Abstract
H-N-H is a motif found in the nuclease domain of a subfamily of bacteria toxins, including colicin E7, that are capable of cleaving DNA nonspecifically. This H-N-H motif has also been identified in a subfamily of homing endonucleases, which cleave DNA site specifically. To better understand the role of metal ions in the H-N-H motif during DNA hydrolysis, we crystallized the nuclease domain of colicin E7 (nuclease-ColE7) in complex with its inhibitor Im7 in two different crystal forms, and we resolved the structures of EDTA-treated, Zn2+-bound and Mn2+-bound complexes in the presence of phosphate ions at resolutions of 2.6 Å to 2.0 Å. This study offers the first determination of the structure of a metal-free and substrate-free enzyme in the H-N-H family. The H-N-H motif contains two antiparallel β-strands linked to a C-terminal α-helix, with a divalent metal ion located in the center. Here we show that the metal-binding sites in the center of the H-N-H motif, for the EDTA-treated and Mg2+-soaked complex crystals, were occupied by water molecules, indicating that an alkaline earth metal ion does not reside in the same position as a transition metal ion in the H-N-H motif. However, a Zn2+ or Mn2+ ions were observed in the center of the H-N-H motif in cases of Zn2+ or Mn2+-soaked crystals, as confirmed in anomalous difference maps. A phosphate ion was found to bridge between the divalent transition metal ion and His545. Based on these structures and structural comparisons with other nucleases, we suggest a functional role for the divalent transition metal ion in the H-N-H motif in stabilizing the phosphoanion in the transition state during hydrolysis.
Keywords: Metal binding in proteins, divalent metal ions, magnesium ion, zinc ion, endonuclease, DNase, DNA hydrolysis mechanism
The conserved sequence of the H-N-H motif was first recognized by the sequence similarity in several intron-encoded homing endonucleases and bacteria toxins (Gorbalenya 1994; Shub et al. 1994). The numbers of proteins discovered containing the H-N-H motif has increased substantially in the past several years; more than one hundred proteins are listed in databases [information found at the Pfam Protein Families database (Bateman et al. 2002)]. The biggest subgroups in the H-N-H family are group I and group II homing endonucleases, which initialize the process of transferring a mobile intervening sequence into a homologous allele that lacks the sequence (Lambowitz and Belfort 1993; Belfort and Perlman 1995; Edgell et al. 2000; Chevalier and Stoddard 2001). These homing endonucleases recognize and cleave DNA site-specifically at target alleles. Amino acid sequences in the H-N-H motif of several examples of homing endonucleases are listed in Figure 1 ▶. They include the group I homing endonucleases I-HmuI (Goodrich-Blair et al. 1990; Goodrich-Blair 1994; Goodrich-Blair and Shub 1996), I-HmuII (Goodrich-Blair 1994; Goodrich-Blair and Shub 1996), I-HmuIII (Goodrich-Blair 1994), I-TevIII (Eddy and Gold 1991), yosQ (Lazarevic et al. 1998), and ORF253 (Foley et al. 2000); and the group II homing endonucleases Avi (Ferat and Michel 1993), Cpc (Ferat and Michel 1993), and PetD (Kuck 1989).
Fig. 1.
Sequence alignment of several H-N-H family proteins in the H-N-H motif region. The dots (.) represent gaps, and numbers represent the insertion of amino acids. The first row shows the consensus sequence of the H-N-H motif with the most conserved Asn and His residues displayed in bold with underline. The H-N-H proteins are classified into four subgroups: (1) bacterial toxins, including colicins and pyocins; (2) Group I homing endonucleases, including I-HmuI (Goodrich-Blair et al. 1990; Goodrich-Blair 1994; Goodrich-Blair and Shub 1996), I-HmuII (Goodrich-Blair 1994; Goodrich-Blair and Shub 1996), I-HmuIII (Goodrich-Blair 1994), yosQ (Lazarevic et al. 1998), I-TevIII (Eddy and Gold 1991), and ORF253 (Foley et al. 2000); (3) Group II homing endonucleases, including Avi (Ferat and Michel 1993), Cpc (Ferat and Michel 1993), and PetD (Kuck 1989); (4) restriction or repair enzymes, including McrA (Hiom and Sedgwick 1991) and S. g. mtMSH (Pont-Kingdon et al. 1995).
The second biggest subgroup of proteins in the H-N-H family are bacteria toxins, such as Escherichia coli colicins: ColE2 (Schaller and Nomura 1976), ColE7 (Chak et al. 1991), ColE8 (Toba et al. 1988), ColE9 (Wallis et al. 1992); and Pseudomonas aeruginosa pyocins S1 and S2 (Sano and Kageyama 1993; Sano et al. 1993). All of these toxins share a highly homologous C-terminal nuclease domain capable of hydrolyzing DNA nonspecifically in target cells. The H-N-H motifs in the nuclease domains of these toxins are more conserved than the ones in homing endonucleases in that they all contain two pairs of histidines separated by an asparagine residue (HHX14NX8HX3H). The crystal structures of the nuclease domains of ColE7 and ColE9 in complex with their inhibitors Im7 or Im9 are the only two structures that have been reported thus far for the H-N-H family proteins (Kleanthous et al. 1999; Ko et al. 1999).
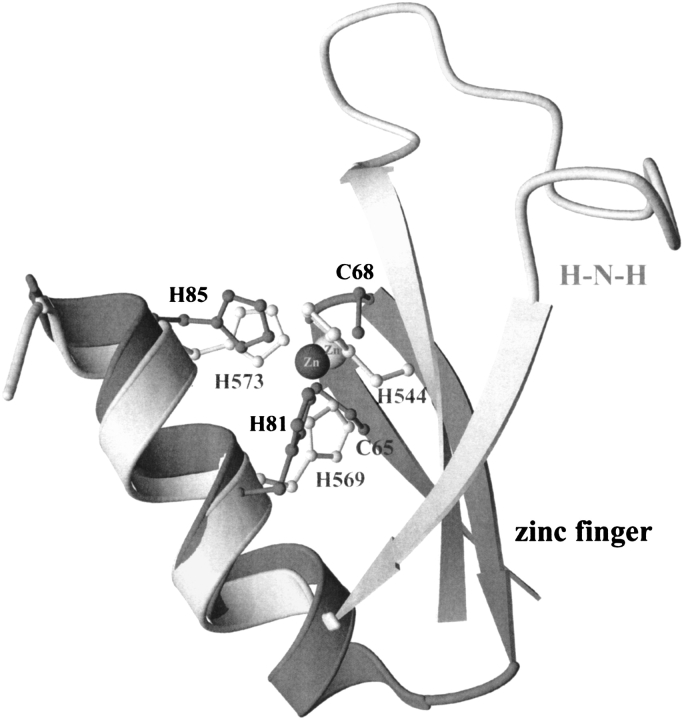
The topology and the coordination of the metal ion in the H-N-H motif share several features similar to those of classic zinc finger motifs (Fig. 2 ▶). Firstly, both of the structural motifs contain an antiparallel-stranded β-sheet linked to a C-terminal α-helix with a zinc ion situated in the center. Secondly, both motifs have two zinc-coordinated histidines separated by three residues, that is, one helical turn, located in the C-terminal α-helix. The major differences in the two motifs are: (1) the relative orientation of β-strands to α-helix; (2) a shorter α-helix and a much longer loop between the two β-strands in the H-N-H motif; and (3) a water molecule instead of a protein residue as the fourth ligand of the zinc ion in the H-N-H motif.
