV-1-poliovirus minireplicons. J Virol. 1991 Jun;65(6):2875–2883. doi: 10.1128/jvi.65.6.2875-2883.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Baron M. H., Baltimore D. Poliovirus replicase: a soluble enzyme able to initiate copying of poliovirus RNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2679–2683. doi: 10.1073/pnas.76.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Dorsky D. I., Crumpacker C. S. Site-specific mutagenesis of a highly conserved region of the herpes simplex virus type 1 DNA polymerase gene. J Virol. 1990 Mar;64(3):1394–1397. doi: 10.1128/jvi.64.3.1394-1397.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc Natl Acad Sci U S A. 1977 Sep;74(9):3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffey M. L., Novotny J., Bruccoleri R. E., Carroll R. D., Stevens J. T., Matthews J. T. Structure-function studies of the herpes simplex virus type 1 DNA polymerase. J Virol. 1990 Oct;64(10):5008–5018. doi: 10.1128/jvi.64.10.5008-5018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y., Hirashima A. Interference with viral infection by defective RNA replicase. J Virol. 1987 Dec;61(12):3946–3949. doi: 10.1128/jvi.61.12.3946-3949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski S. A., Luo M., Morrow C. D. Enzymatic activity of poliovirus RNA polymerase mutants with single amino acid changes in the conserved YGDD amino acid motif. J Virol. 1991 Sep;65(9):4565–4572. doi: 10.1128/jvi.65.9.4565-4572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I., Horwitz M. S., Engler J. A. Mutagenesis of conserved region I in the DNA polymerase from human adenovirus serotype 2. Virology. 1991 Sep;184(1):235–241. doi: 10.1016/0042-6822(91)90840-8. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Purifoy D. J. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4803–4807. doi: 10.1073/pnas.86.13.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B. A., Purifoy D. J., Powell K. L., Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. 1987 Jun 25-Jul 1Nature. 327(6124):716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Marcy A. I., Hwang C. B., Ruffner K. L., Coen D. M. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among alpha-like DNA polymerases is involved in substrate recognition. J Virol. 1990 Dec;64(12):5883–5890. doi: 10.1128/jvi.64.12.5883-5890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C. D., Warren B., Lentz M. R. Expression of enzymatically active poliovirus RNA-dependent RNA polymerase in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6050–6054. doi: 10.1073/pnas.84.17.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S. J., Palant O., Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989 Jan;63(1):216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O. C., Ivanoff L. A., Bienkowska-Szewczyk K., Butt B., Petteway S. R., Jr, Rothstein M. A., Ehrenfeld E. Formation of poliovirus RNA polymerase 3D in Escherichia coli by cleavage of fusion proteins expressed from cloned viral cDNA. Virology. 1987 Dec;161(2):348–356. doi: 10.1016/0042-6822(87)90127-9. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R., Pallansch M. A., Nottay B. K., Kew O. M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987 Oct;160(2):311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- Rueckert R. R., Wimmer E. Systematic nomenclature of picornavirus proteins. J Virol. 1984 Jun;50(3):957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar S., Porter A. G. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J Biol Chem. 1992 May 15;267(14):10168–10176. [PubMed] [Google Scholar]
- Stålhandske P. O., Lindberg M., Pettersson U. Replicase gene of coxsackievirus B3. J Virol. 1984 Sep;51(3):742–746. doi: 10.1128/jvi.51.3.742-746.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Yang C. F., Takeda N., Nomoto A., Wimmer E. Analysis of RNA synthesis of type 1 poliovirus by using an in vitro molecular genetic approach. J Virol. 1987 Sep;61(9):2816–2822. doi: 10.1128/jvi.61.9.2816-2822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaheri A., Pagano J. S. Infectious poliovirus RNA: a sensitive method of assay. Virology. 1965 Nov;27(3):434–436. doi: 10.1016/0042-6822(65)90126-1. [DOI] [PubMed] [Google Scholar]
- Van Dyke T. A., Flanegan J. B. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J Virol. 1980 Sep;35(3):732–740. doi: 10.1128/jvi.35.3.732-740.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. K., Jablonski S. A., Morrow C. D. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J Virol. 1992 Nov;66(11):6806–6812. doi: 10.1128/jvi.66.11.6806-6812.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walgate R. Armadillos fight leprosy. Nature. 1981 Jun 18;291(5816):527–527. doi: 10.1038/291527a0. [DOI] [PubMed] [Google Scholar]
- Wimmer E., Kuhn R. J., Pincus S., Yang C. F., Toyoda H., Nicklin M. J., Takeda N. Molecular events leading to picornavirus genome replication. J Cell Sci Suppl. 1987;7:251–276. doi: 10.1242/jcs.1987.supplement_7.18. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Semler B. L. Processing determinants required for in vitro cleavage of the poliovirus P1 precursor to capsid proteins. J Virol. 1987 Oct;61(10):3181–3189. doi: 10.1128/jvi.61.10.3181-3189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]




