Abstract
We have developed a method to reliably identify partial membrane protein topologies using the consensus of five topology prediction methods. When evaluated on a test set of experimentally characterized proteins, we find that approximately 90% of the partial consensus topologies are correctly predicted in membrane proteins from prokaryotic as well as eukaryotic organisms. Whole-genome analysis reveals that a reliable partial consensus topology can be predicted for ∼70% of all membrane proteins in a typical bacterial genome and for ∼55% of all membrane proteins in a typical eukaryotic genome. The average fraction of sequence length covered by a partial consensus topology is 44% for the prokaryotic proteins and 17% for the eukaryotic proteins in our test set, and similar numbers are found when the algorithm is applied to whole genomes. Reliably predicted partial topologies may simplify experimental determinations of membrane protein topology.
Keywords: Membrane protein, topology, consensus prediction
The vast majority of integral membrane proteins belong to the so-called helix bundle class, that is, their membrane domains are composed of one or more transmembrane α–helices (von Heijne 1999). Recent investigations of complete genomes estimate the fraction of genes encoding helix bundle membrane proteins as 20%–25% in most organisms (Jones 1998; Krogh et al. 2001). Several methods are currently available to predict the topology of helix bundle membrane proteins, and the best methods predict the correct global topology for approximately 65%–70% of all proteins (Ikeda et al. 2001; Möller et al. 2001a). There is thus considerable scope for further improvements in topology prediction.
We previously described how the reliability of a given topology prediction can be estimated by combining the results from five different prediction algorithms (Nilsson et al. 2000), and this approach has been used to reduce the experimental efforts required for topology mapping (Drew et al. 2002). Here, we present an extension of the consensus prediction approach to include cases where only a part of the global topology is covered by the consensus. The new partial consensus topology (PCT) prediction method provides highly reliable topology information for ∼70% of all membrane proteins encoded in a typical bacterial genome and ∼55% of all membrane proteins in a typical eukaryotic genome. Given a partial consensus topology prediction, experimental topology mapping efforts can be focused on the less reliably predicted parts of the global topology.
Results
Global and partial consensus topology predictions
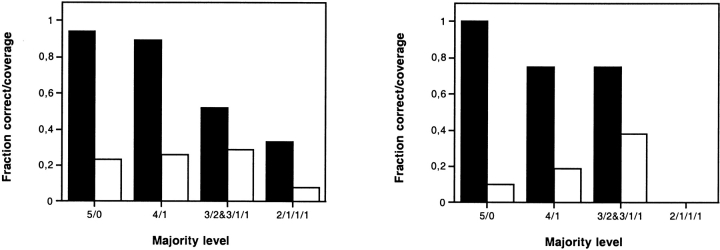
As a basis for an up–to–date comparison of the global and partial consensus topology prediction methods, we first carried out global consensus topology predictions (Nilsson et al. 2000) on new, expanded test sets of prokaryotic and eukaryotic membrane proteins with experimentally known topologies. Figure 1 ▶ shows the fraction of correctly predicted global topologies and the fraction of the test set covered for different majority levels. All five methods agreed for 17 (23%) of the 73 prokaryotic proteins in the test set, and the predicted topology was correct for 16 of these (Fig. 1 ▶, left). For the 23 eukaryotic proteins, all five methods agreed for only two proteins (9%) and both topologies were correct (Fig. 1 ▶, right). In agreement with the results from our previous study of E. coli membrane proteins (Nilsson et al. 2000), the reliability drops with increasing disagreement among the five methods.
Fig. 1.
Fraction correctly predicted global topologies (black bars) and fraction of the test sets covered (white bars) for different levels of agreement among the five prediction methods (5/0, all methods agree; 4/1, four methods agree, etc.). Left: prokaryotic proteins; right: eukaryotic proteins.
