Abstract
Only a minute fraction of all possible protein sequences can exist in the genomes of all life forms. To explore whether physicochemical constraints or a lack of need causes the paucity of different protein folds, we set out to construct protein libraries without any restriction of topology. We generated different libraries (all α-helix, all β-strand, and α-helix plus β-strand) with an average length of 100 amino acid residues, composed of designed secondary structure modules (α-helix, β-strand, and β-turn) in various proportions, based primarily on the patterning of polar and nonpolar residues. We wished to explore that part of sequence space that is rich in secondary structure. The analysis of randomly chosen clones from each of the libraries showed that, despite the low sequence homology to known protein sequences, a substantial proportion of the library members containing α-helix modules were indeed helical, possess a defined oligomerization state, and showed cooperative chemical unfolding behavior. On the other hand, proteins composed of mainly β-strand modules tended to form amyloid-like fibrils and were among the least soluble proteins ever reported. We found that a large fraction of members in non-β-strand–containing protein libraries that are distant from natural proteins in sequence space possess unexpectedly favorable properties. These results reinforce the efficacy of applying binary patterning to design proteins with native-like properties despite lack of restriction in topology. Because of the intrinsic tendency of β-strand modules to aggregate, their presence requires precise topologic arrangement to prevent fibril formation.
Keywords: Binary patterning, protein library design, secondary structure, topology
At present, the structures of more than 15,000 proteins are known (Berman et al. 2000) of which about 700 are nonredundant (Pearl et al. 2000). These known folds can be ordered into different classes and groups. For instance, according to the CATH classification (Pearl et al. 2000), the folds can be sorted into those containing mainly α-helix, mainly β-sheet, both α-helix and β-sheet, and those containing little secondary structure. More than 90% of all known folds belong to the first three classes. In addition, almost 90% of the residues in the known protein structures are involved in some kind of secondary structure (Chothia 1984). From the interpretation of genomic sequencing data, it is likely that the proportion of these four groups will stay similar even when more structures are available, and that the total number of different folds realized in nature may be on the order of 104 (Orengo et al. 1994).
A fundamental question in biochemistry is whether there is anything special about these natural protein folds: Have all possible stable folds been realized by nature or has only a small subset—through diversification and selection—given rise to the repertoire of modern proteins? A number of theoretical studies have addressed these questions (Finkelstein et al. 1995; Yue and Dill 1995; Govindarajan and Goldstein 1996; Helling et al. 2001). Our desire, in contrast, is to attack this problem experimentally by generating novel proteins and subsequently investigating their structures. As most folds will mainly consist of secondary structure modules (Taylor and Orengo 1989), we began by generating a protein library comprised of designed secondary structure elements. Our strategy is thus to explore the combinatorial construction of new folds.
Previously, Hecht and coworkers have constructed a library of four-helix bundle proteins by constraining the pattern of polar and nonpolar residues but not the precise side chains (Kamtekar et al. 1993). They have shown that a substantial proportion of them show cooperative thermal unfolding behavior (Roy and Hecht 2000) and protection of amide protons exchange (Rosenbaum et al. 1999). These results indicate that for this particular fold, namely four-helix bundle proteins, binary patterning is sufficient to achieve some properties of native proteins. In contrast to designing proteins with defined fold, we aim in this study to construct a combinatorial library by not restricting the topology to a certain fold.
We have now built up several protein libraries from secondary structure building blocks to investigate whether proteins with native-like properties can be obtained from them. Using pools of designed α-helix, β-strand, and β-turn modules in different combinations, several protein libraries (all α-helix, all β-strand, and α-helix plus β-strand) were obtained with an average length of 100 amino acid residues, the size of a typical single domain found in natural proteins. By using trinucleotide building blocks (Virnekäs et al. 1994), we could further tailor the allowed amino acids to favor the formation of the desired secondary structure element. By building different libraries (all α-helix, all β-strand, and α-helix plus β-strand) we could investigate the properties of proteins belonging to each library in terms of solubility, aggregation state, conformational stability, and secondary structure formation. Our desire was to explore that part of the sequence space that is biased for secondary structure-rich regions.
Results
Design and synthesis of each secondary structure module
The length of α-helices in known protein structures has a relatively broad distribution, with an average length of 10 residues (Zhu and Blundell 1996). We chose to make two different modules with 10 and five amino acid residues (referred to as α10 and α5 cassettes, respectively) (Figs. 1A, 1B ▶). A length of 10 was chosen, as it corresponds to the observed average length; five-residue helices were designed as short helices that are also often found in globular proteins (Zhu and Blundell 1996). Capping sequences are comprised of residues that interact with unsatisfied N—H groups at the N-terminus of α-helices (N-cap) or unsatisfied C=O groups at the C-terminus (C-cap) (Richardson and Richardson 1988). As the N-cap, Asn, Ser, Asp, Thr, or Gly followed by Pro or Glu occurs with highest frequency (Blundell and Zhu 1995). Therefore, we designed the N-terminus of the helix to have either Pro or Glu with equal probability. The connecting residues (see below) introduce Ser before this Pro/Glu residue, and Ser is one of the favored amino acids at this position (Figs. 1A, 1B ▶). As the C-cap, the preferred residue is Gly (Blundell and Zhu 1995). Again, by the connector design the modules end with Gly, and we consider this as a C-cap.
Fig. 1.

Schematic illustration of the design of each secondary structure cassette. (A) The amino acid sequence of each cassette. Polar and nonpolar residues are indicated as p and n, respectively. The design is described in the text. (B) All cassettes carry a BglII and a BamHI restriction site, as shown. The arrow indicates the direction of the open reading frame. When a BglII site is ligated to a BamHI site, the sequence -GGATCT- is obtained, which is not cleaved by either enzyme, and codes for Gly-Ser. These amino acids are the connecting residues. (C) Amino acid sequences of the clones studied in detail in this paper. Orange (α5), red (α10), blue (β), and green (turn cassette).
Secondary structures can be characterized by particular patterns of polar and nonpolar residues (binary patterning) (West and Hecht 1995). This binary patterning concept was applied to design the sequence of both α10 and α5 cassettes. West and Hecht (West and Hecht 1995) have constructed a database for the pattern of polar (p) and nonpolar (n) residues of a penta-peptide within helices. Our design is mainly based on their results. For α5 cassettes we applied the three most frequently occurring patterns pnnpp, nnppn and nppnp (Fig. 1A ▶). For the α10 cassette the database search of the most frequently found binary patterns of 10 amino acid residues will be less representative, as there are approximately 1000 possible patterns. We therefore designed this cassette based on the pattern of frequently occurring penta-peptides such that the maximum number of the three most frequently occurring penta-peptides are contained, resulting in the pattern ppnnppnnpp (Fig. 1A ▶). Using the trinucleotide technology (Virnekäs et al. 1994), we chose the five amino acids most frequently occurring in helices to code for either n or p (Blundell and Zhu 1995). Thus, Gln, Glu, Lys, Arg and Ala were chosen as p and Ile, Phe, Leu, Met and Ala as n. Ala was allowed in both n and p, as it is known to have the highest propensity to form helices (Pace and Scholtz 1998).
