Abstract
Certain antibodies (Abs) elicited using the cardiac glycoside digoxin (digoxigenin tridigitoxoside) bind preferentially to analogs that differ from digoxin by substitutions on the cardenolide rings, the lactone, or by the presence or absence of attached sugars. Antibody 26–10 binds equally well to digoxin and digitoxin, which differ only by the presence in the former and the absence in the latter of an hydroxyl group at C12. Other antidigoxin Abs, however, can distinguish between these ligands by three orders of magnitude in binding. Inspection of the structure of Fab 26–10 complexed with digoxin shows a gap in complementarity in the region between the digoxin O12 and LCDR3. We proposed that insertions in LCDR3 might result in Abs that bind digitoxin preferentially. We produced libraries of mutants displayed on bacteriophage which were randomized at LCDR3 and contained LCDR3 insertions. Mutants were selected by panning against digoxin and analogs. The mutants bound digitoxin preferentially up to 47-fold greater than digoxin. The mutants that bound well to digitoxin demonstrated a consensus sequence including the substitution of Trp at position L:94. Using site-directed mutagenesis, the binding to digitoxin was shown to be maximized by the combination of an insertion and L:Trp94 mutation, moving the L 94 side chain closer to digoxin. We also selected mutants that bound preferentially to gitoxin, which, like digitoxin, lacks the 12-hydroxyl, increasing relative binding to gitoxin up to 600-fold compared to the unmutated Ab 26–10.
Keywords: Antibody specificity, bacteriophage display, digoxin, insertional mutagenesis
The antibody combining site for antigen is lined with residues contributed by three complementarity-determining regions (CDRs), or loops, for the heavy chain variable region (HCDR 1–3) and three for the light chain variable region (LCDR 1–3). The variable region CDR loops are engrafted upon a conserved framework composed of antiparallel β-pleated sheets (Padlan 1996). Thus, transplanting CDR loop regions from one species onto the framework regions of a different species can result in proper folding for retention of antibody specificity and affinity of the parental CDR donor (Jones et al. 1986; Riechmann et al. 1988). The antigen specificity of antibodies thus relates largely to the conformation of these loops and relies on the identity and position of their amino acid side chains.
Several mechanisms contribute to the ability of the immune system to recognize a large and diverse population of antigens. Many of the steps in B-cell development that lead to antigen binding site structural diversification involve differences in chain length in the complementarity-determining regions (CDRs) in the Ab variable domains. The heavy chain V regions arise following joining of three distinct gene segments (V, D, and JH). Some VH genes contain insertions in CDR1 and CDR2. The D-gene segments vary dramatically in length in humans (4–28 residues) (Morea et al. 1998; Knappik et al. 2000), thus contributing to chain length diversification in CDR3. The VL regions arise from joining of V and JL genes. Diversity in CDR length may also occur in HCDR3 during gene joining owing to N-region diversification (Alt et al. 1987). Somatic mutation occurring following antigen stimulation includes point mutations, insertions, and deletions (de Wildt et al. 1999), and results in selection of B-cell clones displaying receptors with higher affinity for antigen. The occurrence of insertions (and deletions) in CDR loops due to one of the above mechanisms suggests that CDR loops can tolerate significant changes in chain length and yet undergo efficient folding, assembly, and secretion. Thus, epitopes have been engineered into H-chain CDR2 and CDR3 to mimic antigens; such molecules invoke an immune response to the CDR-inserted epitopes (Billetta et al. 1991; Xiong et al. 1997).
Based on the successful expression of antibody variable domains containing CDR loop insertions, insertional mutagenesis has been used ex vivo to alter antibody binding affinity and specificity. The affinity and specificity of estradiol antibodies was improved by HCDR2 insertions of 2–4 residues (Lamminmäki et al. 1999). The affinity of antiarsonate antibodies was increased by HCDR2 insertional mutagenesis (Parhami-Seren et al. 2002). In both cases it was hypothesized that insertions might result in novel contacts with the hapten or antigen, thereby improving binding.
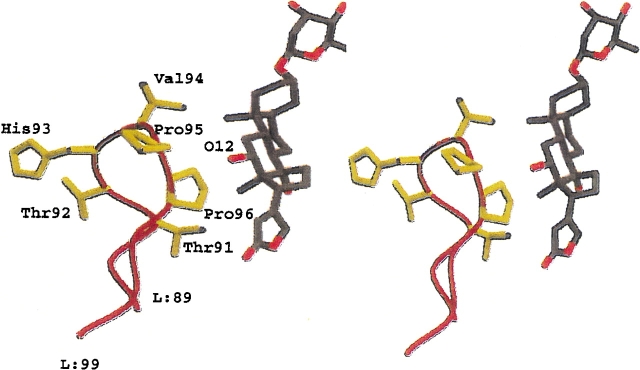
We used as a model the high-affinity antidigoxin antibody 26–10 (Mudgett-Hunter et al. 1982) (Ka = 1.3 × 1010) to insert randomized amino acids in two different CDRs and test selected mutants displayed on bacteriophage for change in specificity. The regions of the combining site chosen for placement of insertions were based on the crystallographically determined structure of the complex between 26–10 Fab and digoxin (Jeffrey et al. 1993). The lactone ring (Fig. 1 ▶) is bound at the base of the binding site cavity. There is extensive shape complementarity between the cardenolide and the Fab; all the interactions are hydrophobic. There are, however, two regions in which the complementarity is imperfect, and there is accessibility to bulk solvent: These are regions around the O12 and O14 of the cardenolide (Fig. 1 ▶). The structure is consistent with binding studies (Schildbach et al. 1991) in which digoxin and digitoxin are bound equally well. These two cardiac glycosides differ only by the presence or absence of the 12-OH (Fig. 1 ▶; Table 1). The region around O12 is surrounded by LCDR3, in which L:Thr91 and L:Pro96 are located at the base of the loop contacting the D ring and lactone (Fig. 2 ▶). The structure of the LCDR3 loop is particularly well suited to insertions, as it is a classic β-hairpin likely to present multiple conformations, with the hapten contact residues 91 and 96 as "anchor points." We therefore randomized amino acid residues in LCDR3 combined with insertions in this region to test the hypothesis that certain mutants would result in improved binding to digitoxin compared to digoxin based on "filling" the cavity in the combining site. Randomized mutant libraries were displayed on bacteriophage and selected by panning on antigen. We also selected mutants based on preferential binding to the C16-OH-substituted analog gitoxin (Fig. 1 ▶; Table 1), which, like digitoxin, lacks a 12-hydroxyl.
Fig. 1.
Structure and numbering of the cardenolide (aglycone/genin ring system). Digoxin (digoxigenin tridigitoxose) coupled to protein was used for immunizations leading to the monoclonal murine Ab 26–10 (Mudgett-Hunter et al. 1982). Structures of selected analogs of digoxin are shown in Table 1.
Table 1.
Structural characteristics of digoxin analogs
| Substitutions at steroid positions | |||
| Analog | 3β | 12β | 16β |
| Digoxin | tridigitoxose | —OH | — |
| Digoxigenin | —OH | —OH | — |
| Digitoxin | tridigitoxose | — | — |
| Gitoxin | tridigitoxose | — | —OH |
| 16-Acetylgitoxin | tridigitoxose | — | —OCOH3 |
Fig. 2.