Fig. 2.
Structural comparison of H-N-H and zinc finger motifs. The ribbon model of the H-N-H motif is lightly shaded, and that of zinc-finger is heavily shaded. The zinc ion in H-N-H motif represented as a darker sphere is coordinated to H544, H569, and H573 in ColE7; the zinc ion in zinc finger motif is coordinated to C65, C68, H81, and H85 in Zif268 (Pavletich and Pabo 1991). The topology of two antiparallel β-strands followed by an α-helix with a zinc ion located in the center is analogous in both motifs. However, the relative orientation of β-strands to the α-helix and the residues bound to the zinc ion are dissimilar; that is, Cys-Cys-His-His in the zinc finger and His-His-His-water in the H-N-H.
The zinc ion in the classic zinc finger has been functionally defined in that it plays a structural role in stabilizing the folded conformation of the protein, required for interactions with nucleic acids (Berg and Godwin 1997; Alberts et al. 1998). However, the role of the zinc ion in the H-N-H motif is still a matter of discussion. The endonuclease activity of ColE7 or ColE9 is activated not only by transitional metal ions, such as Ni2+, Mn2+, and Co2+, but also by alkaline earth metal ions, Mg2+, Ca2+, and Sr2+ (Pommer et al. 1999, 2001; Ku et al. 2002). Spectroscopic, proteolytic, and calorimetric data have shown that equal-molar binding of Zn2+, Ni2+, or Co2+ to the H-N-H motif of ColE9 increases the thermal stability of the protein, and that Zn2+ has a much higher affinity (dissociation constant ∼10−9 M) in comparison to the other transition metal ions (Pommer et al. 1999; Keeble et al. 2002). These results suggest that H-N-H colicin proteins are zinc-dependent enzymes in vivo, and that zinc plays a structural role in stabilizing the enzyme (Pommer et al. 1999). However, structural comparison (Friedhoff et al. 1999b; Miller et al. 1999; Grishin 2001) has revealed that the H-N-H motif has a ββα-Metal fold similar to the active sites of several other nucleases, including the homing endonuclease I-PpoI, Serratia nuclease, and Phage T4 Endonuclease VII (Friedhoff et al. 1999a; Kuhlmann et al. 1999; Raaijmakers et al. 1999). The divalent metal ions in these enzymes have been suggested to play catalytic roles (Lunin et al. 1997; Galburt et al. 1999; Miller et al. 1999), implying that the zinc ion in the H-N-H motif could be involved in DNA hydrolysis. It has been shown that the nuclease domain of ColE9 is inactive in the presence of Zn2+ (Pommer et al. 1999; Keeble et al. 2002); however, our biochemical assays showed that the nuclease domain of ColE7 (referred to as nuclease-ColE7 hereafter) is an active endonuclease in the presence of low concentrations of Zn2+ (Ku et al. 2002). Moreover, the Zn2+-depleted apo-enzyme cannot hydrolyze DNA, although it still binds DNA, indicating that the zinc ion not only plays a structural role but is also involved in catalysis.
To further clarify the role of metal ions in the H-N-H motif, we report here the crystal structures of nuclease-ColE7/Im7 in complex with different divalent metal and phosphate ions in two different crystal forms. The divalent metal ions, originally located in the complex, were removed by ETDA, and afterwards different metal ions, including Zn2+, Mn2+, and Mg2+ were soaked into the complex crystals. The metal-binding sites for Zn2+ and Mn2+ were confirmed in anomalous difference maps. The crystal structures demonstrate unambiguously the transition metal ion binding site and phosphate binding site and suggest possible roles of the transition metal ions in the H-N-H motif.
Results
Nuclease activity of DNase-ColE7 with different metal ions
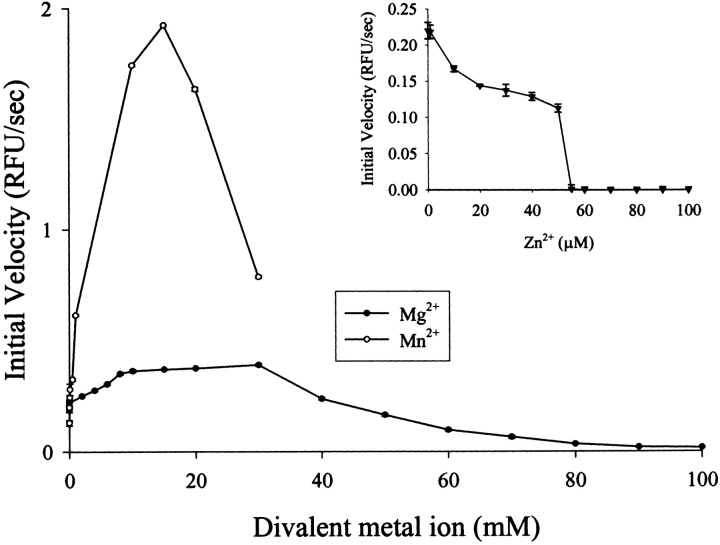
The endonuclease activity of nuclease-ColE7 was measured in the presence of different divalent metal ions by the fluorescent method using 30-bp oligonucleotides covalently bound with fluorescent dye and quencher as substrates (Kelemen et al. 1999; Trubetskoy et al. 2002). The degradation of the fluorophore-labeled oligonucleotides by nuclease-ColE7 released fluorescent emissions monitored by a spectrofluorometer. Figure 3 ▶ shows that the addition of Zn2+ ions to the zinc-bound holoenzyme inhibited DNase activity; however, the addition of Mg2+ in mM range enhanced the activity (Ku et al. 2002). In the present study, we found that the zinc-holoenzyme was further activated by Mn2+ in mM range.
Fig. 3.
Measurement of the DNase activity of nuclease-ColE7 with different metal ions by a fluorescent method using the fluorophore- and quencher- labeled oligonucleotide as a substrate. The zinc-holoenzyme (60 nM) was further activated in the presence of Mn2+ or Mg2+ (mM), but the DNase activity was inhibited when zinc ions were added (μM; Ku et al. 2002).
Structure determination
In order to find out the metal binding sites in the H-N-H colicins, we determined the crystal structures of the nuclease-ColE7/Im7 bound with different metal ions. Nuclease-ColE7 and Im7 were overexpressed by two different vectors with a histidine tag attached at either the N-terminus of nuclease-ColE7 or the C-terminus of Im7. The two complexes of nuclease-ColE7/Im7 were crystallized in unit cells with similar cell dimensions but different space groups. The N-terminal His-tagged complex was crystallized in orthorhombic I222, and the C-terminal His-tagged complex was crystallized in orthorhombic P21212. Both types of crystal were soaked in EDTA buffers to remove divalent metal ions originally associated with the proteins. The EDTA-treated crystals were then transferred to phosphate buffers containing different metal ions. Diffraction data of the I222 and P21212 crystals were collected at the absorption edge of the soaking metal ion using synchrotron radiation or at 1.54 Å using Cu Kα radiation from a rotating anode (Table 1). Crystals soaked in solutions containing EDTA, Mn2+, Zn2+, or Mg2+ diffracted from 2.6 to 2.0 Å resolution at low temperature (∼100 K). The Mg2+-binding site was not found in the Mg2+-soaked crystals, and thus the diffraction statistics for Mg2+-soaked crystals were not included in Table 1.