The lengths of β-strands in known protein structures has a broad distribution, with an average of five (Zhu and Blundell 1996). We thus designed a β-strand module of five residues (referred to as β-cassette). The patterns were chosen to be alternating polar and nonpolar residues, npnpn and pnpnp (Fig. 1A ▶). These patterns are, however, less pronounced in naturally occurring sequences, as β-strands are often fully buried, but these patterns are favored in exposed β strands (West and Hecht 1995; Broome and Hecht 2000). Because we intended to design protein libraries with an average length of 100 residues, most of the secondary structure modules will be partially surface-exposed, and thus the alternating pattern appeared to be suitable. Because Arg is reported to be a good N-capping residue in β-strands (Zhu and Blundell 1996), Arg residues were appropriately placed in the cassette. On the exposed side of the alternating β-strand, Val, Tyr, Thr, and Ser have the highest frequency (Zhu and Blundell 1996). Thus, we chose these four amino acids to encode p. On the buried face the order of preference is Val, Cys, Phe, Ile, and Leu. We omitted Cys to exclude problems occurring with spurious disulfide bridges, and used all of the other four amino acids to encode n.
β-Turns are another important structure module that reverse the peptide chains at the surface of the protein and make it possible to become globular (Wilmot and Thornton 1988). There are seven different types of turns, depending on the hydrogen bonding pattern, and approximately 40% of proteins in the database are type I (Wilmot and Thornton 1988). We have only taken this type I turn into account. In a type I turn, a hydrogen bond is formed between the main-chain position i and i+3. Amino acids at each position between i and i+3 were included based on their known frequency of occurrence (Wilmot and Thornton 1988). For the turn (referred to as turn cassette), Asp, Ser, Asn, and Thr were chosen for position i, Asp, Ser, Thr, and Pro for position i+1, Asp, Ser, Asn, and Ala for position i+2, and Gly for position i+3, provided by the connecting residues (Fig. 1A ▶).
Library construction and characterization
DNA cassettes for all secondary structure modules were prepared from synthetic trinucleotide-encoded oligonucleotides (Virnekäs et al. 1994). All cassettes were designed to have a BglII site upstream and a BamHI site downstream (Fig. 1B ▶). Because these sites are compatible but neither site is regenerated when they are ligated to each other, a unique orientation of the polymerized product is enforced if the ligation is carried out in the presence of both restriction endonucleases. A further benefit is that the ligation product of the two sites is 5′-GGATCT-3′, which codes for Gly-Ser, small and hydrophilic residues ideal for connecting modules (Fig. 1B ▶). The cassettes were amplified by PCR, digested with the restriction enzymes BamHI and BglII, and purified. They were then mixed and polymerized in different combinations. Polymerized fragments were first ligated into an expression vector (derivative of pQE16), yielding typically 3–5 modules, and further extended by consecutive digestion and ligation (see Experimental Procedures) to give four libraries of an average size of 100 residues: α10, α5, and turn cassettes (library 1), α10 and turn cassettes (library 2), α10, α5, β, and turn cassettes (library 3), and β and turn cassettes (library 4). Each of the four libraries is named according to which cassettes it contains (e.g., library α10α5βt is composed of α10, α5, β, and turn cassettes). All libraries were cloned in an expression vector (derivative of pQE16) with a T5 promoter and strong Shine-Dalgarno sequence that at the same time introduces a C-terminal histidine tag used for detection (Lindner et al. 1997) and purification.
DNA sequences of 13 to 15 randomly picked clones from each library showed that between 10 clones and 14 clones (77%–93%) had no frameshift or internal stop codons (Table 1). In addition, all clones contained only the intended secondary structure cassettes. However, the proportion of turn cassettes introduced was lower than expected in all four libraries. This is probably due to the short length of this cassette, resulting in a lower ligation efficiency in the initial in vitro polymerization. Moreover, some of the turn modules were at the end of the protein and thus did not play any role as "turns" (Fig. 1C ▶). The average lengths were 105, 111, 78, and 113 amino acids for the α10t, α10α5βt, βt, and α10α5t libraries, respectively (Table 1). FASTA searches (Pearson 1990) were performed against the SwissProt database (Bairoch and Apweiler 2000). Most protein sequences of members chosen randomly from the four libraries showed only spurious homology to any known protein sequence. Therefore, despite having secondary structure modules derived from a consensus of known proteins as building blocks, most of the library members are unrelated to any sequence of known proteins.
Table 1.
Properties of randomly chosen members from each of the four libraries
| Library | α10t | α10α5βt | βt | α10α5t |
| Length (amino acid residues) | 105 ± 26 | 111 ± 19 | 78 ± 27 | 113 ± 20 |
| Cassette composition | ||||
| α10 | 93% | 40% | 48% | |
| α5 | 38% | 49% | ||
| β | 18% | 95% | ||
| t | 7.0% | 3.6% | 5.5% | 3.4% |
| In frame | 79% (11/14) | 85% (11/13) | 93% (14/15) | 77% (10/13) |
| Expression | 29% (30/103) | 61% (44/72) | 84% (26/31) | 7.9% (10/127) |
To assess the proportion of clones having detectable protein expression, Escherichia coli XL1-blue harboring a plasmid encoding the tRNAs for rare codons (pACYC-RIL) was transformed with the plasmid pool of each of the four libraries, and colony blots were performed using an antihistidine tag single-chain Fv–alkaline phosphatase fusion (Lindner et al. 1997). A large difference between the libraries was found, with library α10α5βt (44/72) and βt (26/31), both containing β-strand modules, giving a higher proportion of positive signals compared to the all-α-helical libraries, library α10α5t (10/127) and library α10t (30/103) (Table 1). If only clones with correct reading frames are considered, 37, 72, 90, and 10% of libraries α10t, α10α5βt, βt, and α10α5t, respectively, have detectable protein expression, which is very high for synthetic libraries (Davidson and Sauer 1994; Prijambada et al. 1996; Cho et al. 2000). As the protein must resist proteolysis in the cytoplasm to be detected, a high proportion of the clones of these totally synthetic libraries must be stable against proteases in the cytosol.