Stereo view of a portion of the combining site of Fab 26–10 complexed with digoxin (Jeffrey et al. 1993). The LCDR3 loop (residues 91–96, Kabat numbering [Kabat et al. 1991]) is on the left. The LCDR3 backbone is in red; side chains are shown in yellow. The hapten, digoxigenin monodigitoxose, is shown in blue on the right. The lactone ring of the hapten is shown at the bottom, at the base of the binding pocket. The 12-hydroxyl of digoxin is shown explicitly. Digitoxin (digitoxigenin tridigitoxoside) lacks a 12-hydroxyl (Table 1). The images were produced using MOLSCRIPT (Kraulis 1991) and Raster3D (Merritt and Bacon 1997).
Antibody 26–10 is indifferent to the presence (digoxin) or absence (digoxigenin) of the digitoxose moiety at the C3 position (Schildbach et al. 1991), corresponding to only limited interactions between the first digitoxose sugar and the Fab. We proposed that the affinity for digoxin could be increased by lengthening the HCDR2 loop in the region of the mouth of the binding site so as to produce new contacts with the first digitoxose. Such antibodies would thus differentiate between digoxin and digoxigenin. We prepared randomized insertion libraries in HCDR2 displayed on bacteriophage, and selected mutants by panning against digoxin and analogs.
We succeeded in increasing the binding to digitoxin relative to digoxin by LCDR3 insertion mutagenesis, dependent in particular on the identity of the residue at LCDR3 position 94, but we did not find antibodies mutated in the HCDR2 region that recognize the first digitoxose sugar of digoxin.
Results
Light chain insertion library (LCI) on 26–10 SA20 background
This library contained randomized residues at positions LCDR3 92–94, as well as two extra random residues within this area. The parental Fab 26–10-SA20 was previously selected from an HCDR3 randomized library (Krykbaev et al. 2001), and had increased specificity for analogs of digoxin with C16 substitutions. After four rounds of panning on gitoxin–BSA, 24 clones were analyzed by sequencing and competition ELISA. Four clones had different sequences but were the same length in LCDR3 as wt 26–10 (Table 2). Three clones contained the insertion of only a single residue, while a majority, 14 mutants, contained two extra residues as initially devised. Three clones had three-residue insertions. Competition ELISA revealed a >100-fold range in relative binding to gitoxin versus digoxin (cf. IGS38 and IGS22). Several mutants, notably IGS38, IGS40, IGS12, IGS29, etc., demonstrated a significant increase in preferential binding to gitoxin compared to the 26–10-SA20 parent (up to 20 times). There was no correlation between the specificity shift and the length of the insertion. Based on competition assays, the majority of clones had affinities for gitoxin are equal to or a few-fold lower than the parent SA20. The specificity shift occurred consequent to a decrease in affinity for digoxin.
Table 2.
Amino acid sequences and specificities of clones selected by gitoxin–BSA panning from the 26-10 mutant SA20 Fab phage-displayed library (Krykbaev et al., 2001), randomized at L chain CDR3 positions 92–94, combined with the insertion of two extra random codons a
| Light-chain CDR3 sequence | IC50 ratiob | ||||||||||
| Fab | 91 | a | b | c | 92 | 93 | 94 | 95 | 96 | gitoxin/digoxin | digitoxin/digoxin |
| 26-10 | T | T | H | V | P | P | 3.0 | 1.4 | |||
| SA20 | T | T | H | V | P | P | 0.08 | 0.58 | |||
| IGS10c | T | A | E | S | P | P | 0.21 | 1.0 | |||
| IGS11 | T | A | G | A | P | P | 0.16 | ||||
| IGS23 | T | K | T | S | P | P | 0.07 | ||||
| IGS25 | T | V | S | H | P | P | 0.086 | ||||
| IGS33 | T | T | K | I | A | P | P | 0.10 | |||
| IGS35 | T | T | L | S | N | P | P | 0.075 | |||
| IGS40 | T | T | K | H | Y | P | P | 0.015 | |||
| IGS4 | T | K | S | Q | L | G | P | P | 0.03 | 0.17 | |
| IGS5 | T | T | D | M | N | G | P | P | 0.26 | 1.0 | |
| IGS8 | T | A | S | S | L | T | P | P | 0.067 | 0.42 | |
| IGS12 | T | Q | Y | D | D | G | P | P | 0.015 | ||
| IGS21 | T | T | W | L | N | G | P | P | 0.058 | ||
| IGS22 | T | S | S | M | Y | G | P | P | 0.47 | ||
| IGS24 | T | T | Y | E | T | S | P | P | 0.036 | ||
| IGS26 | T | G | V | Q | S | S | P | P | 0.035 | ||
| IGS28 | T | V | T | E | M | G | P | P | 0.033 | ||
| IGS29 | T | G | S | W | P | S | P | P | 0.016 | ||
| IGS30 | T | A | W | V | P | G | P | P | 0.027 | ||
| IGS31 | T | S | S | S | N | G | P | P | 0.063 | ||
| IGS32 | T | S | W | N | A | G | P | P | 0.046 | ||
| IGS36 | T | S | D | L | S | G | P | P | 0.09 | ||
| IGS34 | T | T | S | I | D | T | S | P | P | 0.06 | |
| IGS37 | T | T | T | V | E | D | Q | P | P | 0.08 | |
| IGS38 | T | T | D | F | R | F | W | P | P | 0.004 | |
a The amino acid sequence of the mutant SA20 at HCDR3 positions 94–101 is G G D T T S R A L Q. The corresponding wt 26–10 sequence here is G S S G N K W A M D.
b Values reported are ratios of molar concentration of inhibitor required to give 50% inhibition of Fab binding to analog–BSA-coated wells, relative to the respective molar concentrations of digoxin.
c Clones are designated using a three-letter code: I, library constructed with insertion of two extra random codons; G, panning performed on gitoxin–BSA; S, mutants of 26-10 SA20 HCDR3 mutant (sequence G G D T T S R A L Q H at H94–101). The clones are grouped based on the size of the insertion. Randomized sequence positions are in boldface type.
Among the two-residue inserts, glycine occurred frequently (10 out of 14) in the position N-terminal to L:Pro95. The remaining four clones contained serine (three) or threonine at this position. There was a preponderance (9/20) of threonine or serine at the nominal position 91c among clones with inserts of different lengths. In competition assays comparing the binding to digitoxin and digoxin, clones with improved relative binding to gitoxin also showed improved relative binding to digitoxin (both analogs lack the 12-hydroxyl group of digoxin; see Table 1).
Light chain CDR3 insertion library on wt 26–10 background
The same randomized insertion library used in the 26–10-SA20 background (mutated HCDR3) was produced on the background of wt 26–10. Libraries were panned four times separately on digoxin–BSA and on digitoxin–BSA. Nine clones were sequenced following digoxin–BSA panning. Five of these (IDW1, -3, -11, -16, and -17) lacked an insertion. Four (IDW10, -15, -18, and -19) contained a two-residue insertion (Table 3). Competition ELISA did not reveal any significant differences in binding of these mutants between digoxin and digitoxin. The affinities for both digoxin and digitoxin were two- to eightfold less than that of 26–10. All mutants with inserts had phenylalanine N-terminal to the wt proline 95.
