Abstract
MPT63 is a small, major secreted protein of unknown function from Mycobacterium tuberculosis that has been shown to have immunogenic properties and has been implicated in virulence. A BLAST search identified that MPT63 has homologs only in other mycobacteria, and is therefore mycobacteria specific. As MPT63 is a secreted protein, mycobacteria specific, and implicated in virulence, MPT63 is an attractive drug target against the deadliest infectious disease, tuberculosis (TB). As part of the TB Structural Genomics Consortium, the X-ray crystal structure of MPT63 was determined to 1.5-Ångstrom resolution with the hope of yielding functional information about MPT63. The structure of MPT63 is an antiparallel β-sandwich immunoglobulin-like fold, with the unusual feature of the first β-strand of the protein forming a parallel addition to the small antiparallel β-sheet. MPT63 has weak structural similarity to many proteins with immunoglobulin folds, in particular, Homo sapiens β2-adaptin, bovine arrestin, and Yersinia pseudotuberculosis invasin. Although the structure of MPT63 gives no conclusive evidence to its function, structural similarity suggests that MPT63 could be involved in cell-host interactions to facilitate endocytosis/phagocytosis.
Keywords: Mycobacterium tuberculosis, secreted protein MPT63, X-ray crystal structure, TB drug target, cell-host interactions
Tuberculosis (TB) is caused by the bacterial pathogen Mycobacterium tuberculosis (M. tuberculosis), which kills 2–3 million people around the world each year, more than any other infectious disease. The rise in TB incidence over the past two decades is due in part to the TB deaths in HIV-infected patients and in part to the emergence of multidrug resistant strains of the bacteria. This rapid increase in the disease has led the World Health Organization to fund a large effort toward controlling this disease before it becomes a global epidemic (The World Health Organization 1998).
The Structural Genomics Consortium for M. tuberculosis is an international effort that is focusing on solving protein structures of potential drug targets, proteins that have predicted novel folds (Mallick et al. 2000) and proteins that are promising vaccine candidates (http://www.doe-mbi.ucla.edu/TB). One group of potential drug targets is the secreted proteins of M. tuberculosis, which are important to the survival of the bacterium within its host. These proteins have been identified experimentally (Wiker et al. 1991; Young et al. 1991) and computationally (Gomez et al. 2000; Wiker et al. 2000). Secretion of proteins is an important mechanism that bacteria utilize to interact with their surroundings. Secreted proteins are often needed for bacteria to survive in a hostile environment or to colonize a host successfully. Moreover, secreted proteins are often used by bacteria to cause disease, as many virulence factors are either secreted or associated with the bacterial surface. Additionally, secretion of proteins by intracellular pathogens has a central role in determining pathways of antigen presentation and recognition by effector T-cells involved in protective immunity. Thus, a knowledge of proteins secreted by a particular bacterial species should better equip us to fight bacterial infections by countering the bacterium’s offensive weaponry and by more effectively designing vaccines.
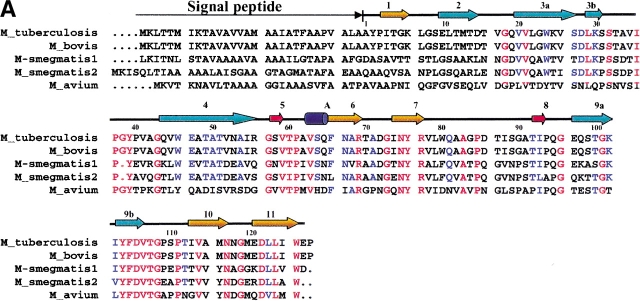
The three most abundant secreted proteins of M. tuberculosis have been proposed to be the 30-kD (antigen 85B, Rv1886c), 32-kD (antigen 85C, Rv0129c), and 16-kD (MPT63, Rv1926c) proteins (Nagai et al. 1991). The functions and structures of antigens 85B and 85C are known (Ronning et al. 2000; Anderson et al. 2001), and are proving to be good vaccine candidates. MPT63 is a secreted protein of unknown function that is specific to mycobacteria. Homologs of MPT63 have been found in M. smegmatis, M. avium, and M. bovis (Fig. 1A ▶), although polyclonal antibodies against MPT63 from M. tuberculosis do not cross-react with proteins of the common environmental mycobacterial species, M. avium (Manca et al. 1997). Interestingly, there is a pseudogene of MPT63 within the M. leprae genome, which is thought not to be translated into protein (Cole et al. 2001).