Solubility in aqueous solution is one of the most important properties of globular proteins. Twelve clones from each of the four libraries were analyzed for the percentage of soluble and insoluble recombinant protein in the cell lysate by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). This analysis showed that the proteins investigated from library α10α5t and α10α5βt have a moderate solubility, with 10% to 60% of each recombinant protein being in the soluble fraction, and this percentage did not differ significantly between the two libraries. For library α10t, severe protein degradation was observed once the cells were lysed, and thus in most cases protein solubility could not be judged. Nevertheless, those proteins that were detectable had a solubility of about 10%, which is an underestimate, due to proteolytic degradation of the soluble fraction. In contrast to the α-helix containing libraries, all proteins tested from library βt were expressed as inclusion bodies. Proteins from library α10α5βt showed no sign of degradation. However, none of the proteins tested from this library bound to nickel-nitrilotriacetic acid (Ni-NTA) agarose beads, except in the presence of 6 M guanidine hydrochloride (GdnHCl). In general, lack of binding to Ni-NTA agarose beads is an indication of the inaccessibility of the histidine tag, and can indicate aggregation of the protein. It is thus possible that proteins from library α10α5βt and βt are expressed as soluble and insoluble aggregates, respectively, making them highly resistant to proteolysis. This would explain why a larger portion of expressing clones could be detected by colony blots in these libraries than in the other two libraries (Table 1).
Structural characteristics of randomly selected library members
Proteins from library α10α5t and α10t.
To understand the structural properties of proteins from each of the four libraries, and to investigate the differences between them, one or two proteins that showed the highest expression levels from preliminary characterized members of each of the four libraries were chosen for further characterization. The amino acid sequences of those are shown in Figure 1C ▶. Figure 2 ▶ shows circular dichroism (CD) spectra of two proteins from library α10α5t (IVex22 and IVex24) and one from library α10t (Iex23). The spectra of all three proteins had a similar dependence on salt concentration. In the absence of or in the presence of low concentrations of NaCl, all proteins exhibit a CD spectrum having a minimum at 200 nm, indicating the existence of a substantial proportion of random coil conformation (Fig. 2 ▶). As the NaCl concentration was increased, the helicity also increased and reached a plateau above 3.5 M or 4.5 M, respectively (see below). At high concentrations of NaCl, the CD spectra show two minima at 208 nm and 222 nm, which is typical for an α-helical protein (Fig. 2 ▶). All three proteins were monomeric at high concentrations of NaCl, as determined by gel filtration experiments (Fig. 3A ▶), which were confirmed by sedimentation equilibrium experiments (data not shown). In 3.5 M NaCl, the elution volumes of these three proteins measured from gel filtration experiments were somewhat smaller than expected for natural proteins of the same number of amino acids (Fig. 3A ▶), indicating that the volumes of the proteins are 5% to 40% larger than that of native globular proteins, typical for the molten globule state (Arai and Kuwajima 2000). The presence of defined oligomerization states is another important property of native proteins and all three proteins examined here are shown to fulfill this criterion in NaCl. A similar behavior to that in NaCl was observed with (NH4)2SO4 (Fig. 2 ▶), again giving rise to a folding transition with increasing (NH4)2SO4 concentration.
Fig. 2.

CD spectra of the arbitrarily selected clones. CD spectra of clones from library α10α5t (IVex22 and IVex24) and α10t (Iex23) at different concentrations of NaCl and (NH4)2SO4. In black is shown the spectrum in 20 mM sodium phosphate, pH 6.5; in gray is the spectrum in the same buffer containing 3.5 M NaCl; and in dashed is the spectrum in buffer containing 1.8, 1.8, and 1.2 M (NH4)2SO4 for Iex23, IVex22, and IVex24, respectively. Protein concentrations were 6, 10, and 6 μM for Iex23, IVex22, and IVex24, respectively. All measurements were performed at room temperature.
Fig. 3.
Hydrodynamic properties of proteins Iex23, IVex22, and IVex24. (A) Gel filtration experiments. The absorbance data were recorded at 230 nm. Buffer conditions were 20 mM sodium phosphate pH 6.5 with 3.5 M NaCl. Protein concentrations were 50 μM. The apparent molecular masses were 9.1, 14.1, and 15.3 kD, and the calculated masses for monomers are 8.7, 10.1, and 12.3 kD for Iex23, IVex22, and IVex24, respectively. The arrows indicate the elution volume of proteins used for calibration. (B) Sedimentation equilibrium experiments. The absorbance data were recorded at 230 nm. Buffer conditions were 20 mM sodium phosphate pH 6.5 with 1.8 M (NH4)2SO4. The concentration of protein Iex23 was 15 μM. The apparent molecular mass was 26.9 kD, and the theoretical mass of the trimer is 26.2 kD. The fitted curve is superimposed over the data points.
Although the gel filtration data in (NH4)2SO4 are not straightforward to interpret because of changes in the elution volume of the standard proteins, sedimentation equilibrium data could be used to determine the oligomerization state. The molecular weight determined from sedimentation equilibrium experiments and the random residuals showed protein Iex23 to be a trimer in (NH4)2SO4 (Fig. 3B ▶), while IVex22 and IVex24 could not be fitted assuming a single species, suggesting coexistence of monomers with dimers or trimers. These three proteins (Iex23, IVex22, and IVex24) were soluble in the absence of any denaturants, and could be successfully concentrated up to 200 μM without any precipitation. The high solubility of the investigated proteins and the solubility tests of additional randomly chosen clones (Table 1) imply that most of the expressed proteins are indeed soluble.
Salts affect the properties of proteins such as stability, solubility, and biological activity (von Hippel and Schleich 1969). For instance, disulfide-bridge lacking ribonuclease T1, which is unfolded in the absence of NaCl, can be refolded in the presence of high concentrations of NaCl, resulting in a shift of the urea denaturation midpoint (Pace et al. 1988) from 0.51 to 2.17 M. (NH4)2SO4 has been shown to induce a conformational change of GroEL from E. coli to a fully functional chaperonin at temperatures up to 75°C (Kusmierczyk and Martin 2000). It has also been observed that high salt concentrations can refold the acid-induced unfolded state of β-lactamase, cytochrome c, and apomyoglobin to a molten-globule state of the protein (Goto et al. 1990). Salts can effect the protein conformation in different ways via preferential hydration and/or preferential ion binding, depending on the salt (Arakawa and Timasheff 1984). Kosmotropes are known to decrease the solubility of the nonpolar residues and to increase the solubility of peptide groups (Nandi and Robinson 1972). They can also screen the repulsive electrostatic interactions between charged residues occurring at low salt concentrations, thereby increasing conformational stability in some cases (Goto et al. 1990). To investigate the effect of salt on our designed proteins, the [θ]222 values were measured as a function of ionic strength in three different salts, NaCl, (NH4)2SO4, and MgSO4. We found a strong correlation between the [θ]222 values and the ionic strength. All three proteins have a net charge between +2 and +10 at pH 6.5, and it is thus likely that helix formation and the ensuing tertiary structure formation is at least partially due to the screening of the repulsive ionic interactions, in addition to the effect the salts have on increasing the hydrophobic effect.