Table 3.
Amino acid sequences and specificities of clones selected by digoxin–BSA or digitoxin–BSA panning from wt 26-10 Fab phage-displayed library, randomized at L chain CDR3 positions 92–94, combined with the insertion of two extra random codons
| Light-chain CDR3 sequence | |||||||||
| Fab | 91 | a | b | 92 | 93 | 94 | 95 | 96 | IC50 ratioa digitoxin/ digoxin |
| 26-10 | T | T | H | V | P | P | 1.4 | ||
| IDW1b | T | K | H | Y | P | P | 0.65 | ||
| IDW3 | T | G | V | H | P | P | 1.25 | ||
| IDW11 | T | T | L | E | P | P | |||
| IDW16 | T | T | D | M | P | P | |||
| IDW17 | T | K | Y | F | P | P | |||
| IDW10 | T | T | P | K | T | F | P | P | 0.6 |
| IDW15 | T | N | P | R | H | F | P | P | 1.1 |
| IDW18 | T | A | P | H | S | F | P | P | 0.9 |
| IDW19 | T | T | P | L | L | F | P | P | 1.4 |
| IDGW7 | T | N | P | R | L | F | P | P | 0.5 |
| IDGW10 | T | M | P | R | Y | F | P | P | 0.4 |
| IDGW12 | T | A | P | V | L | F | P | P | 0.7 |
| IDGW5 | T | R | R | W | P | P | 0.07 | ||
| IDGW15 | T | L | F | W | P | P | 0.2 | ||
| IDGW46 | T | R | S | W | P | P | 0.2 | ||
| IDGW4 | T | N | P | S | R | W | P | P | 0.05 |
| IDGW14 | T | T | P | R | A | W | P | P | 0.06 |
| IDGW37 | T | G | T | G | S | W | P | P | 0.03 |
| IDGW39 | T | D | P | W | H | W | P | P | 0.12 |
| IDGW41 | T | T | P | R | Y | W | P | P | 0.10 |
| IDGW43 | T | N | P | W | T | W | P | P | 0.08 |
| IDGW44 | T | A | D | L | L | W | P | P | 0.09 |
a Values reported are ratios of molar concentration of inhibitor required to give 50% inhibition of Fab binding to analog–BSA-coated wells, relative to the respective molar concentrations of digoxin.
b Clones are designated using letter codes: I, library constructed with insertion of two extra random codons; D, panning performed on digoxin–BSA; DG, panning performed on digitoxin–BSA; W, mutants derived from wt 26-10. Randomized positions are in bold-face type.
Following panning on digitoxin–-BSA, of 13 selected clones, three lacked an insertion, while 10 contained a two-amino acid residue insertion. All clones selected using digitoxin–BSA demonstrated specificity shifted towards digitoxin, two- to threefold (IDGW7, -10, -12) to 20–47-fold (IDGW4, -5, -14, -37, -43, -44). At the position N-terminal to Pro95, only two residues were observed: phenylalanine or tryptophan. Phenylalanine was associated with a modest increase in specificity for digitoxin. However, the presence of tryptophan instead was associated with a much larger specificity shift, particularly in the presence of an insertion (average IC50 = 0.076 versus 1.4 for wt). Specificity shift was observed also in clones with no insertions, but which also contained tryptophan N-terminal to Pro95 (average IC50 = 0.16). Most clones demonstrating preferred specificity favoring digitoxin had affinities for digitoxin approxiately one log lower than 26–10. The affinity for digoxin was decreased even further, resulting in the shifted specificity.
Light chain 5-mer library (LC5) on 26–10-SA20 background
This library was randomized at LCDR3 residues 91 through 94, and also 96, without any insertion included. The library was panned against gitoxin–BSA. Thirty-four digoxin binding clones were sequenced (Table 4). The majority of mutants showed improved relative binding to gitoxin. Some of them (5GS1, -28, -30) showed a further specificity shift to gitoxin of about 1 log compared to the parent SA20. Most clones (e.g., 5GS1, 5GS10, 5GS12, 5GS13, and 5GS28) had affinities for gitoxin equal to the parental SA20 and similar to the affinity of wt 26–10 for digoxin. In all cases, the specificity shift was due to decrease in affinity for digoxin compared to SA20. Clone 5GS38 had affinity for gitoxin reduced by one log compared to SA20, but the digoxin affinity dropped two logs.
Table 4.
Amino acid sequences and specificities of clones selected by gitoxin–BSA panning from the 26-10 mutant SA20aFab phage-displayed library (Krykbaev et al., 2001), randomized at L chain CDR3 positions
| IC50 ratiob | ||||||||
| Light-chain CDR3 sequence | ||||||||
| Fab | 91 | 96 | gitoxin/ digoxin | digitoxin/ digoxin | ||||
| wt | T | T | H | V | P | P | 3.0 | 1.4 |
| SA20 | T | T | H | V | P | P | 0.08 | 0.58 |
| 5GS1c | T | A | W | F | P | I | 0.005 | 0.04 |
| 5GS2 | T | L | E | M | P | V | 0.014 | |
| 5GS3 | T | R | W | G | P | P | 0.05 | |
| 5GS4 | M | T | W | E | P | H | 0.12 | |
| 5GS5 | A | R | V | C | P | T | ||
| 5GS7 | T | L | A | L | P | V | 0.017 | |
| 5GS8 | T | F | N | T | P | A | 0.023 | |
| 5GS10 | T | R | L | F | P | P | 0.01 | |
| 5GS11 | T | R | W | L | P | S | 0.01 | 0.2 |
| 5GS12 | T | R | Y | I | P | V | 0.025 | |
| 5GS13.1d | T | T | N | Y | P | P | 0.02 | |
| 5GS13.2d | T | N | G | L | P | V | 0.024 | |
| 5GS14 | T | R | L | H | P | V | ||
| 5GS16 | H | F | P | E | P | F | ||
| 5GS17 | T | R | A | L | P | I | ||
| 5GS19 | L | L | P | L | P | T | ||
| 5GS20.1d | T | A | Q | L | P | A | 0.022 | |
| 5GS20.2d | T | R | L | L | P | A | 0.022 | |
| 5GS25 | T | L | V | M | P | V | 0.019 | |
| 5GS26 | T | R | H | G | P | P | 0.07 | |
| 5GS27 | S | R | W | L | P | A | 0.023 | |
| 5GS28 | T | L | Q | W | P | P | 0.006 | |
| 5GS30 | S | R | W | L | P | A | 0.028 | |
| 5GS31 | T | R | F | L | P | P | 0.015 | |
| 5GS32 | T | V | F | L | P | V | 0.025 | |
| 5GS33 | T | T | M | G | P | P | 0.09 | |
| 5GS34 | T | R | Y | L | P | S | 0.014 | |
| 5GS35 | C | R | W | L | P | A | 0.21 | |
| 5GS36 | T | A | V | N | P | A | 0.14 | |
| 5GS37 | T | K | W | I | P | V | 0.023 | |
| 5GS38 | T | L | A | T | P | P | 0.008 | |
| 5GS39 | T | K | W | L | P | P | 0.026 | |
| 5GS40 | S | K | W | L | P | A | 0.032 | |
| 5GS41 | T | R | V | L | P | V | ||
| 5GS43 | T | T | S | G | P | P | ||
a Clone SA20 contains the sequence G G D T T S R A L Q at HCDR3 positions H94–101, and was used as a parent clone for this library construction.
b Values reported are ratios of molar concentration of inhibitor required to give 50% inhibition of Fab binding to analog–BSA-coated wells, relative to the respective molar concentrations of digoxin.
c Clones are designated using a three-character code: 5, five randomized residues in L chain sequence; G, panning performed on gitoxin–BSA; S, mutants on G G D T T S R A L Q H94–101 background (SA20).
d 5GS13 and 5GS20 proved to be a mixture of two mutants each, which were subsequently separated, characterized, and designated 5GS13.1, 5GS13.2, and 5GS20.1 and 5GS20.2, respectively.