Fig. 1.

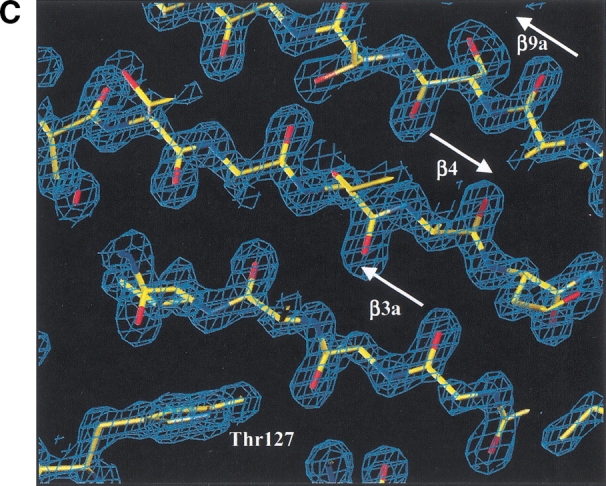
Structure of M. tuberculosis MPT63. (A) Sequence comparison of MPT63 homologs. The top line depicts the M. tuberculosis MPT63 protein in terms of secondary structure elements (residue numbering is for the M. tuberculosis mature sequence). The orange arrows depict the β-strands of the longer β-sheet, the cyan arrows depict the β-strands of the shorter, twisted β-sheet, the red arrows depict the two β-strands of the small β-sheet, and the dark blue cylinder depicts the 310 α-helix; the number of each β-strand is above it. The red letters show conserved residues among the sequences, and blue letters show the partially conserved residues. The numbers directly above the sequence correspond to the residue numbers of mature MPT63. The sequence alignment was made with CLUSTALW and BoxShade. (B) Ribbon diagram of MPT63 shows a β-sandwich and a 310 helix, A. The β-sheets that contribute to the immunoglobulin-like β-sandwich are colored in orange and blue. The small β-sheet is colored in red and 310 helix is in blue. This figure was generated using RIBBONS. (C) The M. tuberculosis MPT63 electron density, a view of the experimental 2Fo-Fc electron density map of three β-sheets (β3a, β4, β9a). The electron density (blue) is contoured at 1.5σ. Oxygen is depicted in red, nitrogen in blue, and carbon in yellow. The figure was generated using O and RayMol.
Immunological studies have been carried out on the mature MPT63 protein. It has been shown to stimulate humoral immune responses in guinea pigs infected with virulent M. tuberculosis (Manca et al. 1997). T-cell epitope mapping showed that MPT63 contained a highly immunodominant region within the first 30 residues of the amino-terminal of the mature protein (Lee and Horwitz 1999). By use of a Tn552‘phoA in vitro transposition system, it was confirmed that MPT63 is a secreted protein (Braunstein et al. 2000), and it has been shown to be a cell envelope-associated protein that may participate in virulence.
MPT63 is a secreted, mycobacteria-specific protein that is implicated in virulence. Thus, MPT63 could be an excellent drug target against TB or a possible vaccine candidate. The X-ray crystal structure of MPT63 has been determined as a β-sandwich, immunoglobulin-like fold.