As described in Materials and Methods, PCTs were also predicted for all sequences in the two test sets, and were compared to the experimentally determined topologies (Table 1). All five methods agreed on one or more partial topologies in 62 of the prokaryotic proteins (85% of the test set), of which 57 were correct (92%). For the eukaryotic proteins, all five methods agreed on nine partial topologies (39% of the test set), of which eight were correct (89%). The average fraction of sequence covered by a PCT in the prokaryotic proteins was 44% [the fraction increases to 58% when comparing with the maximum PCT length possbile, i.e., the region between the first and last transmembrane helix (TMH) in the protein, rather than with the overall length of the protein]. The corresponding coverage values for eukaryotic proteins were 17% and 21%, respectively. We conclude that PCTs provide reliable topology information for both prokaryotic and eukaryotic proteins, but that the coverage is much smaller for the latter. This is consistent with the previously noted tendency that topology prediction is overall less reliable for eukaryotic than for prokaryotic proteins (von Heijne 1997; Ikeda et al. 2001).
Table 1.
PCT predictions for the test set proteins
| Test set | Total number of sequences | Total number of PCTs | Number of correct predictions | Fraction correct | Average fraction of sequence length covered | Average fraction of max. PCT length covered |
| Prokaryotic | 73 | 62 | 57 | 0.92 | 0.44 | 0.58 |
| Eukaryotic | 23 | 9 | 8 | 0.89 | 0.17 | 0.21 |
The average fraction of the maximum possible PCT length covered (column 7) is calculated based on the length of the region between first and last TMH in the experimental topology.
To investigate how many prediction methods are required to yield a highly reliable partial consensus topology, PCT predictions were also performed using all combinations of four of the five methods. The differences in both the fraction of correctly predicted PCTs and the average fraction of sequence length covered by PCTs were only marginal compared to the five-methods results (data not shown), and there is thus no obvious reason to reduce the number of methods included in the generation of PCTs. Because our objective here is to provide highly reliable PCTs that can be used to guide experimental work, we have not investigated the reliability of PCTs based on lower majority levels (e.g., when only four of the five methods agree on a PCT).
Predictions on entire genomes
We carried out global and partial consensus predictions for all putative multispanning membrane proteins in the genomes of eight prokaryotic and five eukaryotic organisms (putative membrane proteins were identified by the TMHMM method as detailed in Materials and Methods). Table 2 shows the fraction of membrane proteins with different majority levels for the global topology prediction. The fraction of proteins for which all five methods agree on the global topology is about 20% in the prokaryotic and about 10% in the eukaryotic genomes, in agreement with the results for the smaller test sets.
Table 2.
Fraction of predicted membrane proteins at different majority levels for 13 genomes
| Organism | 5/0 | 4/1 | 3/2 | 3/1/1 | 2/1/1/1 | No majority |
| Anabaena sp. | 0.18 | 0.21 | 0.12 | 0.17 | 0.17 | 0.15 |
| B. burgdorferi | 0.08 | 0.23 | 0.15 | 0.20 | 0.14 | 0.20 |
| H. pylori | 0.18 | 0.27 | 0.09 | 0.18 | 0.15 | 0.13 |
| M. tuberculosis | 0.20 | 0.22 | 0.11 | 0.15 | 0.16 | 0.16 |
| S. pneumoniae | 0.22 | 0.20 | 0.14 | 0.20 | 0.11 | 0.13 |
| E. coli | 0.23 | 0.24 | 0.09 | 0.16 | 0.12 | 0.16 |
| B. subtilis | 0.27 | 0.26 | 0.11 | 0.14 | 0.09 | 0.13 |
| M. jannaschii | 0.23 | 0.22 | 0.15 | 0.16 | 0.10 | 0.14 |
| S. cerevisiae | 0.07 | 0.19 | 0.09 | 0.18 | 0.22 | 0.25 |
| C. elegans | 0.13 | 0.20 | 0.09 | 0.18 | 0.20 | 0.20 |
| M. musculus | 0.11 | 0.20 | 0.11 | 0.20 | 0.19 | 0.19 |
| D. melanogaster | 0.10 | 0.16 | 0.09 | 0.18 | 0.26 | 0.21 |
| A. thaliana | 0.09 | 0.16 | 0.09 | 0.18 | 0.25 | 0.23 |
A majority level of 5/0 means that all five methods agree on the global topology, 4/1 means that four methods agree, 3/2 means that three methods agree on one global topology, and the remaining two agree on a different global topology, etc. The analyzed collection of membrane proteins with two or more TMHs in each genome was identified by TMHMM (see Materials and Methods).