There was a noticeable preference for the wt residue at position 91 (Table 4). At position 92, most frequent were arginine (14 of 35) and leucine (6/35). Leucine (14 of 34) was commonly observed at position 94; proline (9/34), alanine (7/34), or valine (8/34) at position 96.
Site-directed mutants
Mutants IDGW7, -14, and -15 (see Tables 3 and 5) were chosen as parent sequences to construct site-directed mutants to test the hypothesis that the presence of phenylalanine or tryptophan at the position N-terminal to Pro95 determines the relative specificity for digoxin versus digitoxin of the mutants obtained from insertion libraries on 26–10 (Table 3). In these site-directed mutants, phenylalanine was replaced by tryptophan (IDGW7:F94W), and for mutants IDGW4 and IDGW15 tryptophan was replaced by phenylalanine. As a control, valine in LCDR3 at position 94 was replaced by tryptophan in the wt 26–10. The binding results (Table 5) conformed to the expectation that the residue identity was related to specificity differences due to the presence or absence of the 12-hydroxyl group of the cardiac glycoside. Replacement of wt Val94 by tryptophan resulted in a >10-fold increase in relative binding to digitoxin. The relative binding of clone IDGW7 for digitoxin improved fourfold upon replacement of phenylalanine by tryptophan (IDGW7:F94W). The reverse mutation, W94F in IDGW14, changed the specificity back towards digoxin (40-fold), and the identical mutation in IDGW15 produced a fivefold shift in specificity.
Table 5.
Comparison of the amino acid sequences and specificities of representative mutants from Fab phage-displayed libraries randomized at L chain CDR3 positions 92–94, combined with the insertion of two extra random codons a
| Mutants selected from libraries (see Table 3) | Corresponding site-directed mutantsb | ||||||||||||||||||
| Light-chain CDR3 sequence | Light-chain CDR3 sequence | ||||||||||||||||||
| Fab | 91 | a | b | 92 | 93 | 94 | 95 | 96 | IC50 ratioa digitoxin/ digoxin | Fab | 91 | a | b | 92 | 93 | 94 | 95 | 96 | IC50 ratioc digitoxin/ digoxin |
| 26-10 wt | T | T | H | V | P | P | 3 | 26-10:V94W | T | T | H | W | P | P | 0.25 | ||||
| IDGW7 | T | N | P | R | L | F | P | P | 0.5 | IDGW7:F94W | T | N | P | R | L | W | P | P | 0.12 |
| IDGW12 | T | A | P | V | L | F | P | P | 1 | IDGW12:P91bA | T | A | A | V | L | F | P | P | 1 |
| IDGW14 | T | T | P | R | A | W | P | P | 0.06 | IDGW14:W94F | T | T | P | R | A | F | P | P | 2.25 |
| IDGW15 | T | L | F | W | P | P | 0.18 | IDGW15:W94F | T | L | F | F | P | P | 1 | ||||
a These mutants are among those listed in Table 2, and were obtained following panning on digitoxin–BSA.
b Mutants are defined using the one-letter code by the residue in the parent, followed by the light-chain position number, followed by the resultant mutant residue. The positions for site-directed mutagenesis are given in bold-face type.
c Values reported are ratios of molar concentration of inhibitor required to give 50% inhibition of Fab binding to analog–BSA-coated wells, relative to the respective molar concentrations of digoxin.
Inspection of Table 3 shows that all clones with Phe at position 94 contain Pro at position 92A. To test whether this proline was necessary for the specificity conferred by the presence of Phe94, we constructed a mutant of IDGW12 (Table 5). Replacement of Pro92A by Ala did not change the relative specificity for digitoxin versus digoxin. Therefore, the presence of proline in this subset of mutants cannot be linked to an effect upon specificity.
HCDR2 insertion libraries in wt 26–10
The first library contained randomized insertions of three amino acids between residues H:Ser54 and H:Gly55. The inserted amino acids, as well as residues H:Tyr53, H:Ser54, and H:Gly55, were also randomized. Analyses of selected clones after three rounds of panning showed that the majority of these were wt 26–10. Thus, contamination occurred despite the use of the "nonfunctional" vector pComb3–26–10ALT. Competition ELISA of several randomized clones that were non-wt in sequence showed that none of these had differences in specificity between digoxin and digoxigenin as compared to wt 26–10.
We therefore constructed a second library aimed at eliminating the parental contamination observed for the first library. Three different-length insertions (two, three, and four residues) were introduced into the same region of HCDR2 in a two-step PCR strategy (see Materials and Methods). None of the clones analyzed by sequencing prior to panning were contaminated with the parental 26–10 plasmid. All three insertion lengths were observed, with high sequence variability. After five rounds of panning against digoxin–BGG, 48 clones were tested by ELISA for direct binding to digoxin–BGG and Fab production. The majority of these bound to digoxin–BGG. DNA sequencing of 32 clones demonstrated zero-, one-, three-, and four-residue insertions (Table 6). There was no evidence of wt contamination. The two-residue insertion was not represented. Eight clones with unique sequences in the randomized region had no insertions. There was a preference for Gly at position 55 in the clones without insertions (six of eight). The two clones that had serine at position 55 had a glycine at another HCDR2 position (position 54). Search of the data base using program O (Jones et al. 1991) indicates that most peptide segments, including residues H:52–56, which have a similar conformation to the 26–10 HCDR2, also have Gly at this position. It appears that Gly in the noninsertion mutants is required for proper loop conformation. Analysis of specificity for digoxin versus digoxigenin for all 32 clones indicated that all mutants showed the same relative affinity for digoxin and digoxigenin as the wt 26–10. Because previous studies (Schildbach et al. 1991) had shown that 16-substituted analogs were sensitive to the presence of a sugar moiety at C3, and the insertions in HCDR2 were close to this region, these clones were also tested for the possibility of a specificity shift with respect to the 16-substituted analogs gitoxin and 16-acetylgitoxin. None was observed.
Table 6.