Results
Overall structure
The structure of MPT63 is a β-sandwich consisting of two antiparallel β-sheets similar to an immunoglobulin-like fold, with an additional small, antiparallel β-sheet (β5 and β8; Fig. 1B ▶). The longer-stranded β-sheet is made up of four antiparallel β-strands (β2, β3, β4, and β9). The shorter-stranded β-sheet consists of five β-strands (β1, β6, β7, β10, and β11), four of these β-strands form an antiparallel β-sheet. It is noteworthy that the first strand of this sheet, the amino-terminal β-strand (β1), forms a parallel β-sheet with the carboxy-terminal β-strand (β11). This shorter β-sheet has an anticlockwise twist to it. The 2 β-sheets are linked by a coil of 18 residues, between β-strands 7 and 9, interrupted by a small two-stranded β-sheet (β5 and β8). The only helical secondary structure is a 310 helix of three residues at the start of β-strand 6. The overall topology of MPT63 is that of a β-sandwich with a five-stranded, mainly antiparallel β-sheet packed on top of a four-stranded antiparallel β-sheet, which can be described as an immunglobulin fold with an overlap into a jelly-roll fold. All residues are in the allowed regions of the Ramachandran plot, although Pro51 is the only residue in the generously allowed region and is located in a β-turn and fits the observed electron density well.
Structural similarity to MPT63
MPT63 has structural similarity with immunoglobulin structures and, therefore, the fold of MPT63 can be classified as a member of the immunoglobulin superfamily (Halaby and Mornon 1998). The highest scoring structures from a structural similarity search to MPT63 are shown in Table 1. One should note that the highest Z-score is 6.0, which indicates that there is no strong structural homology to the structure of MPT63. MPT63 has some structural similarity to cell surface-binding proteins such as Homo sapiens β2-adaptin (Owen and Luzio 2000; Owen et al. 2000), bovine arrestin (Hirsch et al. 1999), and Yersinia pseudotuberculosis invasin (Hamburger et al. 1999). It also has structural similarity to eukaryotic fibronectin-binding proteins, major histocompatiblity domains, and T-cell receptors.
Table 1.
Dali structural similarity search based on the Dali server
| Rank | PDB code | Z | RMSD | Sequence | % IDE | Protein | |
| 1 | 1e42–A | 6.0 | 2.7 | 86 | 233 | 6 | β2-adaptin |
| 2 | 1qts–A | 5.8 | 3.9 | 96 | 247 | 8 | ap-2 clathrin adaptor α subunit (α-adaptin c) |
| 3 | 1cfl-A | 5.8 | 3.9 | 91 | 373 | 9 | arrestin (retinal s-antigen, 48-kd protein) |
| 4 | 1im3-D | 5.3 | 3.0 | 83 | 95 | 11 | hla class i histocompatibility antigen |
| 5 | 1iak-B | 4.6 | 4.2 | 84 | 185 | 11 | mhc class ii i-ak hen eggwhite lysozyme peptide |
| 6 | 1agd-B | 4.6 | 3.7 | 76 | 99 | 9 | β-2 microglobulin fragment |
| 7 | 1iar-B | 4.5 | 3.2 | 75 | 188 | 8 | interleukin-4 interleukin-4 receptor α chain fragment |
| 8 | 1iak-A | 4.5 | 3.9 | 80 | 182 | 4 | mhc class ii i-ak hen eggwhite lysozyme peptide |
| 9 | 1cdl-A | 4.5 | 4.2 | 83 | 273 | 8 | cdl fragment (mcdld.l) biological_unit |
| 10 | 1igt-B | 4.4 | 4.3 | 88 | 444 | 7 | igg2a intact antibody–mab231 |
| 11 | 1hdm-B | 4.4 | 4.0 | 84 | 183 | 5 | class ii histocompatibility antigen, m α chain fragment |
| 12 | 1bgl-A | 4.2 | 2.9 | 73 | 997 | 8 | β-galactosidase |
| 13 | 1svb | 4.1 | 3.1 | 75 | 395 | 9 | tick-borne encephalitis virus glycoprotein |
| 14 | 1hxm-D | 4.1 | 4.0 | 84 | 230 | 4 | γ-δ t-cell receptor (t-cell receptor δ cha |
| 15 | 1cqk-A | 4.1 | 3.8 | 79 | 101 | 4 | ch3 domain of mak33 antibody fragment Mutant biological |
| 39 | 1cwv-A | 3.4 | 3.5 | 78 | 484 | 9 | invasin fragment |
| 48 | 1fnh-A | 3.2 | 3.6 | 82 | 269 | 9 | fibronectin fragment |
Column 1 gives rank of structure similarity, column 3 gives the Z-score (number of stand deviations above the mean score of the best domain–domain alignment), column 4 gives RMSD (the root-mean-square deviation) of C (α) atoms from those of MPT63 in the rigid body superimposition. Sequence gives the starting residue number of the structurally equivalent segments. % IDE is the sequence identity between that of the protein and MPT63.