Table 1.
X-ray diffraction statistics for the nuclease-ColE7/Im7 complex
| EDTA | Zn-bound | Zn-bound | Mn-bound | Mn-bound | |
| Data collection and processing | |||||
| Wavelength Å | 1.54 | 1.54 | 1.28004 | 1.89261 | 1.89261 |
| Space group | I222 | I222 | P21212 | I222 | P21212 |
| Cell dimensions Å | a = 63.07 | a = 62.94 | a = 119.73 | a = 62.71 | a = 119.48 |
| b = 74.80 | b = 75.25 | b = 62.41 | b = 74.57 | b = 62.55 | |
| c = 119.05 | c = 118.60 | c = 74.14 | c = 119.64 | c = 74.30 | |
| Resolution Å | 2.6 | 2.0 | 2.0 | 2.3 | 2.2 |
| aRmerge-all data (%) | 7.2 | 6.9 | 6.1 | 9.7 | 5.2 |
| Rmerge-last shell (%) | 25.9 (2.69-2.60) | 33.2 (2.07-2.00) | 27.8 (2.07-2.00) | 29.5 (2.38-2.30) | 9.4 (2.28-2.20) |
| Refinement | |||||
| Resolution range Å | 40.0-2.6 | 40.0-2.0 | 40.0-2.0 | 40.0-2.3 | 40.0-2.0 |
| Non-hydrogen atoms | |||||
| Protein | 1676 | 1687 | 3508 | 1679 | 3503 |
| Solvent molecules | 140 | 225 | 415 | 161 | 232 |
| R-factor (%) | 20.8 | 19.6 | 19.1 | 21.1 | 21.2 |
| bRfree (%) | 29.0 | 25.0 | 24.1 | 28.3 | 26.0 |
| Model quality | |||||
| r.m.s. deviations in bond lengths Å | 0.013 | 0.015 | 0.016 | 0.015 | 0.016 |
| Bond angles (deg) | 1.38 | 1.63 | 1.66 | 1.49 | 1.70 |
| Ramachandran plot (%) | |||||
| Most favored | 88.3 | 93.3 | 91.7 | 92.1 | 89.6 |
| Additionally allowed | 11.2 | 6.7 | 7.5 | 7.3 | 9.3 |
| Generously allowed | 0.6 | 0.0 | 0.8 | 0.6 | 1.1 |
| Disallowed | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
a Rmerge = ∑h∑i | Ih,i − ≤Ih> | /∑h∑iIh,I, where ≤Ih> is the mean intensity of the I observation for a given reflection h.
b Rfree were calculated from 10% reflections of the data set.
The I222 crystals were isomorphous to the previously determined nuclease-ColE7/Im7 (PDB entry: 7cei), which was therefore used as an initial model for refinement. The structure of the P21212 crystals was solved by molecular replacement using the I222 structure (PDB entry: 7cei) as the searching model. The R-factors and the geometric statistics for the final models were within good ranges as listed in Table 1. In the P21212 unit cell, there are two complex molecules per asymmetric unit, and they are separated by (0.49, 0.52, 0.50) of unit cell dimensions, but not exactly by (0.5, 0.5, 0.5). Therefore the space group was shifted from the I-center I222 to the primitive P21212; nevertheless, the protein molecules in the two types of crystals packed in a similar way. The average root mean square (rms) difference between the two complex molecules in the asymmetric unit of P21212 cell is only 0.3 Å (for main-chain atoms), and therefore, for clarity only the structure of the first molecule will be used in the following structural comparison.
Comparison between I222 and P21212 structures
The overall folds of the complex in the two crystal forms are identical (see the ribbon model in Figure 4 ▶). The His-tags were not observed in either I222 or P21212 structures. Im7 contains four α-helices folded in a varied four-helix-bundle structure nearly identical to the previously determined free-form Im7 (Chak et al. 1996). Nuclease-ColE7 has a mixed α/β structure with a metal ion bound in a cleft on the concaved surface. The H-N-H motif (displayed in green in Figure 4 ▶) is located at the C-terminus of nuclease-ColE7 containing two β-strands (βd and βe) and one α-helix (α5) with a metal ion situated in the center of the motif.
Fig. 4.
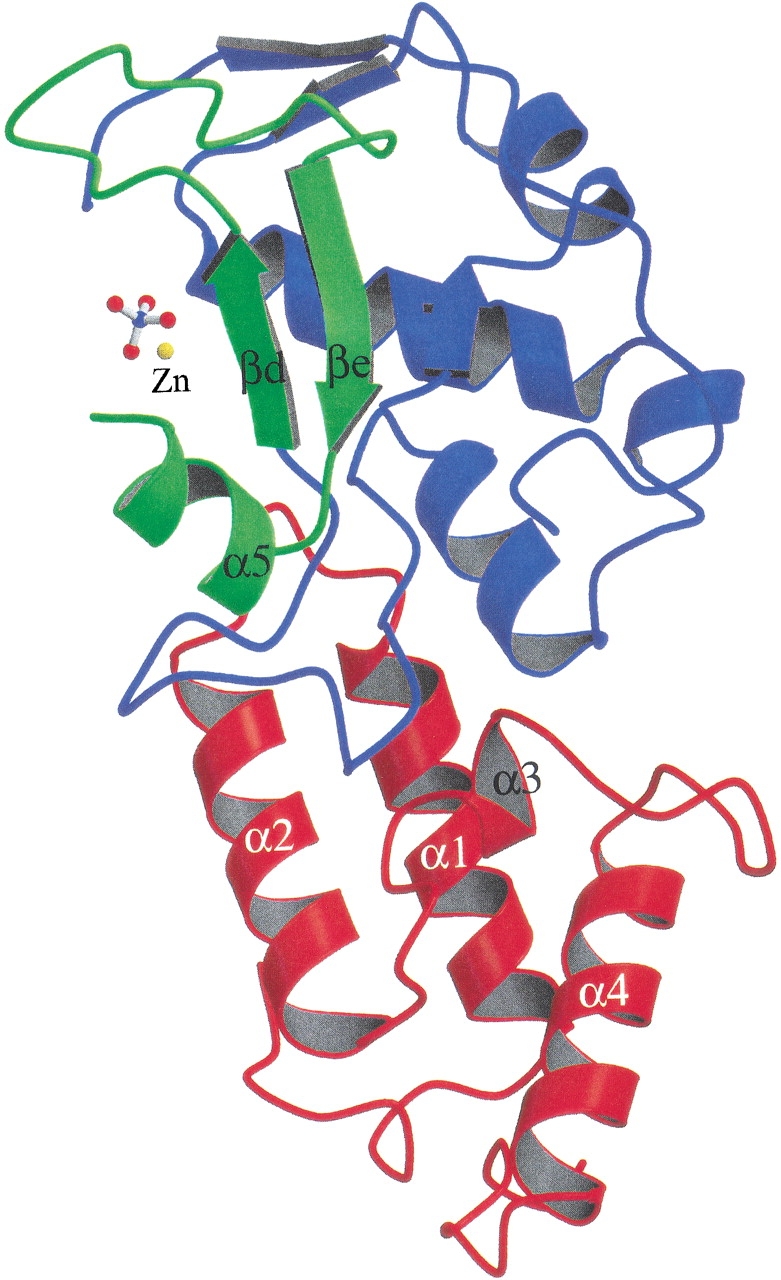
Ribbon model of nuclease-ColE7/Im7 bound with a Zn2+ ion and a phosphate. The four α-helices in Im7 are displayed in red and the nuclease-ColE7 is shown in blue, with only the H-N-H motif in green.