Amino acid sequences of clones selected by panning on digoxin–BGG from a 26-10 Fab insertion libraryarandomized at HCDR2
| HCDR2 residuesb | |||||||
| Fab/mutants | 53 | 54 | a | b | c | d | 55 |
| 26-10 wt | Y | S | G | ||||
| 7, 13, 16, 20, 24, 31, 35, 37, 42, 44, 45 | S | T | G | ||||
| 41, 47 | G | T | G | ||||
| 29, 34 | E | T | G | ||||
| 8, 26 | N | D | G | ||||
| 36 | R | S | G | ||||
| 2, 6, 21, 23 | V | S | G | ||||
| 27 | G | G | S | ||||
| 39 | D | G | S | ||||
| 15 | A | N | P | T | |||
| 28 | G | N | P | T | |||
| 18 | Y | E | P | R | |||
| 14 | S | R | G | H | E | H | |
| 33 | L | T | R | T | T | A | |
| 46 | Y | G | S | F | P | G | |
| 43 | A | T | S | T | P | Q | |
| 30 | A | G | Y | S | R | K | G |
a The HCDR2 insertion library was generated in three separate PCRs performed with oligonucleotides designed to introduce two- and three-residue random amino acid insertions between H:Ser54 and H:Gly55. In addition, Tyr53, Ser54, and Gly55 were included in the randomized segment. The oligonucleotide used to introduce four-residue randomized insertions was designed so that Gly55 was not included in the randomization.
b Wild-type (wt) Fab 26-10 is compared with Fab from mutants selected by panning against digoxin–BGG.
Discussion
We attempted to alter the specificity of an antidigoxin Fab of known three-dimensional structure, based upon inspection of the structure of the complex of digoxin and the 26–10 Fab (Jeffrey et al. 1993). The gap in surface complementarity between digoxin O12 and 26–10 is surrounded by LCDR3, in which L:Thr91 and L:Pro96 are located at the base of the loop, contacting the D ring and lactone (Fig. 2 ▶). The sequence of the LCDR3 loop, including residues 91 through 96, is T T H V P P. Thr91 and Pro96 point into the combining site towards the D ring and lactone of digoxin. The other residues make no contact. Thr92 is partially or mostly buried from solvent. His93 is partially solvent-exposed, while Val94 extends into bulk solvent and is completely solvent-exposed. Pro95 is a cis-proline and packs against H:Asn60. Mutagenesis of L:Val94 at the apex of the LCDR3 loop could increase complementarity by substitution of a bulkier residue to fill the gap. The 12-hydroxyl is not a major determinant of specificity, as Fab 26–10 binds digitoxin, which lacks the 12-hydroxyl group, and digoxin with nearly equal affinity, despite two contacts made between the 12-hydroxyl and L:Pro96. There are no hydrogen bonds made between the 12-hydroxyl and 26–10; thus, the interactions are insufficiently strong to provide a net affinity gain and specifically for O12 (Jeffrey et al. 1993).
To recruit mutants that bind preferentially to digitoxin, we randomized LCDR3 at positions 92–94, combined with a two-residue insertion (Table 3). The shift in specificity towards digitoxin was most pronounced for the seven mutants comprising the lowermost group in Table 2, obtained by panning on digitoxin. The average increase in relative binding to digitoxin was 18.4-fold, with a high of 47-fold.
To further investigate the effects of the residue identity at position 94 on specificity, we produced site-directed mutants at this position in clones with or without LCDR3 insertions, including "forward" (F→W) and "reverse" (W→ F) mutations. The data in Tables 3 and 5, summarizing sequences and binding of LCDR3 insertion mutants obtained by selection using digitoxin–BSA and panning as well as site-directed mutants, indicates that digitoxin binding is favored when Trp is present at position 94 instead of the wt Val. The effect is particularly marked when the substitution is combined with a two-residue insertion. A phenylalanine at position 94 was generally associated with wt-like specificity (Table 2). Inspection of the structures of the 26–10 Fab:digoxin complex (Jeffrey et al. 1993) in the LCDR3 region (Fig. 2 ▶) suggests that the side chain at position 94 packs against VH so that it is immediately adjacent to H:Tyr47 and H:Tyr50, whether the two-residue insertion is present or not. In the wt, Val94 points toward the cardenolide, but is too far from digoxin to make contact.
The mutations L:Val94 to Trp and Phe were modeled into the 26–10 wt structure using Program O (Jones et al. 1991). No modifications were made to the backbone conformation, and the most frequent rotamer was selected for the new side chain. These rotamers did not produce significant clashes with the rest of the 26–10 antigen binding site. In the case of L:Val94→ Trp, the CH2 atom of the Trp side chain lay only 2.3 Å from the O12 of digoxin—much closer than the interatomic contact distance. In the Phe mutant model, the closest approach was 3.5 Å to the O12 atom, an acceptable interatomic contact distance. In both cases the O12 atom was the closest part of digoxin to the L94 side chain. These models suggest that steric hindrance may be the mechanism of recognition of LCDR3 for O12 substituents. The mutagenesis and binding data indicates that binding to digitoxin is maximized when the tryptophan mutation at position 94 is present combined with the insertion. This suggests that the LCDR3 backbone in the mutants containing insertions is altered so as to push the L94 side chain even closer to the hapten. This effect appears to be independent of sequence, although the preponderance of Pro at position 96 among selected mutants suggests that Pro96 might produce the necessary loop conformation.
We hypothesized that antibody specificity for C16-substituted digoxin analogs could be increased further by manipulation of LCDR3. Gitoxin, like digitoxin, lacks the 12-hydroxyl group but contains —OH at position 16 (Fig. 1 ▶). Substitution with a hydroxyl at the 16 position (gitoxin) reduced binding to 26–10 three- to fivefold compared to digoxin (Schildbach et al. 1991). We previously mutagenized HCDR3 of 26–10 to select mutants having improved binding to gitoxin (Krykbaev et al. 2001; Short et al. 2001). The 26–10 mutant SA20 (Krykbaev et al. 2001) binds gitoxin 150-fold more strongly than digoxin, owing to substitutions of the hapten contact residue H:95 (Ser→Gly) and H:100 (Trp→Arg). The mutation at position 100 results in a shift in position of the hapten, allowing more space in the region of the cardenolide C16 due to substitution of the less bulky Arg on the opposite side of the hapten (Short et al. 2001). The addition of the mutation H:Ser95→Gly also allows more space for C16-substituted analogs, owing to removal of the Ser side chain. In the work reported here, we selected mutants of 26–10 SA20 by panning on gitoxin–BSA from libraries randomized at LCDR3, with and without an insertion (Tables 2 and 4). The specificity shifts achieved with the noninsertion LCDR3 library (Table 4) increase relative binding to gitoxin up to 600-fold compared to binding to digoxin, although clear correlations between the sequences obtained and binding were not apparent.
In the case of HCDR2 of 26–10, the closest residue to the digoxin hapten is Val56 (Jeffrey et al. 1993). The HCDR2 loop in this vicinity makes a β-hairpin involving Tyr53 and Ser54. Thus, lengthening the CDR by insertion between 53 and 54 is least likely to radically rearrange the structure. We randomized residues 53–55 inclusive, in addition to inserting two and three residues in this region. H:Tyr53 was included because it packs against the main chain at HCDR1 and may have a role in stabilizing or mediating the HCDR2 conformation. The failure to demonstrate improved binding to digoxin relative to digoxigenin of the mutants might relate to the relatively small number of clones obtained containing two or more residue insertions, thus limiting sampling of the potential conformational space, which might be accessible using a larger library or longer insertions. It is also possible that increasing binding to sugar moieties may require hydrogen bonding for which there are not only distant constraints, but also more precise geometrical requirements than van der Waals contacts. Thus, filling a defined and bounded space at LCDR3 by van der Waals contacts to differentiate O12 binding in a randomized library is likely to be a more frequent event than establishing hydrogen bonding in the case of HCDR2. Moreover, complexes between antibodies and carbohydrates are typically low affinity, thus possibly diminishing the chance of selection of mutants exhibiting the desired additional specificity for the carbohydrate moiety.