1,8-Anilinonaphtalene sulfonate (ANS) is a fluorescent probe that can detect the accessible hydrophobic core area. Detectable binding is consistent with molten-globule states of a protein, whereas it generally shows only weak binding to native or totally unfolded structures (Semisotnov et al. 1991). ANS binding experiments were performed in the presence and absence of NaCl or (NH4)2SO4 (Fig. 4 ▶). Carbonic anhydrase, used as a control, showed almost no binding to ANS, regardless of the salt concentrations used. In contrast, all three proteins tested showed no detectable binding to ANS in the absence of salts, but significant binding in 3.5 M NaCl. The lack of binding in the absence of NaCl is indicative of the lack of secondary or tertiary structure, consistent with the CD data under theses conditions (Fig. 2 ▶). On the other hand, these three proteins are not only forming helices but are relatively compact, as judged from the gel filtration experiments in the presence of high concentrations of NaCl (Figs. 2, 3A ▶ ▶). In addition, binding to ANS under these conditions indicates that all three proteins have an accessible hydrophobic core, consistent with a state similar to molten-globule. Similar results were obtained in (NH4)2SO4, but giving lower ANS-fluoresence signal than that in NaCl (Fig. 4 ▶), indicating the presence of a less accessible hydrophobic core, probably due to the oligomerization of the proteins and/or a more compact structure.
Fig. 4.
ANS binding experiments of proteins Iex23, IVex22, and IVex24. Relative fluorescence intensity of 25 mM ANS in 20 mM sodium phosphate, pH 6.5, in the presence or absence of salts at 480 nm with excitation at 370 nm. The concentration of NaCl was 3.5 M and the concentrations of (NH4)2SO4 were 1.8, 1.8, and 1.2 M for Iex23, IVex22, and IVex24, respectively. The relative signals with carbonic anhydrase (CA) were identical regardless of the (NH4)2SO4 concentrations. Protein concentrations were 0.5 μM.
Urea equilibrium unfolding experiments were performed in the presence of both NaCl and (NH4)2SO4. In the presence of NaCl, none of the three showed cooperative unfolding behavior (Fig. 5 ▶). In contrast, clear cooperative unfolding behavior was observed in (NH4)2SO4 (Fig. 5 ▶) (Roy and Hecht 2000). Especially Iex23, which is trimeric under these conditions showed a pretransition baseline up to 1 M urea.
Fig. 5.

Urea equilibrium unfolding experiments of proteins Iex23, IVex22, and IVex24. Experiments were performed in 20 mM sodium phosphate, pH 6.5 and 3.5 M NaCl (open circles), and in 20 mM sodium phosphate, pH 6.5 and 1.8, 1.8 and 1.2 M (NH4)2SO4 (filled circles) for Iex23, IVex22, and IVex24, respectively. Measurements were performed at room temperature, and at 222 nm. In the presence of NaCl, protein concentrations were 6, 10, and 10 μM for Iex23, IVex22, and IVex24, respectively. In the presence of (NH4)2SO4, protein concentrations were 7, 6, and 9 μM for Iex23, IVex22, and IVex24, respectively.
Taken all the results together, observations from the CD spectroscopy, gel filtration, ANS binding and urea equilibrium unfolding experiments are all consistent with properties of the molten-globule state of a protein (Arai and Kuwajima 2000) in NaCl, but some properties are closer to native proteins in (NH4)2SO4.
Proteins from library α10α5βt and βt.
Two proteins from library α10α5βt (IIex8 and IIex10; Fig. 1C ▶) were chosen for further characterization. None of the proteins tested from this library bound to Ni-NTA agarose beads, except in the presence of 6 M GdnHCl, and thus they were purified under denaturing conditions. After elution from the Ni-NTA agarose, the purified proteins were dialyzed against buffer without GdnHCl. More than 90% of the protein remained soluble after dialysis.
Although the proteins from library α10α5t and library α10t (Iex23, IVex22, and IVex24) were soluble in all concentrations of NaCl, the two proteins IIex8 and IIex10 precipitated above 0.5 and 1.5 M NaCl, respectively, at a protein concentration of 5 μM (Fig. 6A ▶). NaCl can increase the strength of hydrophobic interactions between the proteins. As the only difference in sequence between the all α-helix libraries and library α10α5βt is the presence of the β-strand modules, it is likely that these proteins have exposed hydrophobic patches, which are due to the β-strand modules.
Fig. 6.
Properties of arbitrarily selected clones from library α10α5βt (IIex8 and IIex10) compared to members from other libraries. (A) Relative solubility of proteins Iex23 (open circles), IVex22 (open squares), IVex24 (open diamonds), IIex8 (filled circles), and IIex10 (filled squares) as a function of NaCl concentrations. Buffer conditions were 20 mM sodium phosphate, pH 6.5 and the protein concentrations were 5 μM. Solubility was normalized according to the absorbance of each protein in a buffer with 150 mM NaCl. (B) CD spectra of IIex8 and IIex10 in 20 mM sodium phosphate, pH 6.5 at different NaCl concentrations: With IIex10, an increase in NaCl concentrations resulted in the precipitation of the protein (Fig. 6A ▶), and, therefore, that spectra is not shown. Protein concentrations were 17 and 7 μM for IIex8 and IIex10, respectively. All measurements were performed at room temperature.
CD spectra of these two proteins were indicative of the presence of a random coil conformation (Fig. 6B ▶). Library α10α5βt shares the same α-helix module with all α-helical libraries, of which the representative members showed α-helix formation by increasing the ionic strength. We tested the effect of the addition of NaCl on the secondary structure formation of IIex8 and IIex10, and observed an increasing CD signal at around 220 nm (Fig. 6B ▶); however, it was too weak to be distinguished whether the increase is from α-helix or β-sheet formation.