The P21212 structures on average had lower temperature factors than those of I222 structures. For example, in the Zn-bound P21212 structure, the average B-factors were 30.5 Å2 for nuclease-ColE7 and 23.8 Å2 for Im7, compared to the higher values of 42.7 Å2 for nuclease-ColE7 and 29.0 Å2 for Im7 in the Zn-bound I222 structure. In the structures of nuclease-ColE7, the N-terminal end (residues 448–460), the surface loop (residues 486–490), and the long loop between the two β-strands in the H-N-H motif (residues 446–560) had higher B-factors (>40 Å2) in both crystal forms, indicating that these regions were more flexible. Superimposition of only the structures of the nuclease-ColE7 in P21212 and I222 gave an average rms difference of 0.30 Å for the main-chain atoms. However, a larger shift was observed in the long loop region (residues 550–554) within the H-N-H motif with a maximum rms difference of 4.7 Å at residue 552. This region corresponded to the residues with higher temperature factors; therefore, excluding the flexible surface loops, the two structures are almost identical.
Metal ions and phosphate binding in H-N-H motif
The complex crystals were first soaked in EDTA to remove the associated divalent metal ions. X-ray absorption spectra were recorded for the crystals before and after EDTA soaking. The absorption at zinc edge was observed before EDTA soaking and disappeared after ETDA soaking (data not shown). The previously solved complex structure with the zinc ion deleted was used as the initial model for refinement of the EDTA-treated crystal (Ko et al. 1999). A Fourier map (Fig. 5A ▶) revealed the absence of a strong peak in the center of the H-N-H motif, but instead showed two water molecules located in the original metal-binding sites. One water molecule was hydrogen-bonded to His545 (ND1). The second water molecule was hydrogen-bonded to the first water molecule and located approximately at the original metal-binding site. The electron densities of the two histidine residues, His573 and His544, were ill-defined, indicating that the side-chain conformations of the two residues were not fixed by metal ions. This result demonstrated that the divalent metal ions initially associated with the complex had been removed by EDTA.
Fig. 5.
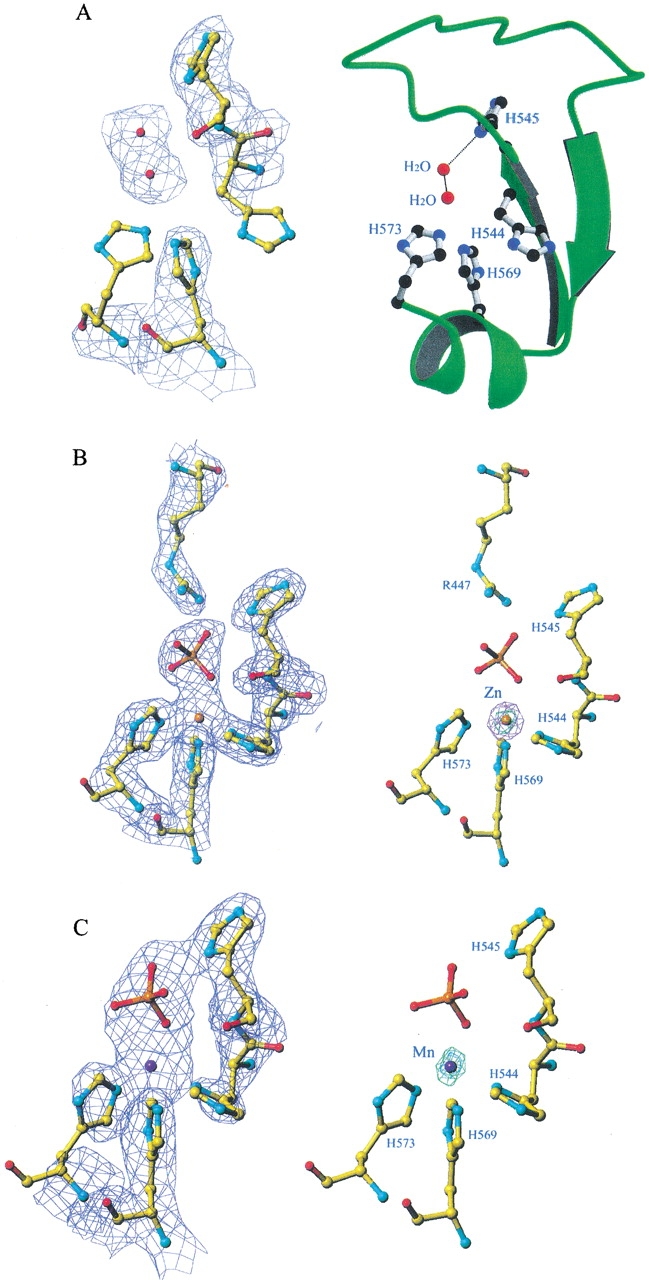
Electron density maps and structural models for the EDTA-treated and metal-ion-bound nuclease-ColE7 at the center of the H-N-H motif. (A) The Fourier map (2Fo-Fc) of the EDTA-treated nuclease-ColE7 (left panel) shows that the zinc ion originally associated with nuclease-ColE7 was removed by EDTA. Two residues, H573 and H544, have ill-defined electron density. The metal-binding site was substituted by a water molecule in the center of the H-N-H motif. The ribbon model of the H-N-H motif with several histidine side chains and water molecules is displayed in the right panel. (B) The Fourier and anomalous difference maps for the Zn2+-bound nuclease-ColE7. The Fourier map (2Fo-Fc) of the P21212 Zn-bound structure (left panel) was calculated at 2.0 Å resolution and contoured at 1σ. The anomalous difference map shown in the same orientation is displayed in the right panel and was contoured at 30 σ (green) and 15 σ (purple), respectively. (C) The Fourier (left) and anomalous difference (right) maps of the I222 Mn-bound structure were calculated at 2.3 Å resolution and contoured at 1σ. The anomalous difference map was contoured at 17 σ (green).
The EDTA-treated crystals were then soaked in phosphate buffers containing Zn2+, Mn2+, or Mg2+ ions. The X-ray absorption spectra were recorded first to find out the absorption edges of Zn2+ and Mn2+ ions in crystals. Diffraction data were collected at the absorption edge of the metal ion correspondingly, to generate the largest anomalous differences between Friedel’s pairs. In Figure 5 ▶, B and C show the Fourier and anomalous difference maps of Zn2+-bound (in P21212) and Mn2+-bound (in I222) structures at the center of the H-N-H motif. The electron densities of a metal ion and a phosphate ion were observed in the Fourier (2Fo-Fc) maps. The locations of metal-binding sites were further confirmed unambiguously by the anomalous difference maps in which Zn2+ has a strong peak, with a peak height of 30 σ, and Mn2+ also has a strong peak, with a peak height of 17 σ. No other metal ion binding sites were identified in the electron density maps for the Zn2+- and Mn2+-bound crystals. This result demonstrated that the transition metal ions of Zn2+ and Mn2+ bind at the same site in the H-N-H motif, coordinating to the three histidine residues (His544, His569, and His573) and one phosphate ion.