Electrostatic molecular surface potential of MPT63
The electrostatic surface potential of MPT63 reveals that one side of the protein has two important features, a negatively charged cavity and a positively charged channel, characteristics that suggest a binding site for peptides, lipids, carbohydrates, or a cofactor. The residues that contribute to the positively charged channel that runs down the smaller twisted β-sheet are the side chains of Arg 83 and Arg 91, and the N atoms of Gln 79 and Ala 129. The main contributions to the negatively charged cavity are the side chains Asp 29 and Glu 126. The opposite side of the protein is mainly hydrophobic except for positively charged nodes due to the side chains of lysines, and negatively charged nodes due to the side chains of arginines and glutamines.
Discussion
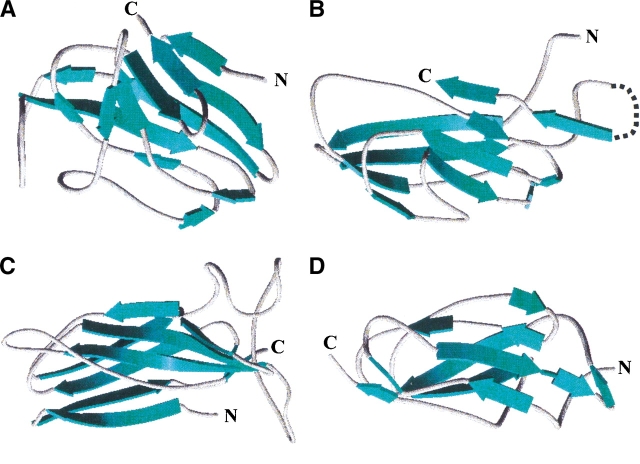
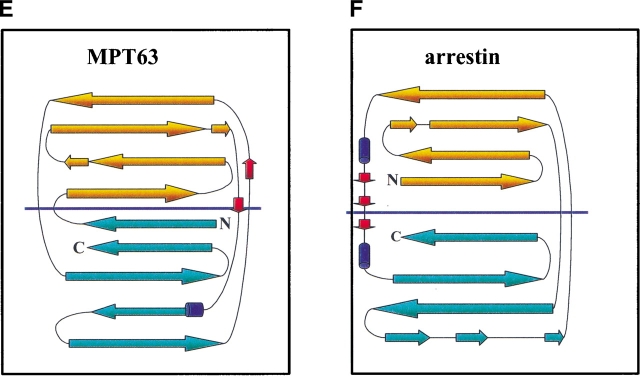
One of the questions posed by the TB structural genomics consortium is whether one can predict function from structure. BLAST analysis of MPT63 found no close sequence relatives, and in addition, MPT63 was predicted to have a novel fold by an automated fold assignment program using binary hypothesis testing (Mallick et al. 2000). The observed structural similarity of MPT63 to immunoglobulin-like folds and, in particular, cell surface-binding proteins, suggests that MPT63 could be involved in cell-host interactions to facilitate endocytosis or phagocytosis. The highest scoring structural analog (Table 1), is Homo sapiens β2-adaptin appendage domain, a dimeric protein that binds to clathrin buds and has been implicated in endocytosis (Owen and Luzio 2000). The structure of adaptin reveals a platform and a sandwich domain, the latter to which MPT63 has structurally similarity. The third highest scoring position, bovine arrestin, is a member of a family of intracellular proteins that inhibit receptor activity. They bind to the cytoplasmic surface of the receptor, thereby occluding the interaction with G-proteins. Some arrestins also serve as adaptor molecules to the cellular protein trafficking machinery, facilitating the receptor’s sequestration by clathrin-mediated endocytosis (Hirsch et al. 1999). Further down the list is Y. pseudotuberculosis invasin, a bacterial protein that mediates entry into eukaryotic cells by binding to members of the β1 intergin family (Hamburger et al. 1999). The protein is thought to activate a reorganization of the host cytoskeleton to form pseudopods that envelop the bacterium. Invasin and adaptin have very similar structures, and both have platform and sandwich domains, and they are also similar to the fibronectin type III repeats (Hamburger et al. 1999; Hirsch et al. 1999). Alhough these three proteins have structural similarity to MPT63, the structures basically have similar topologies