PCTs were likewise predicted for all putative multispanning membrane proteins in the five genomes (Table 3). For the prokaryotic genomes, PCTs were obtained for 54%–77% of the sequences. For the eukaryotic genomes, the number was somewhat lower (45%–65%). The mean fraction of the sequences that are covered by these predictions is 23%–41% for the prokaryotic and 14%–26% for the eukaryotic genomes.
Table 3.
Partial consensus topology predictions for membrane proteins from 13 genomes
| Organism | Total number of MPs with ≥2 TMHs | Total number of PCTs | Number of MPs with ≥1 PCT | Fraction MPs with ≥1 PCT | Average fraction of sequence length covered by PCT |
| Anabaena sp. | 792 | 531 | 490 | 0.62 | 0.26 |
| B. burgdorferi | 154 | 92 | 83 | 0.54 | 0.23 |
| H. pylori | 217 | 152 | 140 | 0.65 | 0.30 |
| M. tuberculosis | 492 | 368 | 336 | 0.68 | 0.29 |
| S. pneumoniae | 365 | 290 | 263 | 0.72 | 0.35 |
| E. coli | 769 | 612 | 556 | 0.72 | 0.38 |
| B. subtilis | 828 | 680 | 640 | 0.77 | 0.41 |
| M. jannaschii | 199 | 147 | 134 | 0.67 | 0.34 |
| S. cerevisiae | 829 | 412 | 374 | 0.45 | 0.14 |
| C. elegans | 4059 | 2690 | 2470 | 0.61 | 0.26 |
| M. musculus | 3912 | 2707 | 2534 | 0.65 | 0.25 |
| D. melanogaster | 1619 | 997 | 906 | 0.56 | 0.19 |
| A. thaliana | 2892 | 1577 | 1408 | 0.49 | 0.16 |
The analyzed collection of membrane proteins with two or more TMHs in each genome was identified by TMHMM (see Materials and Methods).
Discussion
We describe here an extension of our earlier consensus method for membrane protein topology prediction (Nilsson et al. 2000), and show that it is possible to identify reliably predicted subparts of the global topology. By considering partial topologies, the fraction of proteins for which highly reliable topology information can be derived is substantially increased. Thus, highly reliable partial consensus topologies (PCTs) are predicted for around 70% of all membrane proteins in a typical prokaryotic genome, and for around 55% of all membrane proteins in a typical eukaryotic genome (Table 3).
A PCT prediction can be valuable in the context of experimental topology mapping, where it can help to identify regions in the protein where the topology is very likely to be correctly predicted, making it possible to focus the experimental efforts on the remaining, less reliably predicted, parts. The average fraction of sequence length covered by a PCT in the prokaryotic proteins of our test set is 44% (Table 1), implying that the experimental efforts may be significantly reduced for a typical prokaryotic membrane protein. For eukaryotic proteins, the corresponding fraction is much lower (17%). Similar tendencies are found when the algorithm is applied to whole genomes (Table 3).
Over our test set, the PCT predictions have a reliability of approximately 90% for both prokaryotic and eukaryotic sequences (Table 1). This is roughly the same reliability that we find for the global topology predictions when all five methods agree (Fig. 1 ▶). It is interesting to note that while it is well established that topology prediction is more difficult for eukaryotic proteins than for prokaryotic proteins (von Heijne 1997; Ikeda et al. 2001), the reliabilities of the partial consensus predictions on eukaryotic and prokaryotic proteins seem to be roughly equal (although the coverage is much smaller for the latter group). Because of the small number of eukaryotic proteins in the test set, these reliability and coverage estimates should be regarded as preliminary.
Consensus techniques have previously proven successful for, for example, globular protein fold recognition (Lundström et al. 2001) and secondary structure prediction (Cuff et al. 1998). Algorithms for consensus prediction of membrane protein topology have also been described (Promponas et al. 1999; Ikeda et al. 2001; Möller et al. 2001b). However, our method focuses more specifically on identifying global or partial topologies of high reliability and thus increases the usefulness of topology predictions.
In summary, we have shown that partial consensus topologies can be predicted with high reliability for many membrane proteins for which no global consensus topology can be predicted. Such partial topology predictions may be used to guide experimental topology determination efforts.