Discussion
The roles of divalent metal ions in the H-N-H motif in DNA hydrolysis
Divalent metal ions play important roles in DNA hydrolysis. The most widely known examples are probably the requirement of Mg2+ for restriction enzymes in cleavage of DNA substrates (Pingoud and Jeltsch 1997). Detailed structural analysis of type II restriction endonucleases show that they catalyze the DNA cleavage via similar two-metal-ion or three-metal-ion catalysis pathways (Kovall and Matthews 1999). Mg2+ ions play important catalytic roles in DNA hydrolysis catalyzed by restriction enzymes, and they may function as: (1) general bases to activate the nucleophilic water molecules for an in-line attack of the scissile phosphate; (2) Lewis acids to stabilize the negative-charged pentavalent phosphorous transition state; or (3) general acids that the Mg2+-bound water donating a proton to the hydroxyl leaving group. As have Mg2+ ions, divalent transition metal ions have been identified in the active sites of endonucleases, such as Zn2+ ions in P1 nuclease from Penicillium citrinum (Romier et al. 1998) and Endonuclease IV from Escherichia coli (Hosfield et al. 1999). In these zinc-dependent endonucleases, a three-metal-ion catalysis pathway was proposed with zinc ions functioning either as general bases activating nucleophilic water molecules or stabilizing scissile phosphate/hydroxyl-leaving groups.
In our previous study, we found that a water molecule was bound to the zinc ion in the H-N-H motif of nuclease-ColE7 (Ko et al. 1999). In the present study, we show that a phosphate ion replaces the water molecule and forms a bridge between the zinc ion and a histidine residue. The Mn2+ ion binds at the same site and coordinates to the same residues as that of zinc ion. A similar result was reported for Ni2+-bound nuclease-ColE9/Im9 in which a phosphate ion is also bridged between the Ni2+ ion and a histidine residue (Kleanthous et al. 1999). However, we did not find any Mg2+ binding sites in either apo-enzyme or zinc-holoenzyme crystals soaked with Mg2+ ions. The original metal-binding site in the H-N-H motif of the apo-enzyme soaked with Mg2+ ions was occupied with water molecules similar to that of EDTA-treated crystals (data not shown), suggesting that the Mg2+ ion likely does not bind at the center of the H-N-H motif in ColE7. This result was expected, since a magnesium ion does not favor the binding site of a transition metal ion coordinated with three histidine residues (Dudev and Lim 2001).
A previous structural comparison shows that the zinc ion in the H-N-H motif matches well with the Mg2+ ion in active sites of I-PpoI and Serratia nuclease (Kuhlmann et al. 1999). The Mg2+ in I-PpoI and Serratia nuclease are involved in stabilization of the phosphoanion transition state and in providing a proton to the 3′ oxygen-leaving group. Therefore we suspect that the zinc ion located in the H-N-H motif has a similar function for the stabilization of the phosphoanion transition state. Because Mg2+ ion further enhanced the nuclease activity of the zinc-bound holoenzyme, it is possible that there is a second metal-binding site for Mg2+ in colicins, but it is likely that Mg2+ is only bound upon DNA binding. It has been proposed that nuclease ColE9 hydrolyses DNA by two distinct catalytic mechanisms in the presence of only Mg2+ or Ni2+ (Pommer et al. 2001; Walker et al. 2002). However, alkaline earth metal ions (Mg2+ in mM) and transition meal ions are both present in cells, and most likely ColE7 is a zinc-enzyme in vivo, because Zn2+ binds colicins with highest affinity and Zn2+ is one of the most abundant transition metal ions in cells. Therefore, we propose here that nuclease-ColE7 functions as a zinc-enzyme and that Mg2+ ions are also involved in DNA hydrolysis in vivo.
H545 is likely the general base
In nuclease-ColE7, there are three histidine residues, H544, H569, and H573, directly bound to the zinc ion. These three residues are not strictly conserved among H-N-H proteins, but the residues that are aligned with these histidines are either polar or charged and capable of binding to metal ions. Therefore, the residues aligned with these histidines are likely to be responsible for metal ion binding. The roles of the two most conserved residues, H545 and N560, in the H-N-H motif, are less clear, however. A structural comparison between the H-N-H motif and the active sites in I-PpoI and Serratia nuclease sheds light on the possible functions of H545 (Kuhlmann et al. 1999). The superimposition of the ββα-fold between nuclease-ColE7 and I-PpoI shows that H545 in ColE7 locates at a similar position as H98 in I-PpoI (Fig. 6A ▶). H98 in I-PpoI serves as a general base to activate a water molecule, which functions as a nucleophile to attack the scissile phosphate group. In our structures, a phosphate ion is bound between H545 and a zinc ion, and the distance between H545 (ND1 atom) and phosphate oxygen is 2.6 Å. Therefore it is very likely that H545 functions as a general base. This could explain the strict requirement for a histidine at this position in the entire H-N-H family of proteins.
Fig. 6.
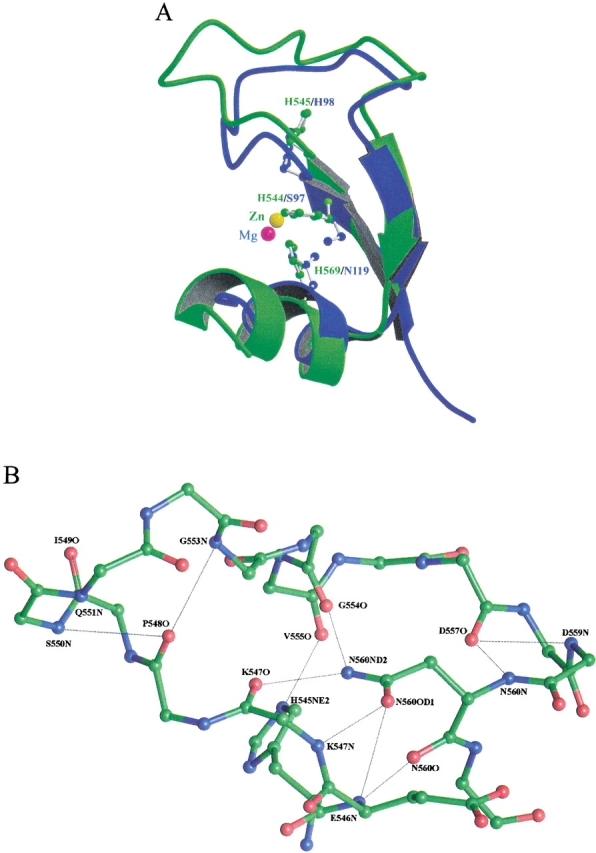
(A) The superimposition of the H-N-H motif in nuclease-ColE7 with the ββα-fold active site of the homing endonuclease I-PpoI (Galburt et al. 1999). The H-N-H motif displayed in green is bound with a zinc ion (yellow sphere), and the I-PpoI displayed in blue is bound with a magnesium ion (red sphere). The general base H98 in I-PpoI is matched at the same position as His545 in nuclease-ColE7. (B) Close-up view of the detailed hydrogen bond network around N560 in the H-N-H motif of the P21212 Zn-bound nuclease-ColE7. The side chain of N560 makes hydrogen bonds to the main-chain atoms of E546, K547, and G554. The side chain of H545 makes a hydrogen bond to the main-chain carbonyl atom of V555.