rather than similar globular structures, as shown in Figure 2A ▶–D. The topology of MPT63 and arrestin are the most similar (Fig. 2E,F ▶). The most strikingly different feature of the structure of MPT63 is the curvature of the smaller β-sheet of MPT63, which is much more pronounced than that of adaptin, arrestin, and invasin. On the basis of these structural similarities to proteins with a common functional theme, one can speculate that this protein may be involved in host-bacterial interactions involved in endocytosis or phagocytosis, possibly during bacterial internalization.
Fig. 2.
Ribbon diagrams of MPT63 and three of the proteins with similar structures and topology diagrams of MPT63 and arrestin. The β-strands are shown in blue for each structure and each figure was generated in WebLab ViewerPro version 3.7. (A) M. tuberculosis MPT63; (B) Homo sapiens β2-adaptin (residues 86–233), the dotted line is an extended β-sandwich that has no structural similarity to MPT63; (C) bovine arrestin (residues 91–373); (D) Y. pseudotuberculosis invasin (residues 78–484). Topology diagram of (E) MPT63 and (F) arrestin. The β-sheets that contribute to the immunoglobulin-like β-sandwich are colored in orange and blue. The amino- (N) and carboxy- (C) termini are also shown.
The function of MPT63 cannot be clearly specified by its structural features as it has an extremely common immunoglobulin-like fold that occurs in ∼24% of the structures in the Protein Data Bank. The β-sandwich fold that MPT63 resembles is at the core of many proteins with diverse functions. This topology is distributed among such functionally distinct and phylogenetically disparate molecules as vertebrate immune system proteins, plant cytochromes, bacterial pathogenesis and molecular chaperones, and eukaryotic transcription factors and cell adhesion molecules (Shapiro et al. 1995). Many of these functionally distinct proteins may have evolved in parallel, neither diverging from a common ancestor nor converging onto a common function (Halaby and Mornon 1998). The diverse nature of the β-sandwich fold may simply be an energetically favorable and kinetically readily accessible fold, which would account for the structure of MPT63 not providing a conclusive function for MPT63.
The immunogenic epitopes of MPT63 have been shown to be the first 30 residues of the mature protein (Lee and Horwitz 1999), these residues are located in the first three β-strands of the structure (β1, β2, and β3). Interestingly, the first β-strand (β1) extends the antiparallel β-sheet into a parallel β-sheet, thus providing a starting point for probing the proposed cell-host interactions of MPT63.
Notes
The coordinates and structure factors for the crystal structure of MPT63 have been deposited with the Protein Data Bank (RCSB, http:/www.rcsb.org/pdb) as entry 1LM1.