Materials and methods
Test set of proteins with experimentally verified topology
A nonredundant test set of multispanning membrane proteins with experimentally determined topology was extracted from the database compiled by Möller et al. (2000), from the MPtopo database (Jayasinghe et al. 2001), and from the recent literature. From the Möller database, only multispanning proteins of ‘trust levels’ A–C were included, that is, proteins for which reliable experimental topology information is available. From the MPtopo database, only multispanning proteins from the 3D_helix and 1D_helix subsets were used, that is, proteins where either the three-dimensional structures have been determined, or where the approximate positions of the transmembrane helices (TMHs) have been identified by other experimental techniques (gene fusions, proteolytic cleavages, etc.). If a sequence occurred both in the MPtopo and the Möller databases, only the entry from the Möller database was included. All proteins annotated to contain a cleavable signal peptide were removed.
The resulting test set was split into a prokaryotic subset and a eukaryotic subset. Both test sets were then homology-reduced using an implementation of the Hobohm algorithm (Hobohm et al. 1992) with a pairwise global sequence similarity threshold of 30%. ClustalW (Thompson et al. 1994) was used for the pairwise, global sequence alignments. The numbers of sequences in the final prokaryotic and eukaryotic test sets were 73 and 23, respectively (see Supplementary Information).
Genome databases
The genomes analyzed were from Anabaena sp. PCC7120 (Kaneko et al. 2001; ftp://ftp.kazusa.or.jp/pub/cyano/Anabaena/chromo/), Borrelia burgdorferi B31 (Fraser et al. 1997; ftp://ftp.tigr.org/pub/data/b_burgdorferi/), Helicobacter pylori 26695 (Tomb et al. 1997; ftp://ftp.tigr.org/pub/data/h_pylori/), Mycobacterium tuberculosis CDC1551 (Cole et al. 1998; ftp://ftp.tigr.org/pub/data/m_tuberculosis/), Salmonella pneumoniae (Tettelin et al. 2001; ftp://ftp.tigr.org/pub/data/s_pneumoniae/), Mus musculus (ftp://ftp.ensembl.org/pub/current_mouse/data/fasta/pep/), Drosophila melanogaster (Adams et al. 2000; ftp://ftp.ncbi.nih.gov/genbank/genomes/D_melanogaster/), Arabidopsis thaliana (Theologis et al. 2000; ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/), Escherichia coli (Blattner et al. 1997; http://bmb.med.miami.edu/EcoGene/EcoWeb/), Saccharomyces cerevisiae (ftp://genome-ftp.stanford.edu/pub/yeast/yeast_ORFs/), Caenorhabditis elegans (Stein et al. 2001; (ftp://ftp.sanger.ac.uk/pub/wormbase/),Bacillus subtilis 168 (Kunst et al. 1997; ftp://ftp.pasteur.fr/pub/GenomeDB/SubtiList/), and Methanococcus jannaschii DSM2661 (Bult et al. 1996; ftp://ftp.tigr.org/pub/data/m_jannaschii/). For each genome, TMHMM2.0 (Sonnhammer et al. 1998) was used to identify putative membrane proteins with a minimum of two predicted TMHs. The resulting data sets were then analyzed by the PCT prediction procedure described below.
Prediction methods
Five topology prediction methods—TMHMM2.0 (Sonnhammer et al. 1998; Krogh et al. 2001), HMMTOP2.0 (Tusnady and Simon 1998Tusnady and Simon 2001), MEMSAT1.8 (Jones et al. 1994), PHD2.1 (Rost et al. 1996), and TOPPRED1.0 (von Heijne 1992; Claros and von Heijne 1994)—were used in their single-sequence mode (i.e., no information from homologous proteins was included). All methods produce a prediction of both the number and location of the TMHs, and the in/out location of the N-terminus relative to the membrane. All user-adjustable parameters were kept at their default values, with the exception of TOPPRED predictions for eukaryotic proteins, where the organism parameter was set to ‘eukaryote.’ The output from the different topology prediction programs was converted into a standard format for further analysis.