The second most conserved residue is N560, which is almost always strictly conserved in the H-N-H motif with only a few exceptions. The side chain of N560 makes a hydrogen bond network with the main-chain atoms in the long loop region, with the amide group being fully occupied (Fig. 6B ▶). The ND2 atom makes two hydrogen bonds with the carbonyl groups of G554 and K547, and the OD1 atom also makes two hydrogen bonds with the amino groups of K547 and E546. It appears that N560 has no unoccupied positions in polar atoms to participate in hydrolysis. Therefore, it is likely that N560 only plays a structural role to fix the backbone structure in the loop region so that the general base residue H545, which hydrogen bonds to the carbonyl group of V555, is appropriately held.
A structural model for nuclease-ColE7-bound DNA
A structural model for nuclease-ColE7 bound with DNA was constructed based on the crystal structure of the I-PpoI/DNA complex (Fig. 7 ▶). The H-N-H motif in nuclease-ColE7 was superimposed with the two β-strands and one α-helix in the active site of I-PpoI, with an rms difference of only 1.2 Å for the main-chain atoms. Without further adjustment, the nuclease-ColE7 appears to bind DNA snugly. The concave surface of nuclease-ColE7 faces the DNA backbone with the zinc ion positioned close to the phosphate backbone at the minor groove, appropriate for its role in stabilizing the phosphoanion transition state. The α-helix, α2, binds DNA at the neighboring major groove, and there are indeed several basic residues (R496, K497, K498, K505) located in this helix.
Fig. 7.
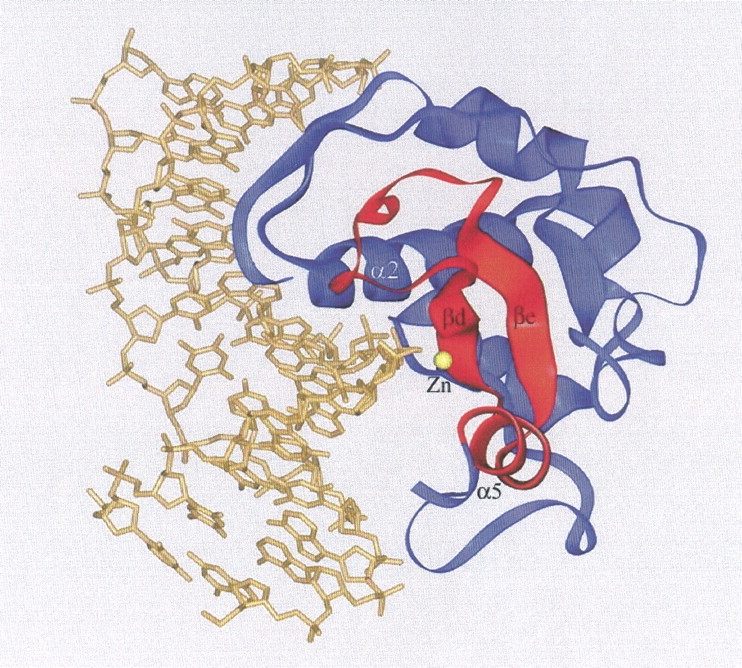
Structural model of nuclease-ColE7 bound to DNA. The model was constructed by overlapping the two β-strands and one α-helix in the H-N-H motif (displayed in red) with the similar fold in the active site of the I-PpoI/DNA complex (PDB entry: 1A73). The cleft of the nuclease-ColE7 faces the DNA with a zinc ion situated close to the phosphate backbone and an α-helix (α2) binding at the major groove.
One puzzling question remained: What is the general acid in this DNA hydrolysis reaction? In I-PpoI or Serratia nuclease, the magnesium-bound water molecule serves as the general acid to provide a proton for the 3′ oxygen-leaving group. In the nickel-bound nuclease-ColE9 structure, the Ni2+ ion was bound with two histidine residues and one water molecule; therefore, it was proposed that this nickel-bound water molecule functions as a general acid (Pommer et al. 2001; Walker et al. 2002). However, in our zinc-bound holoenzyme, the zinc ion was in a tetrahedral not an octahedral geometry, and Zn2+ was not bound with a water molecule. We did not find any residue surrounding the zinc ion that seems likely to function as a general acid. One possibility is that the zinc ion acquires another water molecule and coordinates to five ligands after it is bound to DNA. Alternatively, it is likely that a magnesium ion also participates in this reaction and that the magnesium-bound water molecule donates a proton to the 3′ oxygen-leaving group. We are eagerly pursuing the crystal structures of the nuclease-ColE7/DNA complex, which will reveal unambiguously the arrangement of substrate binding and cleavage catalyzed by H-N-H proteins.
Materials and methods
Construction of expression vectors
The expression vectors pQE-30 and pQE-70 (QIAGEN) containing a six-histidine affinity tag at either the N- or C-terminus of the cloning site were used to clone the nuclease domain-ceiE7 of the ColE7 operon. The detailed procedure for the construction of the recombinant plasmid pQE-30 was previously described (Ko et al. 1999). The pQE-30 overexpressed proteins contain a histidine tag of MRGSHHHHHHGS at the N-terminus of nuclease-ColE7 (residues 447 to 573) and full-length Im7.
The overexpression vector for nuclease-ColE7/Im7 in pQE70 was constructed following a similar procedure (Ko et al. 1999). A 673-bp DNA fragment encoding nuclease-ColE7 and Im7 from the plasmid pHK001 (Chak et al. 1991) was amplified by PCR using a pair of oligonucleotide primers: 5′GCTAAAGGCATGCTAG ATAAGGAGAGTA3′ and 5′CATTCATAGATCTGCCC TGTT TAAATCC3′. The cutting sites of SphI and BglII were generated at either end of the PCR-amplified fragment. PCR reactions were carried out in 100-μL volumes using one unit of Taq polymerase with 30 cycles of 94°C for 1 min, 55°C for 30 sec and 72°C for 1 min. The PCR product was then cleaved with SphI and BglII and ligated into the expression vector pQE-70 preincubated with the same restriction enzymes. The resulting recombinant plasmid was transformed into E.coli M15. The overexpressed protein complex contained the nuclease-ColE7 (residues 444 to 576) and a full-length Im7 with a six-histidine tag attached at the C-terminus.