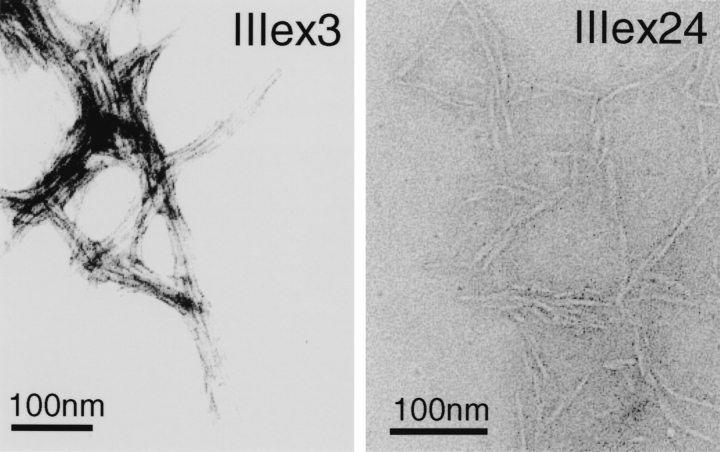
Twelve proteins from library βt were expressed and analyzed by SDS-PAGE. The behavior of these proteins on SDS-PAGE was very different from that of proteins from the other three libraries. They did not enter the SDS-polyacrylamide gel, except in the presence of 8 M urea in both the sample buffer and the gel. It was also important not to boil the sample prior to loading on the gel. This phenomenon is often found with integral membrane proteins (Gould 1994). Two proteins from library βt (IIIex3 and IIIex24; Fig. 1C ▶) were purified via IMAC under denaturing conditions using 8 M urea. Among those that showed relatively high expression, IIIex24 was chosen as it was found to contain two α10 cassettes (Fig. 1C ▶), yet showed similar behavior on the SDS-polyacrylamide gel electrophoresis. After 1 d, the proteins precipitated even in 8 M urea. These precipitates could be solubilized in formic acid and were then confirmed by electrospray mass spectrometry to have the expected mass of IIIex3 and IIIex24. To our surprise, these proteins could neither be solubilized in 8 M urea nor in 6 M GdnHCl, once they had precipitated. Figure 7 ▶ shows electron micrographs of the precipitates from IIIex3 and IIIex24. They show clear evidence of the formation of amyloid-like fibrils. The formation of amyloid fibrils is known to be associated with diseases such as Alzheimer’s disease and spongiform encephalopathies (Kelly 1996). However, amyloid formation is not only restricted to these disease-associated proteins but appears to occur for a wide variety of proteins, including acylphosphatase (Chiti et al. 1999), the SH3 domain (Guijarro et al. 1998), and the bacterial cold-shock protein CspB (Gross et al. 1999), suggesting that this may be a common property of many proteins. Fibrils are generally reported to dissociate in the presence of denaturants (Fink 1998). West et al. (1999) found that a combinatorial library of β-strands, in which each module has alternating polar and nonpolar residues, tends to form soluble amyloid fibrils. In contrast, the fibrils that are shown in Figure 7 ▶ were not soluble even in high concentrations of denaturants, and thus may be a good model for studying fibril formation and its architecture.
Fig. 7.
Electron micrographs showing the fibril formation of proteins IIIex3 and IIIex24. For details, see text.
Discussion
Our aim was to create protein libraries biased to form stretches of secondary structure that are thus poised to form compact domains with tertiary structure of the typical size found in natural proteins. Rather than to select single members for their binding (Keefe and Szostak 2001) or solubility properties (Waldo et al. 1999), we wanted to first characterize unselected members of the libraries for several reasons. Primarily, we wanted to validate our module design by assessing their propensity to form the intended structure elements. Second, we wished to investigate the differences in the properties among the four libraries built from different combinations of modules. Third, we wanted to probe if and with what frequency native-like properties can be observed among arbitrarily selected clones.
The protein libraries that contained the designed secondary structure modules α10, α5, and turn gave rise to members that were soluble, monomeric, helical, and relatively compact in high concentrations of NaCl. However, their binding to ANS indicates that these proteins have a molten-globule like structure. In contrast, cooperative unfolding by denaturants was observed in (NH4)2SO4, and binding to ANS was significantly lower than that in NaCl, indicating that these arbitrarily selected proteins already show some evidence of foldedness (Roy and Hecht 2000). On the other hand, as soon as β-strand cassettes are integrated into the protein, their behavior changes dramatically, making them much more aggregation-prone. Therefore, while with α-helix modules it is relatively easy to obtain proteins with many characteristics of natural globular proteins, the presence of β-strands seems to require precise topological arrangement to prevent aggregation.
The behavior of the β-containing libraries was highly influenced by the presence of α-modules. The α- and β-module-containing proteins belonging to library α10α5βt are aggregated but soluble, while those from library βt are among the least soluble proteins ever reported. This is consistent with the view that fibril formation is correlated with β-sheet formation (Sunde et al. 1997), and the hypothesis that fibrils are essentially "infinite β-sheets" (Fink 1998; Xu et al. 2001). Apparently, our design inadvertently fulfills this requirement unusually well. It follows that in all-β-proteins a number of mechanisms must be operative to prevent this fibril formation, including topology enforcing turns (underrepresented in our libraries) and deviations from the regular spacing of the β-strands in the sequence.
Theoretical studies based on a lattice model suggested that many polypeptide sequences containing only two residue types, polar and nonpolar, could form proteins with native-like properties (Dill 1990). In addition, several experimental studies have demonstrated that proteins with native-like properties can be obtained by simplifying the sequence to polar and nonpolar residues (Kamtekar et al. 1993; Davidson and Sauer 1994; Marshall and Mayo 2001). Because of the restricted nature of our cassettes (only 4–5 amino acids allowed at most positions) and the absence of any selection applied, we cannot reasonably expect to obtain the finely tuned packing required for natively folded proteins. Nevertheless, this restrictive design has led to an extraordinary percentage of soluble, expressed proteins, at least in the non-β-containing libraries, which confirms the efficacy of applying binary patterning and residues with high α-helix or β-strand propensity in protein design.