Materials and methods
Overexpression and purification of MPT63
A recombinant plasmid producing the mature MPT63 (residues 30 – 159) in pQE30 (QIAGEN) was constructed as described previously (Manca et al. 1997). The amino-terminal His6-tagged fusion protein was expressed in Escherichia coli XL1-Blue cells by using a standard induction protocol. The E. coli cells were grown aerobically at 37°C in LB medium containing 100 μg/mL ampicillin. Protein expression was induced by addition of 1 mM isopropyl-beta-D-thiogalactoside (IPTG) at an OD600 of ∼1.0, and the cells were harvested 4 h after induction. The cells were centrifuged at 5000 rpm for 10 min, then washed with resuspension buffer (50 mM Hepes at pH 7.8, 300 mM NaCl). After addition of resuspension buffer that contained phenylmethylsulfonyl fluoride (PMSF) and hen egg lyzosome, the cells were disrupted by sonication and then centrifuged at 13,000 rpm for 40 min before filtration (0.22 μm) to remove cell debris. The cell lysate was then loaded on to a Ni2+-charged HisTrap column (5 mL) and washed with 50 mM Hepes (pH 7.8), 300 mM NaCl, and 10 mM imidazole. The protein was eluted with an imidazole 10–500 mM linear gradient (100 mL); the purified protein is eluted between 200 and 300 mM imidazole. The fractions were collected and concentrated in a centricons (15 mL) before dialysis against 50 mM Tris/HCl (pH 7.4), and 0.5 M NaCl. The protein was further purified on a S200 gel filtration column and concentrated to 28 mg/mL.
The selenomethionine-MPT63 was grown as described above, except that the cells were grown in M9 minimal medium supplemented with amino acids supplements (leucine, isoleucine, valine, 50 mg/L; phenylalanine, lysine, threonine, 100 mg/L; and selenomethionine; 75 mg/L) (Van Duyne et al. 1993). The selenomethionine-MPT63 was purified under identical conditions as the native protein and concentrated in a centricon to 60 mg/mL.
Crystallization and structure determination of MPT63
The crystals were grown by hanging drop-vapor diffusion against a reservoir containing 26.5% PEG 4000, 0.1 M Tris/HCl (pH 7.0), and 15% glycerol over a period of 1 wk at room temperature. The crystals were mounted and collected under cyroconditions, which is the same condition as above. Crystals were of space group P6522 with one monomer per asymmetric unit; unit cell dimensions were 43.1 × 43.1 × 228.8 Å. The selenomethionine (SeMet)-MPT63 was prepared as described previously and crystallized under identical conditions to the native protein.
The structure of MPT63 was determined by the multiwavelength anomalous diffraction (MAD) phasing method. Three data sets were collected for the SeMet protein in the P6522 crystal form at wavelengths near the Se absorption edge at the synchrotron light source at Brookhaven National Laboratories on a charge-coupled device detector. Data were processed using DENZO and SCALEPACK (Otwinowski and Minor 1996), and multiwavelength anomalous diffraction (MAD) phasing proceeded by the standard methods of heavy atom location (SHELDX; http://shelx.uni-ac.gwdg.de/SHELX/), maximum likelihood phase refinement ML-PHARE; Collaborative Computational Project 1994) and density modification (DM; Cowtan and Main 1998). Phase extension to 1.5 Å permitted automated model building for all but five residues of the protein with ARP/wARP (Perrakis et al. 1999), which also traced the atomic model apart from seven residues. The remaining seven residues were traced with the program O (Jones et al. 1991). The model was refined (Table 2) with the program SHELXL, and then was checked by both Verify3D (http://www.doe-mbi.ucla.edu/Services/Verify_3D/) and ERRAT (http://www.doe-mbi.ucla.edu/Services/ERRAT/).
Table 2.