Protein purification
The purification procedures for the two different His-tagged nuclease-ColE7/Im7 complexes were similar to the procedure described previously (Ko et al. 1999). Eight liters of LB M15 cell culture containing either the pOE-30 or pOE-70 expression vectors were incubated and induced by IPTG at A600nm of 0.6 O.D. at 37°C. Crude cell extracts were purified by chromatography on a Ni-NTA resin affinity column (QIAGEN), followed by a CM column (CM Sepharose Fast Flow, Pharmacia). Protein samples were concentrated by Centriplus 3K (Amicon) ultrafiltration and stored at a concentration of 15 mg/mL in H2O, at −70°C. The concentration of the purified protein was determined by DC protein assay (BioRad).
DNase activity assay
A 30-bp oligonucleotide labeled with Hetrachloro-6-carboxyfluorescein and a quencher of BHQ-1™ (DNaseAlert™ QC system DNA Substrate, Ambion) was used to measure the endonuclease activity of nuclease-ColE7. The enzyme was mixed with the oligonucleotide, and the increased fluorescence emission intensity resulting from the DNA cleavage was measured on a NUNC™ black 96-well plate with a Fluoroskan Ascent plate reader. The detailed procedure for the measurement was as described (Ku et al. 2002). The holo-nuclease-ColE7 was obtained by the supplement of zinc ions to the protein samples during the purification processes. Excess zinc ions were removed by a desalting column (Amersham) followed by dialysis of the protein samples against Milli-Q water. The activity assays were carried out in different concentrations of Mn2+ or Mg2+ using the zinc-bound holoenzymes.
Crystallization and X-ray data collection
The two different types of crystals of the nuclease-ColE7/Im7 were obtained by the hanging-drop vapor diffusion method. The protein complex from pQE-30 with the His-tag at the N-terminus of nuclease-ColE7 was crystallized in the I222 space group as described (Ko et al. 1999). The protein complex overexpressed from pQE-70 was crystallized using similar conditions. Drops of a solution containing 20 mg/mL of protein complex, 50 mM phosphate buffer (pH 6.3), 0.35 M ammonium acetate, and 11% PEG4000 were set up against a reservoir of 22.5% PEG4000. Plates of crystal appeared within about 4 d at room temperature. The metal ions originally associated in the complex were removed by soaking the crystals in the mother liquid containing 5–10 mM EDTA for 16 h. After the crystals were washed with phosphate buffer to remove EDTA, different metal ions, including Zn2+, Mn2+, and Mg2+, were soaked into the complex crystals for 16 h–40 h.
Diffraction data were collected at the absorption edge of the correspondent metal ion using synchrotron radiation (Synchrotron Radiation Research Center, BL17B2, Hsin-Chu, Taiwan) or at 1.54 Å using Cu Kα radiation from a rotating anode. X-ray absorption spectra were measured first to determine the absorption edge for Zn2+ or Mn2+ in crystals. X-ray radiation at inflection points of λZn = 1.29004 Å and λMn = 1.89261 Å was used to collect full sets of diffraction data for the Zn2+-soaked or Mn2+-soaked crystals at ∼100 K. Diffraction data were recorded on a Mac Science image plate (synchrotron data) or on an MSC R-AXIS-IV imaging plate (in-house source). In all cases, data were processed with DENZO and SCALEPACK (Otwinowski and Minor 1997).
Structure determination and refinement
The Zn2+-bound P21212 structure was solved by molecular replacement using the previously determined I222 structure (PDB entry: 7cei) as the searching model. Cross-rotation and translation function [CNS (Brunger et al. 1998)] unambiguously identified two unique complexes in the asymmetric unit using the diffraction data from 15–4.0 Å. Rigid-body refinement gave an R-factor of 38.6%. This model was subjected to manual rebuilding with Turbo-Frodo, alternating with torsional angle-simulated annealing, standard positional and individual isotropic B value refinement, and automatic water placement and deletion with the program CNS (Brunger et al. 1998). Fourier (2Fo-Fc) and anomalous maps in the metal-binding site were calculated at the final stage of refinement. Metal ions and phosphate ions were added into the model before the last cycle of refinement. The final refinement statistics are listed in Table I. The coordinates of the Zn2+-bound P21212 complex structure have been deposited in the Protein Data Bank with the accession code 1MZ8.
Acknowledgments
We thank Yuch-Cheng Jean for his kind assistance and suggestions during data collection at synchrotron beamline 17B, in the Synchrotron Radiation Research Center, Hsinchu, Taiwan. This work was supported by research grants from the National Science Council of the Republic of China and Academia Sinica to H.S.Y. (NSC90-2320-B001-024).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0220602
References
- Alberts, I.L., Nadassy, K., and Wodak, S.J. 1998. Analysis of zinc binding sites in protein structures. Protein Sci. 7 1700–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Barney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L.L. 2002. The Pfam protein families database. Nucleic Acid Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort, M. and Perlman, P.S. 1995. Mechanisms of intron mobility. J. Biol. Chem. 270 30237–30240. [DOI] [PubMed] [Google Scholar]
- Berg, J.M. and Godwin, H.A. 1997. Lessons from zinc-binding peptides. Annu. Rev. Biophys. Struct. 26 357–371. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crystallography and NMR System (CNS): A new software system for macromolecular structure determination. Acta Cryst. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Chak, K.-F., Kuo, W.-S., Lu, F.-M., and James, R. 1991. Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol. 137 91–100. [DOI] [PubMed] [Google Scholar]
- Chak, K.-F., Safo, M.K., Ku, W.-Y., Hsieh, S.-Y., and Yuan, H.S. 1996. The crystal structure of the ImmE7 protein suggests a possible colicin-interacting surface. Proc. Natl. Acad. Sci. 93 6437–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, B.S. and Stoddard, B.L. 2001. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acid Res. 29 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudev, T. and Lim, C. 2001. Metal selectivity in metalloproteins: Zn2+ vs. Mg2+. J. Phys. Chem. 105 4446–4452. [Google Scholar]
- Eddy, S.R. and Gold, L. 1991. The phage T4 nrdB intron: A deletion mutant of a version found in the wild. Genes & Dev. 5 1032–1041. [DOI] [PubMed] [Google Scholar]
- Edgell, D.R., Belfort, M., and Shub, D.A. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferat, J.L. and Michel, F. 1993. Group II self-splicing introns in bacteria. Nature 364 358–361. [DOI] [PubMed] [Google Scholar]
- Foley, S., Bruttin, A., and Brussow, H. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilius bacteriophages. J. Virol. 74 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedhoff, P., Franke, I., Krause, K.L., and Pingoud, A. 1999a. Cleavage experiments with deoxythymidine 3′,5′-bis-(p-nitrophenylphosphate) suggest that the homing endonuclease I-PpoI follows the same mechanism of phosphodiester bond hydrolysis as the non-specific Serratia nuclease. FEBS Lett. 443 209–214. [DOI] [PubMed] [Google Scholar]
- Friedhoff, P., Franke, I., Meiss, G., Wende, W., Krause, K.L., and Pingoud, A. 1999b. A similar active site for non-specific and specific endonucleases. Nat. Struct. Biol. 6 112–113. [DOI] [PubMed] [Google Scholar]