Most previous studies based on binary patterning aimed to design a protein with a simple and/or a specific fold (Kamtekar et al. 1993; Marshall and Mayo 2001). Our aim, in contrast, was to acquire proteins with native-like properties without restricting the topology of the protein to explore new topologies. Despite the fact that the members of our libraries are distant from natural sequences in protein sequence space, we estimated that about one in six members of the all-α-helix libraries exhibit properties of the molten globule state. Although natural proteins, in general, have a distinct global free energy minimum that allows them to fold into one unique structure, molten-globule–like properties probably lack a distinct global minimum, and thus do not have specific tertiary structure (Arai and Kuwajima 2000) even though they may have a particular topology. How can a protein evolve to have a distinct global minimum? We think the answer is functional selection (binding and/or catalytic activity), as the way natural proteins have evolved, based on their functional requirements. In addition, Yomo et al. (1999) have suggested in their studies that functional selection can lead the protein to acquire an ability to fold into a unique structure. Moreover, as single mutations can convert native proteins into one with molten-globule properties (Creighton and Ewbank 1994), the converse introduction of point mutations by error-prone PCR and/or DNA shuffling (Stemmer 1994) combined with a selection technology (Forrer et al. 1999; Schaffitzel et al. 1999) can very likely also shift the properties of many of our library members more to those of natural globular proteins. The chain topology is mainly defined by the structural arrangement of secondary structure modules (Taylor and Orengo 1989), and thus our library is an excellent starting point for investigating the possibility of generating a series of novel proteins with new folds.
In conclusion, the unexpectedly favorable properties of a large fraction of members in the designed libraries reported here make them a unique starting point for directed evolution and structure determination of a variety of proteins, very distant in sequence space from natural proteins.
Material and methods
Library construction
Oligonucleotides for each cassette were designed as described in the text and synthesized using trinucleotide codon building blocks (Virnekäs et al. 1994) (Figs. 1A, 1B ▶). All oligonucleotides encoding secondary structure modules were PCR amplified, purified, and digested with BamHI and BglII (Fermentas). The digested products were purified either by polyacrylamide gel electrophoresis followed by electro-elution or by gel filtration (Superdex 75, Pharmacia). Using a pool of α10, α5, β, and turn cassettes, four different mixtures, (1) a mixture of α10, α5 and turn cassettes in a 1:1:2 molar ratio; (2) α10 and turn cassettes in a 1:2 molar ratio; (3) α10, α5, β, and turn cassettes in a 1:1:1:2 molar ratio; and (4) β and turn cassettes in a 1:2 molar ratio were prepared. They were polymerized at 37°C by addition of T4 ligase (2.5 units/35 μL; Fermentas) and the two restriction enzymes BamHI and BglII (25 units each/35 μL). The polymerized products thus have a BglII (upstream) and BamHI (downstream) site. They were ligated into a derivative of the expression vector pQE16 (Qiagen) (referred to as pQE16rev), which had been cut with BamHI and BglII, and into which a T5 promoter and Shine-Dalgarno sequence, a His6 tag at the C-terminus, and a stop codon had been introduced (Fig. 8A ▶). To eliminate the inserts ligated in the wrong orientation, ligation products were PCR amplified with biotinylated primers and digested with EcoRI and BamHI. After digestion, those with biotin were removed using streptavidin-coated magnetic beads (Roche) as only those with incorrectly orientated inserts possess biotin at either end (Fig. 8A ▶). The resulting DNA fragments were ligated into the same vector, pQE16rev, cut with EcoRI and BamHI. The length of the library at this point was, on average, 30 amino acids.
Fig. 8.


Scheme of the library construction procedure. (A) After polymerization of the modules (see Experimental Procedures), these fragments (hatched arrows) were ligated into the vector pQE16rev digested with BamHI and BglII. Inserts can be ligated in both orientations, as they have BamHI and BglII sites at each end. To remove those with incorrectly oriented ones, the ligation product was directly PCR amplified with biotinylated primers (open arrows). Subsequently, PCR products were digested with EcoRI and BamHI, followed by a removal of biotinylated DNA fragments. After digestion, PCR products with correctly oriented insert (right) do not contain biotin at either end, whereas incorrectly oriented ones (left) do, which are then removed by streptavidin-coated magnetic beads (see Experimental Procedures). (B) After the removal of incorrectly oriented inserts, fragments were ligated into a vector pQE16rev digested with BamHI and EcoRI. E. coli XL1-blue was transformed with the resulting product and plasmids were extracted (pQE16rev+insert). Plasmid pools were first digested with XbaI to linearize the plasmid, dephosphorylated, and then divided into two aliquots. One was cut with BamHI (left) and the other with BglII (right). Fragments were then ligated together to generate longer ORFs. This in vitro digestion and ligation step was repeated three times to achieve an average length of 300 bp.
E. coli XL1-blue (Stratagene) was transformed with the library and plasmids were extracted (Fig. 8B ▶). Plasmid pools were first digested with XbaI to linearize the plasmid, dephosphorylated, and then divided into two aliquots, one of which was cut with BamHI and the other with BglII. Fragments were ligated together to generate longer ORFs. The correctly ligated products were purified by agarose gel electrophoresis followed by the QIAquick gel extraction kit (Qiagen). This alternating in vitro digestion and ligation step was repeated three times to achieve an average length of 300 bp (Fig. 8B ▶). Finally, we obtained 10 ng of DNA, which corresponds to about 109 copies of DNA. As the theoretical diversity of the library is 1040, far larger than copies of DNA we obtained, the practical diversity of the library before transformation was estimated to be 109.
Colony blot
The constructed DNA libraries were PCR amplified, digested with EcoRI and BamHI, and ligated into the expression vector pQE16rev, which had been cut with EcoRI and BamHI. Ligation products were then transformed into E. coli XL1-blue harboring the plasmid pACYC-RIL (Stratagene), blotted onto nitrocellulose filters, and probed with an antihistidine tag single-chain antibody fragment alkaline–phosphatase fusion protein (Lindner et al. 1997). The filter was developed with a chromogenic substrate (5-bromo-4-chloro-indolyl-phosphate toluidine salt and nitro blue tetrazolium). When E. coli XL1-blue harboring the plasmid pACYC-RIL without any constructs was blotted, no positive signal was observed. Some of the colonies that gave positive signals on the blot were grown in liquid media (LB, 37°C), and protein expression was confirmed by a Western blot.
Protein expression and purification
E. coli BL21codonplusRIL (Stratagene), carrying the plasmid encoding the recombinant protein, was grown in LB media at 27°C with ampicillin and chloramphenicol to OD600 = 0.5 to 1.0 and induced by addition of 1 mM IPTG followed by an additional 3-h incubation at 27°C. Purification of the proteins was achieved with immobilized metal ion affinity chromatography (IMAC) using Ni-NTA agarose (Qiagen) and subsequent cation exchange chromatography on CM-sepharose (Pharmacia) when necessary. IMAC under denaturing conditions was carried out according to the manufacturer’s protocol. Under native conditions, the cell pellet was first subjected to a periplasmic extraction (to remove the periplasmic proteases) by suspending and incubating the cells on ice in 0.1 M Tris-HCl buffer pH 8, 0.5 M sucrose, and 1 mM EDTA and subsequently washing with the same buffer without EDTA several times. This procedure significantly increased the yield of the protein after purification. The cells were then lysed by sonication and IMAC was carried out according to the manufacturer’s protocol. Ion exchange chromatography (CM-sepharose: Pharmacia) was done in 20 mM sodium phosphate buffer pH 6.5 and elution was achieved with a NaCl gradient from 0.1 to 1 M. The protein yield was 1–10 mg L−1. Protein concentrations were determined by amino acid analysis.