Partial consensus topology prediction algorithm
The partial consensus topology prediction method is based on our previous observation that the reliability of a predicted topology can be estimated from the number of prediction methods that agree on the global topology (i.e., that give the same number of predicted TMHs and the same predicted orientation for the N–terminus). Specifically, that study (Nilsson et al. 2000) indicated that very high reliability can be assigned to topologies where five different prediction methods give the same prediction.
Here, we tested the assumption that this relationship holds also for cases where all five prediction methods agree on the topology of only a part of the protein. These cases are referred to as partial consensus topologies (PCTs).
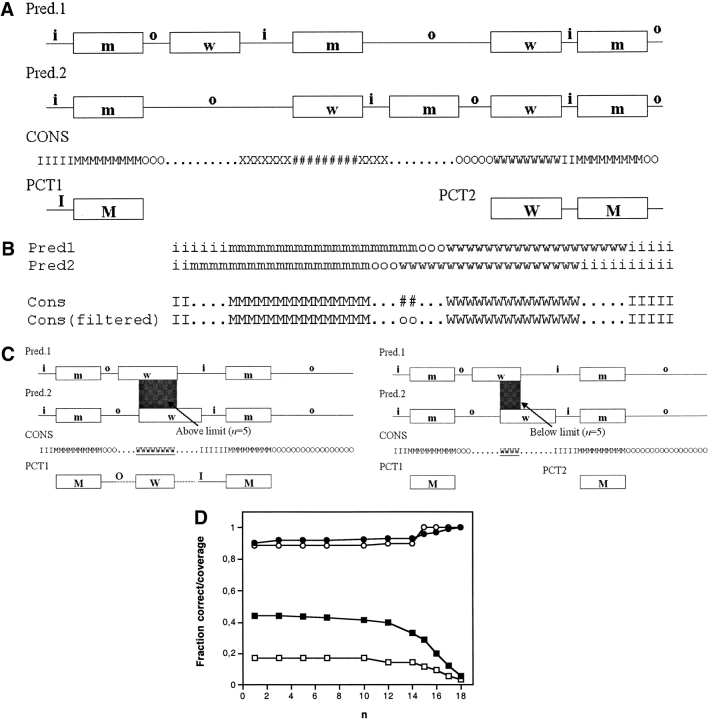
The PCT algorithm is described in Figure 2A ▶. In the first step, if all methods agree on the topology at a certain position in the sequence, a consensus topology prediction is assigned to this position (inside loop, outside loop, in–to–out helix, and out–to–in helix states are designated i, o, m, and w, respectively). If all methods do not agree at a certain position, no consensus is assigned (designated ‘.’). To aid in the construction of the final partial consensus prediction, we define two additional symbols that represent positions for which the predicted topology states are incompatible with each other. Thus, when loop states with opposite locations (i and o) are predicted at the same position, we define this as a loop clash (X). In the same manner, a TMH clash (#) is defined for positions where two TMHs with opposite directions (m and w) are predicted.
Fig. 2.
Partial consensus topology prediction procedure (for simplicity shown only for two prediction methods). (A) For each residue position, a consensus topology state (i, o, m, w) is assigned only if all five prediction methods agree. Otherwise, no consensus is assigned (designated as ‘.’). If two loops with opposite locations (o and i) appear at the same position, a loop clash (X) is assigned. In the same manner, a TMH clash (#) is assigned when two TMHs with opposite orientations (m and w) appear at the same position. After the filtering step described in B, the consensus topology is scanned from the N–terminus, and the beginning of the first PCT is defined by the first TM region (m or w states) of at least n consecutive residues in the consensus topology (where the default value is n = 5). The PCT is then extended towards the C–terminus until either a consensus TMH of less than n consecutive residues is encountered, or a loop– or TM–clash occurs. In either case, the end of the PCT is defined by the most C–terminally located TMH in the consensus topology. The process of PCT construction is then repeated until the C-terminal end of the protein is reached. (B) Removal of TMH clashes which are due to slight misalignments of the TMHs. When a TMH clash occurs between two partially overlapping consensus TMHs with opposite orientations, the clash state is replaced by a loop state of the appropriate type. To emphasize that the resulting loop was obtained in the filtering procedure (and not in the majority–vote procedure), it is indicated in lowercase. (C) The figure illustrates how the choice of n–value affects the PCT prediction results. The result can differ substantially when a consensus TMH that is longer (left) or shorter (right) than n is encountered. (D) Average fraction of sequence length covered by PCTs (squares) and average fraction of correctly predicted PCTs (circles) for different values of n for the prokaryotic (black symbols) and eukaryotic (white symbols) test sets.