X-ray diffraction data collection and atomic refinement for MPT63 from M. tuberculosis
| Data set | Peak | Inflection | High remote | Native |
| Wavelength (Å) | 0.9792 | 0.9795 | 0.9782 | 0.9792 |
| Resolution range (Å) | 100 - 2.2 | 100 - 2.2 | 100 - 2.2 | 100 - 1.5 |
| Unique reflections (total) | 1231 (12099) | 1231 (12115) | 1212 (11686) | 2537 (20972) |
| Completeness (%)a | 99.8 [100.0] | 99.9 [100.0] | 98.5 [100.0] | 97.3 [92.7] |
| Rmergeb | 5.1 [7.3] | 5.0 [7.6] | 4.5 [5.9] | 6.2 [19.0] |
| I/σ(I)a | 46.7 [65.7] | 63.5 [42.4] | 44.4 [43.9] | 30.00 [14.28] |
| # Se sites/monomer | 3 | |||
| Phasing resolution range (Å) | 19.5 - 2.3 | — | 19.5 - 2.3 | |
| Rcullisc,d acentric/centric | 0.78/0.72 | — | 0.67/0.60 | |
| Rcullise anomalous | 0.35 | 0.36 | 0.54 | |
| Phasing Powerc,f:acentric/centric | 1.08/0.84 | — | 1.57/1.25 | |
| Figure of meritg | 0.66 | |||
| Model refinement | ||||
| Resolution range (Å) | 20 - 1.5 | |||
| No. of reflections (working/free) | 19363/986 | |||
| No. of protein atoms | 967 | |||
| No. of water molecules | 194 | |||
| Rwork/Rfreeh, % | 19.2/24.6 | |||
| Bond lengths (Å) | 0.037 | |||
| Bond angles (°) | 1.483 |
The first three columns refer to the Se derivative and the fourth column refers to the native protein extended to 1.5 Angstrom resolution.
a Statistics for the highest resolution shell are given in brackets.
b Rmerge = ∑I-<I>/∑
c The inflection data set was treated as a reference for phasing.
dRcullis = ∑ɛ/∑|FPH-Fp|, where ɛ = lack of closure.
eRcullis = ∑ɛ/∑|F+-F−|, where ɛ = lack of closure.
f Phasing power = <FH/ɛ>.
g Value is given after density modification.
hRwork = ∑|Fobs-Fcalc|/∑Fobs,Rfree was computed identically except where all reflections belong to a test set of 5% randomly selected data.
Protein structural similarity was searched for using DALI (Holm and Sander 1993), and aligned with combinatorial extension (CE; Shindyalov and Bourne 1998). Electrostatic and surface area calculations as shown in Figure 2 ▶, A and B, were performed with GRASP (Sharp et al. 1991).
Acknowledgments
This work has been supported by a grant from the National Institutes of Health for the TB Structural Genomics Consortium, and HHMI. We thank Dr. Dan Anderson and Mari Gingery for useful discussions, Brookhaven National Laboratory for the use of beamline X8C of the National Synchrotron Light Source, and Joel Berendzen, Li Wei Hung, and Leonid Flaks.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0219002.