Spectroscopic measurements
CD measurements were carried out with a Jasco J-715 spectropolarimeter. All measurements were performed at room temperature, after equilibrating the protein sample overnight in appropriate buffer conditions as described in the text. The data were normalized to molar ellipticity with a pathlength of 0.1 cm.
ANS binding was measured with an PTI Alpha Scan spectrofluorimeter at 25°C. Emission scans between 400 and 600 nm were recorded with excitation at 370 nm. The peak of the emission spectra shifts from 520 to 480 nm when ANS is bound to the protein (Semisotnov et al. 1991). Therefore, relative binding refers to the signal at 480 nm in the presence of the protein relative to the one in absence of the protein in the same buffer.
The solubility of proteins were determined by measuring the absorbance at 230 nm after incubating in an appropriate buffer for overnight followed by centrifugation at 20,000 × g for 20 min.
Gel filtration experiments
Gel filtration experiments were performed with a SMART system using a Superdex 75 column (Pharmacia) at 20°C. The absorbance data were recorded at 230 nm. Buffer conditions were as described in the text. Standards used for molecular weight calibrations were bovine albumin (66 kD) (Sigma), carbonic anhydrase (29 kD) (Sigma), and protein D (17.4 kD) (Yang et al. 2000).
Analytical ultracentrifuge
Sedimentation equilibrium experiments were carried out with an XL-A analytical ultracentrifuge (Beckman) at 40,000 rpm at 20°C. The absorbance data were recorded at 230 nm. A single species model was assumed and data were fit to the equation: cr = c0exp[[Mw,app(1 − 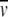 2ρ) (r22 − r02)ω2]/2RT] where Mw,app is the apparent molecular mass,
2ρ) (r22 − r02)ω2]/2RT] where Mw,app is the apparent molecular mass, 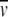 2 is the partial specific volume, c is molar concentration, r is radial distance, ω is rotation speed, and ρ is the solvent density.
2 is the partial specific volume, c is molar concentration, r is radial distance, ω is rotation speed, and ρ is the solvent density.  2 values for each protein in the presence of (NH4)2SO4 were calculated according to the equation described in reference (Arakawa and Timasheff 1985).
2 values for each protein in the presence of (NH4)2SO4 were calculated according to the equation described in reference (Arakawa and Timasheff 1985).
Electron microscopy
Electron microscopy was performed with a Philips EM301 instrument. Protein pellets were washed first with 6 M GdnHCl, and then with water several times. They were absorbed onto a Formvar-coated grid and then negatively stained with 2% uranyl acetate.
Acknowledgments
We thank the Protein Analysis Unit of the Biochemisches Intitut for mass spectroscopy and amino acid analysis, Dr. Richard M. Thomas for sedimentation equilibrium experiments, and Dr. Ernst Wehrli for electron microscopy and Morphosys AG for the trinucleotide-containing oligonucleotides. We are indebted to Dr. Stephen Marino for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0215102.
References
- Arai, M. and Kuwajima, K. 2000. Role of the molten globule state in protein folding. Adv. Protein Chem. 53 209–282. [DOI] [PubMed] [Google Scholar]
- Arakawa, T. and Timasheff, S.N. 1984. Mechanism of protein salting in and salting out by divalent cation salts: Balance between hydration and salt binding. Biochemistry 23 5912–5923. [DOI] [PubMed] [Google Scholar]
- ———. 1985. Calculation of the partial specific volume of proteins in concentrated salt and amino acid solutions. Methods Enzymol. 117 60–65. [DOI] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell, T.L. and Zhu, Z.Y. 1995. The α-helix as seen from the protein tertiary structure: A 3-D structural classification. Biophys. Chem. 55 167–184. [DOI] [PubMed] [Google Scholar]
- Broome, B.M. and Hecht, M.H. 2000. Nature disfavors sequences of alternating polar and non-polar amino acids: Implications for amyloidogenesis. J. Mol. Biol. 296 961–968. [DOI] [PubMed] [Google Scholar]
- Chiti, F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G., and Dobson, C.M. 1999. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. 96 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, G., Keefe, A.D., Liu, R., Wilson, D.S., and Szostak, J.W. 2000. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J. Mol. Biol. 297 309–319. [DOI] [PubMed] [Google Scholar]
- Chothia, C. 1984. Principles that determine the structure of proteins. Annu. Rev. Biochem. 53 537–572. [DOI] [PubMed] [Google Scholar]
- Creighton, T.E. and Ewbank, J.J. 1994. Disulfide-rearranged molten globule state of α-lactalbumin. Biochemistry 33 1534–1538. [DOI] [PubMed] [Google Scholar]
- Davidson, A.R. and Sauer, R.T. 1994. Folded proteins occur frequently in libraries of random amino acid sequences. Proc. Natl. Acad. Sci. 91 2146–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, K.A. 1990. Dominant forces in protein folding. Biochemistry 29 7133–7155. [DOI] [PubMed] [Google Scholar]
- Fink, A.L. 1998. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 3 R9–R23. [DOI] [PubMed] [Google Scholar]
- Finkelstein, A.V., Badretdinov, A., and Gutin, A.M. 1995. Why do protein architectures have Boltzmann-like statistics? Proteins 23 142–150. [DOI] [PubMed] [Google Scholar]
- Forrer, P., Jung, S., and Plückthun, A. 1999. Beyond binding: Using phage display to select for structure, folding and enzymatic activity in proteins. Curr. Opin. Struct. Biol. 9 514–520. [DOI] [PubMed] [Google Scholar]
- Goto, Y., Calciano, L.J., and Fink, A.L. 1990. Acid-induced folding of proteins. Proc. Natl. Acad. Sci. 87 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, G.W. 1994. Membrane protein expression systems: A user’s guide. Portland Press, London.