Mutations resulting in insertions and deletions in antibody L and H chain regions occur in vivo, and are an additional mechanism for diversification of antibodies in addition to somatic point mutations. It was shown that insertions and deletions occur in functional antibodies expressed as B cell receptors and thus are subject to antigen selection (de Wildt et al. 1999). They occur mainly in CDRs, resulting in changes in the CDR loop main chain conformation that potentially affect antibody affinity and specificity. Engineering CDR loop insertions in vivo provides an additional route to altering antibody binding, particularly where the target for antigen selection of random mutants of phage-displayed antibodies is based on tertiary structures of antibody–antigen complexes.
Materials and methods
Vector construction
To prepare the phagemid vector for cloning of 26–10 LCDR3 libraries, two restriction sites were introduced into the pComb3 vector containing the 26–10 mutant SA20 (Krykbaev et al. 2001) via site-directed mutagenesis: a HindIII site at the 3′ end of the LCDR3 region, and an EcoRV site within LCDR3. HindIII was subsequently used for cloning of PCR fragments containing library sequences. The EcoRV restriction site was introduced for the purpose of eliminating the background presence of parent SA20 sequences among the library clones. Subsequently, after ligation of library-containing DNA fragments into the vector, EcoRV digestion was used to linearize all contaminating parental SA20 sequences, thus reducing their transformation efficiency. In addition, the EcoRV site contained mutations replacing the hapten contact residue LPro96 with Ile, potentially lowering binding affinity to digoxin and its analogs, and further reducing the chances of selecting the parental SA20 sequence after several rounds of panning. The HindIII and EcoRV sites were introduced to create the pComb3–26–10-SA20EH vector by overlap extension mutagenesis (Higuchi et al. 1988). A pair of oligonucleotides, designated LCMC: 5′-CCGTTTGATTTCAAGCTTGGTGCCTCCACCGATATCAGGAACATGTGTAGTTTG-3′ and LCD: 5′-TTGCTAACATACTGCGTAATA-3′, respectively, was used to create a 664-bp PCR fragment, corresponding to positions 1687–2351 of the pComb3–26–10 vector (HindIII and EcoRV sites underlined), and the pair of oligonucleotides LCM: 5′-CAAACTACACATGTTCCTGATATCTTCGGTGGAGGCACCAAGCTTGAAATCAAACGG-3′ and LCR1: 5′-GTGAATTGTAATACGACTCAC TAT-3′ was used to synthesize a 467-bp PCR fragment, corresponding to positions 2284–2751. The 664- and 467-bp fragments were then combined and PCR performed to create a fusion 1064-bp DNA fragment, corresponding to positions 1687–2751 of the pComb3–26–10–1 vector and containing introduced HindIII and EcoRV sites. The 1064-bp DNA fragment was digested with NheI and XbaI and the resulting 928-bp fragment ligated into the pComb3–26–10-SA20, thereby replacing its original sequence.
LCDR3 library construction
Two types of libraries of randomly mutated sequences at 26–10 LCDR3 were constructed by PCR: LC5—5-mer randomized sequences at positions L91 through L94 and L96, and LCI—5 randomized amino acid residues inserted at positions L92–94, including two extra amino acid residues in this region ("insertion library"). To create the LC5 library the following pair of primers was used: LCDA: 5′-GTCATAATGAAATACCTATTGCC-3′ and LCL5: 5′-CCGTTTGATTTCAAGCTTGGTGCCTCCACCGAACGTSNNAGGSNNSNNSNNSNNTTGAGAGCAGAAATA AATTCC-3′. To create the insertion library LCI a pair of primers was used: LCDA, and LCLI: 5′-CCGTTTGATTTCAAGCTTGGTGCCTCCACCGAACGTCGGAGGSNNSNNSNNSNNSNNAGTTTG AGAGCAGAAATAAATTCC-3′. HindIII sites are underlined. LCDA represents the upper strand sequence at positions 1941–1951 of the pComb3–26–10 construct (Barbas III et al. 1991), and LC5 and LCI represent lower strand sequence at positions 2277–2351. N represents an equimolar mixture of all four dNTPs, and S represents an equimolar mixture of dCTP and dGTP. Two combinations of primers were used in a PCR (Taq Polymerase from Perkin-Elmer) with the pComb3–26–10 template. The 410-bp product of this reaction extends from the beginning of the L chain of 26–10 through LCDR3 and the adjacent HindIII site at nucleotide position 2334. This 410-bp fragment was digested with AatII and HindIII, and the resulting 326-bp fragment band was excised from a 2% NuSieve agarose gel (FMC) and purified using a gel extraction kit (Qiagen). The purified fragment was ligated into the AatII/HindIII-digested pComb3–26–10-SA20EH vector, and introduced by electroporation into Escherichia coli XL1 Blue cells. Infection of bacterial cell cultures of each library of LCDR3 mutants with VCSM13 helper phage (XL1-Blue and VCSM13 from Stratagene) generated two libraries of phage with surface-displayed Fab containing a five-amino acid randomized segment (positions L91–94 and 96) in LCDR3 or two random amino acid residues inserted within the randomized L92–94 region. After electroporation, phage were recovered and concentrated by polyethylene glycol/NaCl precipitation from bacterial supernatants. Bacteriophage yield was quantitated by titration on lawns of XL1-Blue bacteria and phagemid quantitated by postinfection XL1-Blue F' colony formation on LB/agar/carbenicillin plates (Sambrook et al. 1989; Barbas III et al. 1991). To create an insertion library on a wt 26–10 HCDR3 background, the total library DNA of the LCI library was purified after electroporation and digested with MfeI and XhoI, which are located in the H-chain region. The large 4733-bp vector fragment containing library sequences was gel purified and ligated with the 562-bp MfeI/XhoI fragment of wt 26–10 sequence, thus replacing the SA20 HCDR3 sequence GGDRASRALQ with the wt HCDR3 sequence GSSGNKWAMD.
HCDR2 insertion library
Two different strategies were followed for the generation of 26–10 HCDR2 insertion libraries. In the first, three randomized codons (NNS)3 were inserted between H:Ser54 and H:Gly55, which were also randomized, together with H:Tyr53. The expression vector pComb3–26–10ALT used for the construction of the initial library is a derivative of pComb3–26–10 (Short et al. 1995) mutated in heavy chain framework 3 using the oligonucleotides 1ALT, 5′-GCCTACATGGAGCTCCGAAGCTTGACATAAGAGGATTCTTGAGTC-3′ and CH1RB, 5′-GGACAGGGATCCAGAGTTCCAGGTCAAGGT-3′, to prevent library contamination from parental 26–10. The mutations introduced a unique HindIII site to linearize and eliminate unwanted self-ligated plasmids, as well as stop codons that replace residues H:Ser84 and H:Ala88 of parental 26–10, rendering the resulting construct inactive. A 393-bp fragment was amplified by PCR using the oligonucleotides CDRH2IFN, 5′-CAGACCCATGGAAAGAGCCTTGATTACATTGGATATATTTCTCCT(NNS)6GTTACTGGCTACAACCAG-3′ and CH1RB. The amplification product was digested with NcoI and BamHI restriction endonucleases (underlined) and a 376-bp fragment was excised from 2% NuSieve agarose gel (FMC) and purified by electroelution. This fragment was ligated into the respective sites of the pComb3–26–10ALT vector used to transform XL-1 Blue electrocompetent cells, followed by infection with VCSM13 helper phage (Stratagene).