After this initial step, a filtering procedure is used to remove “spurious” TMH clashes caused by slight misalignments of predicted TMHs (Fig. 2B ▶). In this procedure, a TMH clash which is flanked by consensus TMHs with opposite directions is replaced by a loop state. Such clashes occur frequently for proteins containing closely spaced TMHs.
The final step is the construction of the partial consensus topology (Fig. 2A ▶). Starting from the N-terminus of the protein, the N-terminal end of the first PCT is defined by the first TMH (m or w states) of at least n residues in the consensus topology (where n is an adjustable parameter; the default value used here is n = 5). The PCT is then extended towards the C–terminus until either a consensus TMH of less than n residues is encountered, or a loop clash or TMH clash occurs. In either case, the end of the PCT is defined by the most C–terminally located m or w state in the consensus. The process is then repeated until the C–terminal end of the protein is reached. A protein may thus contain more than one PCT.
The significance of the n–value is illustrated in Figure 2C ▶, where the resulting PCT prediction differs depending on whether the consensus TMH is longer or shorter than the value of n.
To be included in a PCT, a consensus TMH has to be at least a minimum number of residues n in length. The larger the n–value, the smaller the risk that an incorrectly predicted consensus TMH is included in the PCT. However, a high n–value also decreases the average length of a PCT. To determine the optimal n–value, the evaluation step above was performed for different length thresholds. Figure 2D ▶ shows the fraction of correctly predicted PCTs and the average fraction of sequence length covered by a PCT for different values of n. For the prokaryotic proteins, both the fraction of sequence length covered and the fraction of correctly predicted PCTs is relatively constant for n = 1–12. For n > 12 residues, the fraction of sequence length covered drops significantly, whereas there is only a minor increase in the fraction of correct PCTs. The trend is basically the same for the eukaryotic proteins, though we consider these results less reliable because of the small test set. In summary, the results do not vary appreciably for n–values < 10, and the default value n = 5 has been used for all results reported here.
Method evaluation
To assess the performance of the PCT prediction algorithm, it was applied to the prokaryotic and eukaryotic test sets of proteins with experimentally determined topologies described above. For a given PCT, the corresponding region in the experimentally determined topology was checked, and if both the number and directions of TMHs in this region agreed with the PCT, the prediction was considered to be correct.
Electronic supplemental material
A list of the proteins included in the training set is provided as Supplementary Material. Additional data describing, for each test set protein, the number of transmembrane helices and the in/out location of the N–terminus are available as well.
Acknowledgments
This work was supported by grants from the Foundation for Strategic Research, the Swedish Research Council, and Karolinska Institutet.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
PCT, partial consensus topology
TMH, transmembrane helix
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0226702.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277 1453–1474. [DOI] [PubMed] [Google Scholar]
- Bult, C.J., White, O., Olsen, G.J., Zhou, L., Fleischmann, R.D., Sutton, G.G., Blake, J.A., FitzGerald, L.M., Clayton, R.A., Gocayne, J.D., et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273 1058–1073. [DOI] [PubMed] [Google Scholar]
- Claros, M.G. and von Heijne, G. 1994. TopPred II: An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10 685–686. [DOI] [PubMed] [Google Scholar]
- Cole, S.T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S.V., Eiglmeier, K., Gas, S., Barry III, C.E., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 537–544. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M., and Barton, G.J. 1998. JPred: A consensus secondary structure prediction server. Bioinformatics 14 892–893. [DOI] [PubMed] [Google Scholar]
- Drew, D., Sjöstrand, D., Nilsson, J., Urbig, T., Chin, C., de Gier, J.W., and von Heijne, G. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. 99 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, C.M., Casjens, S., Huang, W.M., Sutton, G.G., Clayton, R., Lathigra, R., White, O., Ketchum, K.A., Dodson, R., Hickey, E.K., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390 580–586. [DOI] [PubMed] [Google Scholar]