References
- Anderson, D.H., Harth, G., Horwitz, M.A., and Eisenberg, D. 2001. An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase. J. Mol. Biol. 307671–681. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., Griffin, T.I., Kriakov, J.I., Friedman, S.T., Grindley, N.D., and Jacobs, Jr., W.R., 2000. Identification of genes encoding exported Mycobacterium tuberculosis proteins using a Tn552’phoA in vitro transposition system. J. Bacteriol. 1822732–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S.T., Eiglmeier, K., Parkhill, J., James, K.D., Thomson, N.R., Wheeler, P.R., Honore, N., Garnier, T., Churcher, C., Harris, D., et al. 2001. Massive gene decay in the leprosy bacillus. Nature 4091007–1011. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project. 1994. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan, K. and Main, P. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr. D. Biol. Crystallogr. 54487–493. [DOI] [PubMed] [Google Scholar]
- Gomez, M., Johnson, S., and Gennaro, M.L. 2000. Identification of secreted proteins of Mycobacterium tuberculosis by a bioinformatic approach. Infect. Immun. 682323–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaby, D.M. and Mornon, J.P. 1998. The immunoglobulin superfamily: An insight on its tissular, species, and functional diversity. J. Mol. Evol. 46389–400. [DOI] [PubMed] [Google Scholar]
- Hamburger, Z.A., Brown, M.S., Isberg, R.R., and Bjorkman, P.J. 1999. Crystal structure of invasin: A bacterial integrin-binding protein. Science 286291–295. [DOI] [PubMed] [Google Scholar]
- Hirsch, J.A., Schubert, C., Gurevich, V.V., and Sigler, P.B. 1999. The 2.8 Å crystal structure of visual arrestin: A model for arrestin’s regulation. Cell 97257–269. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233123–138. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110–119. [DOI] [PubMed] [Google Scholar]
- Lee, B.Y. and Horwitz, M.A. 1999. T-cell epitope mapping of the three most abundant extracellular proteins of Mycobacterium tuberculosis in outbred guinea pigs. Infect. Immun. 672665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick, P., Goodwill, K.E., Fitz-Gibbon, S., Miller, J.H., and Eisenberg, D. 2000. Selecting protein targets for structural genomics of Pyrobaculum aerophilum: Validating automated fold assignment methods by using binary hypothesis testing. Proc. Natl. Acad. Sci. 972450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca, C., Lyashchenko, K., Wiker, H.G., Usai, D., Colangeli, R., and Gennaro, M.L. 1997. Molecular cloning, purification, and serological characterization of MPT63, a novel antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 6516–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, S., Wiker, H.G., Harboe, M., and Kinomoto, M. 1991. Isolation and partial characterization of major protein antigens in the culture fluid of Mycobacterium tuberculosis. Infect. Immun. 59372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1996. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol.. 276A307–326. [DOI] [PubMed] [Google Scholar]
- Owen, D.J. and Luzio, J.P. 2000. Structural insights into clathrin-mediated endocytosis. Curr. Opin. Cell. Biol. 12467–474. [DOI] [PubMed] [Google Scholar]
- Owen, D.J., Vallis, Y., Pearse, B.M., McMahon, H.T., and Evans, P.R. 2000. The structure and function of the β 2-adaptin appendage domain. EMBO J. 194216–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6458–463. [DOI] [PubMed] [Google Scholar]
- Ronning, D.R., Klabunde, T., Besra, G.S., Vissa, V.D., Belisle, J.T., and Sacchettini, J.C. 2000. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7141–146. [DOI] [PubMed] [Google Scholar]
- Shapiro, L., Kwong, P.D., Fannon, A.M., Colman, D.R., and Hendrickson, W.A. 1995. Considerations on the folding topology and evolutionary origin of cadherin domains. Proc. Natl. Acad. Sci. 926793–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov, I.N. and Bourne, P.E. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11739–747. [DOI] [PubMed] [Google Scholar]
- Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L., and Clardy, J. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229105–124. [DOI] [PubMed] [Google Scholar]
- Wiker, H.G., Harboe, M., and Nagai, S. 1991. A localization index for distinction between extracellular and intracellular antigens of Mycobacterium tuberculosis. J. Gen. Microbiol. 137875–884. [DOI] [PubMed] [Google Scholar]
- Wiker, H.G., Wilson, M.A., and Schoolnik, G.K. 2000. Extracytoplasmic proteins of Mycobacterium tuberculosis - mature secreted proteins often start with aspartic acid and proline. Microbiology 1461525–1533. [DOI] [PubMed] [Google Scholar]
- The World Health Organization. 1998. The World Health report 1998: Life in the 21st century - a vision for all. Geneva, Switzerland.
- Young, D., Garbe, T., Lathigra, R., Abou-Zeid, C., and Zhang, Y. 1991. Characterization of prominent protein antigens from mycobacteria. Bull. Int. Union Tuberc. Lung Dis. 6647–51. [PubMed] [Google Scholar]