For the second library an additional step was added to the randomization strategy. The library was generated from a combination of two, three, and four random amino acid insertions introduced at the same HCDR2 site as described above. This was accomplished in three separate PCR reactions performed with one of the following oligonucleotides: CHRH2INS2, 5′-CAGACCCATGGAAAGAGCCTTGATTACATTGGATATATTTCTCCT (NNS)5 GTTACTGGCTACAACCAG-3′, CDRH2INS3, 5′-CAGACCCATGGAAAGAGCCTTGATTACATTGGATATATTTCTCCT(NNS)6GTTACTGGCTACAACCAG-3′, CDRH2INS4, 5′-CAGACCCATGGAAAGAGCCTTGATTACATTGGATATATTTCTCCT(NNS)6GGTGTTACTGGCTACAACCAG-3′, respectively, and CH1RB on a modified 26–10 template. Note that for the two- and three-residue insertions, position 55 is randomized, while for the four-residue insertion represented by the oligonucleotide CDR2 H2INS4, position 55 is not randomized, being wt Gly (GTT). The modified template was generated in a previous PCR step performed on pComb3–26–10 with the use of the oligonucleotide 2ALT, 5′-CAGAGCCATGGAAAGAGCCTTGATTACATTGGATATATTTCTCCTGATATCTGATAAACTGGCTAC-3′ designed to introduce a unique EcoRV restriction site and two stop codons (underlined). The mismatches were in positions that would be corrected by all three primers carrying the randomized codons at the final PCR step. The PCR-generated fragments were purified, mixed, and digested with NcoI and BamHI followed by ligation to the respective sites of pComb3–26–10ALT. All subsequent manipulations were the same as in the first library.
Panning on cardiac glycoside BSA conjugates
Digoxin, gitoxin, and digitoxin were conjugated to bovine serum albumin (BSA) using a periodate/borohydrate coupling procedure (Short et al. 1995). All samples were adjusted to 1 mg protein/mL in PBSA and further diluted to 10 μg/mL in 0.1 M NaHCO3, pH 8.6, to coat ELISA wells, or 40 μg/mL to coat biopanning wells. In the case of HCDR2 insertion libraries, clones were enriched by successive rounds of selection on digoxin-BGG (5 μg/mL)-coated microtiter plates (Becton Dickinson).
Before panning, libraries were sequenced in the respective CDR regions to estimate their degree of randomization. Enrichment of bacteriophage expressing specifically bound Fab mutants was achieved by four successive rounds of binding to glycoside–BSA-coated microtiter wells (Costar 3690). Washing, elution, reinfection, and growth were done according to the procedure of Barbas and Lerner (1991). Bacterial colonies were isolated and screened for Fab production and antigen binding by ELISA. The DNA sequences of cardiac glycoside ELISA-positive clones were determined using the dideoxy sequencing method.
Fab production
Each colony of mutant phagemid was grown in Superbroth (SB) media containing carbenicillin at 37°C to an OD 600 of 1.0. Isopropyl-B-thiogalactopyranoside (Sigma) was added to the culture and incubation continued overnight at 30°C. Fab were collected from either culture supernatants or periplasmic extracts according to Barbas (Barbas III and Burton 1996). The yield of Fab was determined by ELISA titration (see below) on goat antimouse Fab-coated plates using enzymatically prepared 26–10 Fab of known concentration as a standard. Fab produced by E. coli cells containing phagemid with mutant 26–10 sequences terminate at Arg228 in the hinge region of 26–10 and contain a gene 3-encoded Thr and Ser just 5′ of the stop codon.
ELISA immunoassay
A direct binding ELISA was used for initial screening of binding to congeners of digoxin of clones obtained after panning and removal of gene 3. The assay was performed as previously described (Krykbaev et al. 2001). Wells of 96-well microtiter plates (Falcon) were coated with BSA alone, cardiac glycoside–BSA or goat antimouse Fab (Sigma) in 0.1 M bicarbonate, pH 8.6, and blocked with 3% BSA in TBSA. Bacterial cultures were induced by 1 mM IPTG at logarithmic phase and collected overnight at 30° C with shaking. Bacterial supernatants were collected, added to ELISA plates, and incubated for 2 h at room temperature, followed by a 30-min incubation at room temperature with peroxidase-labeled Fab-specific goat antimouse IgG. ABTS (Amersham/Pharmacia) was used as a substrate for peroxidase. Color development was measured at 405 nM in a Bio-Tek ELISA reader.
Competition ELISA immunoassay
The specificity (relative affinity) of Fab for different cardiac glycosides was determined using a competition ELISA assay in a 96-well format. Wells were coated with either digoxin–BSA or gitoxin–BSA, and contained a constant amount of Fab and serial dilutions in TBSA of cardiac glycosides (10−3 M stock solution in pyridine). This resulted in a range of 10−5–10−12 M concentrations of inhibitor in the final mixture. The amount of Fab used was equal to the amount needed to reach a half maximum binding point when titrated on congener–BSA-coated plates in the absence of competing congener. Mutant Fabs were tested in ELISA with digoxin, digitoxin, 16-substituted congeners, and digoxigenin as competitors. The values reported are ratios of molar concentrations of inhibitor required to give 50% inhibition of Fab binding to congener–BSA-coated wells, relative to the respective molar concentration of digoxin. The ratios of inhibitory concentrations remained the same whether digoxin– or gitoxin–BSA was applied to coat the wells in competition ELISA, provided the appropriate amount of Fab was used as determined by titration on the same congener–BSA-coated plates.
Site-directed mutagenesis of LCDR3 mutants
Variants of clones selected from the LCDR3 insertion library LCI on a wt HCDR3 background (IDGW7:F94W [mutants are denoted by the position number, preceded by the wt or parental residue and followed by the mutated residue] IDGW12:P91bA, IDGW14: W94F, IDGW15:W94F) (Table 4) and the mutant of the wt 26–10 (2610:V94W) were produced in essentially the same way as for the library construction (Winter et al. 1994). The LCDA primer was used in a PCR reaction together with five different primers representing lower DNA strands in the 2282–2344 region that included the HindIII site and contained the desired mutations. The resulting 430-bp PCR product was digested with AatII/HindIII and ligated with the AatII/HindIII digested and gel-purified large fragment of LC5 library DNA (wt 26–10 with a HindIII site introduced 3′ to the LCDR3). Sequences of primers used are listed below:
IDGW7: 5′-GATTTCAAGCTTGGTGCCTCCACCGAACGTCGGAGGCCACAGTCTTGGATTAGTTTGAGAGCAGAAATA 3′; IDGW12: 5′-GATTTCAAGCTTGGTGCCTCCACCGAACGTCGGAGGGAACAATACTGCTGCAGTTTGAGAGCAGAA ATA-3′; IDGW14: 5′-GATTTCAAGCTTGGTGCCTCCACCGAACGTCGGAGGAAATGCTCGTGGTGTAGTTTGAGAGCAGAAATA-3′; IDGW15: 5′-GATTTCAAGCTTGGTGCCTCCACCGAACGTCGGAGGGAAGAACAAAGTTTGAGAGCAGAAATA-3′; 2610: 5′-GATTTAAGCTTGGTGCCTCCACCGAACGTCGGAGGCCAATGTGTAGTTTGAGAGCAGAAATA-3′. HindIII sites are underlined.