- Galburt, E.A., Chevalier, B., Tang, W., Jurica, M.S., Flick, K.E., Monnat, R.J., and Stoddard, B.L. 1999. A novel endonuclease mechanism directly visualized for I-PpoI. Nat. Struct. Biol. 6 1096–1099. [DOI] [PubMed] [Google Scholar]
- Goodrich-Blair, H. 1994. The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res. 22 3715–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich-Blair, H. and Shub, D.A. 1996. Beyond homing: Competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell 84 211–221. [DOI] [PubMed] [Google Scholar]
- Goodrich-Blair, H., Scarlato, V., Gott, J.M., Xu, M.-Q., and Shub, D.A. 1990. A self-splicing group I intro in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell 63 417–424. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A.E. 1994. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 3 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin, N.V. 2001. Treble clef finger—A functionally diverse zinc-binding structural motif. Nucleic Acids Res. 29 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom, K. and Sedgwick, S.G. 1991. Cloning and structural characterization of the mcrA locus of Escherichia coli. J. Bacteriol. 173 7368–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield, D.J., Guan, Y., Haas, B.J., Cunningham, R.P., and Tainer, J.A. 1999. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double nucleotide flipping at basic sites and three-metal-ion catalysis. Cell 98 397–408. [DOI] [PubMed] [Google Scholar]
- Keeble, A.H., Hemmings, A.M., James, R., Moore, G.R., and Kleanthous, C. 2002. Multistep binding of transition metals to the H-N-H endonuclease toxin colicin E9. Biochemistry 41 10234–10244. [DOI] [PubMed] [Google Scholar]
- Kelemen, B.R., Klink, T.A., Behlke, M.A., Eubanks, S.R., Leland, P.A., and Raines, R.T. 1999. Hypersensitive substrates for ribonucleases. Nucleic Acid Res. 27 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous, C., Kuhlmann, U.C., Pommer, A.J., Ferguson, N., Radford, S.E., Moore, G.R., James, R., and Hemmings, A.M. 1999. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 6 243–252. [DOI] [PubMed] [Google Scholar]
- Ko, T.-P., Liao, C.-C., Ku, W.-Y., Chak, K.-F., and Yuan, H.S. 1999. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure 7 91–102. [DOI] [PubMed] [Google Scholar]
- Kovall, R.A. and Matthews, B.W. 1999. Type II restriction endonucleases: Structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol. 3 578–583. [DOI] [PubMed] [Google Scholar]
- Ku, W.-Y., Liu, Y.-W., Hsu, Y.-C., Liao, C.-C., Liang, P.-H., Yuan, H.S., and Chak, K.-F. 2002. The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acid Res. 30 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck, U. 1989. The intron of a plasmid gene from a green alga contains an open reading frame for a reverse transcriptase-like enzyme. Mol. Gen. Genet. 218 257–265. [DOI] [PubMed] [Google Scholar]
- Kuhlmann, U.C., Moore, G.R., James, R., Kleanthous, C., and Hemmings, A.M. 1999. Structural parsimony in endonuclease active site: Should the number of homing endonuclease families be redefined? FEBS Lett. 463 1–2. [DOI] [PubMed] [Google Scholar]
- Lambowitz, A.M. and Belfort, M. 1993. Introns as mobile genetic elements. Annu. Rev. Biochem. 62 587–622. [DOI] [PubMed] [Google Scholar]
- Lazarevic, V., Soldo, B., Dusterhoft, A., Hilbert, H., Mauel, C., and Karamata, D. 1998. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc. Natl. Acad. Sci. 95 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunin, V.Y., Levdikov, V.M., Shlyapnikov, S.V., Blagova, E.V., Lunin, V.V., Wilson, K.S., and Mikhailov, A.M. 1997. Three-dimensional structure of Serratia marcescens nuclease at 1.7 Å resolution and mechanism of its action. FEBS Lett. 412 217–222. [DOI] [PubMed] [Google Scholar]
- Miller, M.D., Cai, J., and Krause, K.L. 1999. The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol. 288 975–987. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode, pp. 307–326. Academic Press, NY. [DOI] [PubMed]
- Pavletich, N.P. and Pabo, C.O. 1991. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252 809–817. [DOI] [PubMed] [Google Scholar]
- Pingoud, A. and Jeltsch, A. 1997. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem. 246 1–22. [DOI] [PubMed] [Google Scholar]
- Pommer, A.J., Kuhlmann, U.C., Cooper, A., Hemmings, A.M., Moore, G.R., James, R., and Kleanthous, C. 1999. Homing in on the role of transition metals in the HNH motif of colicin endonuclease. J. Biol. Chem. 274 27153–27160. [DOI] [PubMed] [Google Scholar]
- Pommer, A.J., Cal, S., Keeble, A.H., Walker, D., Evans, S.J., Kuhlmann, U.C., Cooper, A., Connolly, B.A., Hemmings, A.M., Moore, G.R., et al. 2001. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 314 735–749. [DOI] [PubMed] [Google Scholar]
- Pont-Kingdon, G.A., Okada, N.A., Macfarlane, J.L., Beagley, C.T., Wolstenholme, D.R., Cavalier-Smith, T., and Clark-Walker, G.D. 1995. A coral mitochondrial mutS gene. Nature 375 109–111. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, H., Vix, O., Toro, I., Golz, S., Kemper, B., and Suck, D. 1999. X-ray structure of T4 endonuclease VII: A DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 18 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier, C., Dominguez, R., Lahm, A., Dahl, O., and Suck, D. 1998. Recognition of single-strand DNA by nuclease P1: High resolution crystal structures of complexes with substrate analogs. Proteins 32 414–424. [PubMed] [Google Scholar]
- Sano, Y. and Kageyama, M. 1993. A novel transposon-like structure carries the genes for pyocin AP41, a Pseudomonas aeruginosa bacteriocin with a DNase domain homology to E2 group colicins. Mol. Gen. Genet. 237 161–170. [DOI] [PubMed] [Google Scholar]
- Sano, Y., Matsui, H., Kobayashi, M., and Kageyama, M. 1993. Molecular structures and functions of Pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 175 2907–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, K. and Nomura, M. 1976. Colicin E2 is a DNA endonuclease. Proc. Natl. Acad. Sci. 68 3989–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub, D.A., Goodrich-Blair, H., and Eddy, S.R. 1994. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. TIBS 19 402–404. [DOI] [PubMed] [Google Scholar]
- Toba, M., Masaki, H., and Ohta, T. 1988. Colicin E8, a DNase which indicates an evolutionary relationship between colicins E2 and E3. J. Bacteriol. 170 3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubetskoy, V.S., Hagstrom, J.E., and Budker, V.G. 2002. Self-quenched covalent fluorescent dye-nucleic acid conjugates as polymeric substrates for enzymatic nuclease assays. Anal. Biochem. 300 22–26. [DOI] [PubMed] [Google Scholar]
- Walker, D.C., Georgiou, T., Pommer, A.J., Walker, D., Moore, G.R., Kleanthous, C., and James, R. 2002. Mutagenic scan of the H-N-H motif of colicin E9: Implications for the mechanistic enzymology of colicins, homing enzymes and apoptotic endonucleases. Nucleic Acid Res. 30 3225–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, R., Moore, G.R., Kleanthous, C., and James, R. 1992. Molecular analysis of the protein-protein interaction between the E9 immunity protein and colicin E9. Eur. J. Biochem. 210 923–930. [DOI] [PubMed] [Google Scholar]