- Hobohm, U., Scharf, M., Schneider, R., and Sander, C. 1992. Selection of representative protein data sets. Protein Sci. 1 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, M., Arai, M., Lao, D., and Shimizu, T. 2001. Transmembrane topology prediction methods: A re-assessment and improvement by a consensus method using a dataset of experimentally-characterised transmembrane topology. In Silico Biol. 2 1–15. [PubMed] [Google Scholar]
- Jayasinghe, S., Hristova, K., and White, S.H. 2001. MPtopo: A database of membrane protein topology. Protein Sci. 10 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. 1998. Do transmembrane protein superfolds exist? FEBS Lett. 423 281–285. [DOI] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33 3038–3049. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., Nakamura, Y., Wolk, C.P., Kuritz, T., Sasamoto, S., Watanabe, A., Iriguchi, M., Ishikawa, A., Kawashima, K., Kimura, T., et al. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8 227–253. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- Kunst, F., Ogasawara, N., Moszer, I., Albertini, A.M., Alloni, G., Azevedo, V., Bertero, M.G., Bessieres, P., Bolotin, A., Borchert, S., et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390 249–256. [DOI] [PubMed] [Google Scholar]
- Lundström, J., Rychlewski, L., Bujnicki, J., and Elofsson, A. 2001. Pcons: A neural-network-based consensus predictor that improves fold recognition. Protein Sci. 10 2354–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller, S., Kriventseva, E.V., and Apweiler, R. 2000. A collection of well characterised integral membrane proteins. Bioinformatics 16 1159–1160. [DOI] [PubMed] [Google Scholar]
- Möller, S., Croning, M., and Apweiler, R. 2001a. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17 646–653. [DOI] [PubMed] [Google Scholar]
- Möller, S., Schroeder, M., and Apweiler, R. 2001b. Consistent integration of non-reliable heterogeneous information resources applied to the annotation of transmembrane proteins. Comput. Chem. 26 41–49 [DOI] [PubMed] [Google Scholar]
- Nilsson, J., Persson, B., and von Heijne, G. 2000. Consensus predictions of membrane protein topology. FEBS Lett. 486 267–269. [DOI] [PubMed] [Google Scholar]
- Promponas, V.J., Palaios, G.A., Pasquier, C.M., Hamodrakas, J.S., and Hamodrakas, S.J. 1999. CoPreTHi: A Web tool which combines transmembrane protein segment prediction methods. In Silico Biol. 1 159–162. [PubMed] [Google Scholar]
- Rost, B., Fariselli, P., and Casadio, R. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5 1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E., von Heijne, G., and Krogh, A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6 175–182. [PubMed] [Google Scholar]
- Stein, L., Sternberg, P., Durbin, R., Thierry-Mieg, J., and Spieth J. 2001. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 29 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin, H., Nelson, K.E., Paulsen, I.T., Eisen, J.A., Read, T.D., Peterson, S., Heidelberg, J., DeBoy, R.T., Haft, D.H., Dodson, R.J., et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293 498–506. [DOI] [PubMed] [Google Scholar]
- Theologis, A., Ecker, J.R., Palm, C.J., Federspiel, N.A., Kaul, S., White, O., Alonso, J., Altafi, H., Araujo, R., Bowman, C.L., et al. 2000. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408 816–829. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb, J.F., White, O., Kerlavage, A.R., Clayton, R.A., Sutton, G.G., Fleischmann, R.D., Ketchum, K.A., Klenk, H.P., Gill, S., Dougherty, B.A., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388 539–547. [DOI] [PubMed] [Google Scholar]
- Tusnady, G.E. and Simon, I. 1998. Principles governing amino acid composition of integral membrane proteins: Application to topology prediction. J. Mol. Biol. 283 489–506. [DOI] [PubMed] [Google Scholar]
- ———. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17 849–850. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225 487–494. [DOI] [PubMed] [Google Scholar]
- ———. 1997. Principles of membrane protein assembly and structure. Progr. Biophys. Mol. Biol. 66 113–139. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Recent advances in the understanding of membrane protein assembly and structure. Q. Rev. Biophys. 32 285–307. [DOI] [PubMed] [Google Scholar]