Acknowledgments
This work was supported by NIH Grants HL47415 (NHLBI) and CA24432 (NCI) to M.N.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Ab, antibody
mAb, monoclonal antibody
BGG, bovine gamma globulin
BSA, bovine serum albumin
CDR, complementarity-determining region
ELISA, enzyme-linked immunosorbent assay
Fab, antigen-binding fragment of antibody
PBSA, 0.14 M NaCl, 0.0027 M KCl, 0.01 M Na2HPO4, 0.0018 M KH2PO4, pH 7.4, with 0.02% NaN3
PCR, polymerase chain reaction
V, variable region
wt, wild type
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0223402.
References
- Alt, F.W., Blackwell, T.K., and Yancopoulos, G.D. 1987. Development of the primary antibody repertoire. Science 238 1079–1087. [DOI] [PubMed] [Google Scholar]
- Barbas III, C.F. and Burton, D.R. 1996. Selection and evolution of high-affinity human anti-viral antibodies. Trends Biotechnol. 14 230–234. [DOI] [PubMed] [Google Scholar]
- Barbas III, C.F., and Lerner, R.A. 1991. Combinatorial immunoglobulin libraries on the surface of phage (PhAbs): Rapid selection of antigen-specific Fabs. In Methods: A companion to methods in enzymology, Vol. 2 (eds. R.A. Lerner and D.R. Burton), pp. 119–124. Academic Press, Orlando, FL.
- Barbas III, C.F., Kang, A.S., Lerner, R.A., and Benkovic, S.J. 1991. Assembly of combinatory antibody libraries on phage surfaces: The gene III site. Proc. Natl. Acad. Sci. 88 7978–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billetta, R., Hollingdale, M.R., and Zanetti, M. 1991. Immunogenicity of an engineered internal image antibody. Proc. Natl. Acad. Sci. 88 4713–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wildt, R.M.T., van Venrooij, W.J., Winter, G., Hoet, R.M.A., and Tomlinson, I.M. 1999. Somatic insertions and deletions shape the human antibody repertoire. J. Mol. Biol. 294 701–710. [DOI] [PubMed] [Google Scholar]
- Higuchi, R., Krummel, B., and Saiki, R.K. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 16 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey, P.D., Strong, R.K., Sieker, L.C., Chang, C.Y., Campbell, R.L., Petsko, G.A., Haber, E., Margolies, M.N., and Sheriff, S. 1993. 26–10 Fab:digoxin complex—Affinity and specificity due to shape complementarity. Proc. Natl. Acad. Sci. 90 10310–10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P.T., Dear, P.H., Foote, J., Neuberger, M.S., and Winter, G. 1986. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321 522–525. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47 110–119. [DOI] [PubMed] [Google Scholar]
- Kabat, E.A., Reed-Miller, M., Perry, H.M., and Gottesman, K.S. 1991. Sequences of proteins of immunological interest, 5th ed. Department of Health and Human Services, Public Health Service, National Institutes of Health, Bethesda, MD.
- Knappik, A., Ge, L., Honegger, A., Pack, P., Fischer, M., Wellnhofer, G., Hoes, A., Wölle, J., Plückthun, A., and Virnekäs, B. 2000. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 290 57–85. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Krykbaev, R.A., Liu, W.R., Jeffrey, P.D., and Margolies, M.N. 2001. Phage display-selected sequences of the heavy-chain CDR3 loop of the anti-digoxin antibody 26–10 define a high affinity binding site for position 16-substituted analogs of digoxin. J. Biol. Chem. 276 8149–8158. [DOI] [PubMed] [Google Scholar]
- Lamminmäki, U., Paupérioj, S., Westerlund-Karlsson, A., Karvinen, J., Virtanen, P.L., Lövgren, T., and Saviranta, P. 1999. Expanding the conformational diversity by random insertions to CDRH2 results in improved anti-estradiol antibodies. J. Mol. Biol. 291 589–602. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Bacon, D.J. 1997. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277 505–524. [DOI] [PubMed] [Google Scholar]
- Morea, V., Tramontano, A., Rustici, M., Chothia, C., and Lesk, A.M. 1998. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 275 269–294. [DOI] [PubMed] [Google Scholar]
- Mudgett-Hunter, M., Margolies, M.N., Ju, A., and Haber, E. 1982. High-affinity monoclonal antibodies to the cardiac glycoside, digoxin. J. Immunol 129 1165–1172. [PubMed] [Google Scholar]
- Padlan, E.A. 1996. X-ray crystallography of antibodies. Adv. Protein Chem. 49 57–133. [DOI] [PubMed] [Google Scholar]
- Parhami-Seren, B., Viswanathan, M., and Margolies, M.N. 2002. Selection of high affinity p-azophenylarsonate Fabs from heavy-chain CDR2 insertion libraries. J. Immunol. Methods 259 43–53. [DOI] [PubMed] [Google Scholar]
- Riechmann, L., Clark, M., Waldmann, H., and Winter, G. 1988. Reshaping human antibodies for therapy. Nature 332 323–327. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schildbach, J.F., Panka, D.J., Parks, D.R., Jager, G.C., Novotny, J., Herzenberg, L.A., Mudgett-Hunter, M., Bruccoleri, R.E., Haber, E., and Margolies, M.N. 1991. Altered hapten recognition by two anti-digoxin hybridoma variants due to variable region point mutations. J. Biol. Chem. 266 4640–4647. [PubMed] [Google Scholar]
- Short, M.K., Jeffrey, P.D., Kwong, R.F., and Margolies, M.N. 1995. Contribution of antibody heavy chain CDR1 to digoxin binding analyzed by random mutagenesis of phage-displayed 26–10. J. Biol. Chem. 170 28541–28550. [DOI] [PubMed] [Google Scholar]
- Short, M.K., Jeffrey, P.D., Demirjian, A., and Margolies, M.N. 2001. A single HCDR3 residue in the anti-digoxin antibody 26–10 modulates specificity for 16-substituted digoxin analogs. Protein Eng. 14 287–296. [DOI] [PubMed] [Google Scholar]
- Winter, G., Griffiths, A.D., Hawkins, R.E., and Hoogenboom, H.R. 1994. Making antibodies by phage display technology. Annu. Rev. Immunol. 12 433–455. [DOI] [PubMed] [Google Scholar]
- Xiong, S., Gerloni, M., and Zanetti, M. 1997. Engineering vaccines with heterologous B and T cell epitopes using immunoglobulin genes. Nat. Biotechnol. 15 882–886. [DOI] [PubMed] [Google Scholar]