- Govindarajan, S. and Goldstein, R.A. 1996. Why are some proteins structures so common? Proc. Natl. Acad. Sci. 93 3341–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, M., Wilkins, D.K., Pitkeathly, M.C., Chung, E.W., Higham, C., Clark, A., and Dobson, C.M. 1999. Formation of amyloid fibrils by peptides derived from the bacterial cold shock protein CspB. Protein Sci. 8 1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro, J.I., Sunde, M., Jones, J.A., Campbell, I.D., and Dobson, C.M. 1998. Amyloid fibril formation by an SH3 domain. Proc. Natl. Acad. Sci. 95 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling, R., Li, H., Melin, R., Miller, J., Wingreen, N., Zeng, C., and Tang, C. 2001. The designability of protein structures. J. Mol. Graph. Model. 19 157–167. [DOI] [PubMed] [Google Scholar]
- Kamtekar, S., Schiffer, J.M., Xiong, H., Babik, J.M., and Hecht, M.H. 1993. Protein design by binary patterning of polar and nonpolar amino acids. Science 262 1680–1685. [DOI] [PubMed] [Google Scholar]
- Keefe, A.D. and Szostak, J.W. 2001. Functional proteins from a random-sequence library. Nature 410 715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J.W. 1996. Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol. 6 11–17. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk, A.R. and Martin, J. 2000. High salt-induced conversion of Escherichia coli GroEL into a fully functional thermophilic chaperonin. J. Biol. Chem. 275 33504–33511. [DOI] [PubMed] [Google Scholar]
- Lindner, P., Bauer, K., Krebber, A., Nieba, L., Kremmer, E., Krebber, C., Honegger, A., Klinger, B., Mocikat, R., and Plückthun, A. 1997. Specific detection of his-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFv-phage fusions. Biotechniques 22 140–149. [DOI] [PubMed] [Google Scholar]
- Marshall, S.A. and Mayo, S.L. 2001. Achieving stability and conformational specificity in designed proteins via binary patterning. J. Mol. Biol. 305 619–631. [DOI] [PubMed] [Google Scholar]
- Nandi, P.K. and Robinson, D.R. 1972. The effects of salts on the free energies of nonpolar groups in model peptides. J. Am. Chem. Soc. 94 1308–1315. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Jones, D.T., and Thornton, J.M. 1994. Protein superfamilies and domain superfolds. Nature 372 631–634. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. and Scholtz, J.M. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, C.N., Grimsley, G.R., Thomson, J.A., and Barnett, B.J. 1988. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J. Biol. Chem. 263 11820–11825. [PubMed] [Google Scholar]
- Pearl, F.M., Lee, D., Bray, J.E., Sillitoe, I., Todd, A.E., Harrison, A.P., Thornton, J.M., and Orengo, C.A. 2000. Assigning genomic sequences to CATH. Nucleic Acids Res. 28 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W.R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183 63–98. [DOI] [PubMed] [Google Scholar]
- Prijambada, I.D., Yomo, T., Tanaka, F., Kawama, T., Yamamoto, K., Hasegawa, A., Shima, Y., Negoro, S., and Urabe, I. 1996. Solubility of artificial proteins with random sequences. FEBS Lett. 382 21–25. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. and Richardson, D.C. 1988. Amino acid preferences for specific locations at the ends of α helices. Science 240 1648–1652. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, D.M., Roy, S., and Hecht, M.H. 1999. Screening combinatorial libraries of de novo proteins by hydrogen-deuterium exchange and electrospray mass spectrometry. J. Am. Chem. Soc. 121 9509–9513. [Google Scholar]
- Roy, S. and Hecht, M.H. 2000. Cooperative thermal denaturation of proteins designed by binary patterning of polar and nonpolar amino acids. Biochemistry 39 4603–4607. [DOI] [PubMed] [Google Scholar]
- Schaffitzel, C., Hanes, J., Jermutus, L., and Plückthun, A. 1999. Ribosome display: An in vitro method for selection and evolution of antibodies from libraries. J. Immunol. Methods 231 119–135. [DOI] [PubMed] [Google Scholar]
- Semisotnov, G.V., Rodionova, N.A., Razgulyaev, O.I., Uversky, V.N., Gripas, A.F., and Gilmanshin, R.I. 1991. Study of the "molten globule" intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 31 119–128. [DOI] [PubMed] [Google Scholar]
- Stemmer, W.P. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370 389–391. [DOI] [PubMed] [Google Scholar]
- Sunde, M., Serpell, L.C., Bartlam, M., Fraser, P.E., Pepys, M.B., and Blake, C.C.F. 1997. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273 729–739. [DOI] [PubMed] [Google Scholar]
- Taylor, W.R. and Orengo, C.A. 1989. Protein structure alignment. J. Mol. Biol. 208 1–22. [DOI] [PubMed] [Google Scholar]
- Virnekäs, B., Ge, L., Plückthun, A., Schneider, K.C., Wellnhofer, G., and Moroney, S.E. 1994. Trinucleotide phosphoramidites: Ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res. 22 5600–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel, P.H. and Schleich, T. 1969. Structure and stability of biological macromolecules, pp. 417–574. Marcel Dekker, New York.
- Waldo, G.S., Standish, B.M., Berendzen, J., and Terwilliger, T.C. 1999. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17 691–695. [DOI] [PubMed] [Google Scholar]
- West, M.W. and Hecht, M.H. 1995. Binary patterning of polar and nonpolar amino acids in the sequences and structures of native proteins. Protein Sci. 4 2032–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M.W., Wang, W., Patterson, J., Mancias, J.D., Beasley, J.R., and Hecht, M.H. 1999. De novo amyloid proteins from designed combinatorial libraries. Proc. Natl. Acad. Sci. 96 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmot, C.M. and Thornton, J.M. 1988. Analysis and prediction of the different types of β-turn in proteins. J. Mol. Biol. 203 221–232. [DOI] [PubMed] [Google Scholar]
- Xu, G., Wang, W., Groves, J.T., and Hecht, M.H. 2001. Self-assembled monolayers from a designed combinatorial library of de novo β-sheet proteins. Proc. Natl. Acad. Sci. 98 3652–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F., Forrer, P., Dauter, Z., Conway, J.F., Cheng, N., Cerritelli, M.E., Steven, A.C., Plückthun, A., and Wlodawer, A. 2000. Novel fold and capsid-binding properties of the λ-phage display platform protein gpD. Nat. Struct. Biol. 7 230–237. [DOI] [PubMed] [Google Scholar]
- Yomo, T., Saito, S., and Sasai, M. 1999. Gradual development of protein-like global structures through functional selection. Nat. Struct. Biol. 6 743–746. [DOI] [PubMed] [Google Scholar]
- Yue, K. and Dill, K.A. 1995. Forces of tertiary structural organization in globular proteins. Proc. Natl. Acad. Sci. 92 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z.Y. and Blundell, T.L. 1996. The use of amino acid patterns of classified helices and strands in secondary structure prediction. J. Mol. Biol. 260 261–276. [DOI] [PubMed] [Google Scholar]