Abstract
Identifying mechanisms of pathogen transmission is critical to controlling disease. Social organization should influence contacts among individuals and thus the distribution and spread of disease within a population. Molecular genetic markers can be used to elucidate mechanisms of disease transmission in wildlife populations without undertaking detailed observational studies to determine probable contact rates. Estimates of genealogical relationships within a bovine tuberculosis-infected white-tailed deer (Odocoileus virginianus) population indicated that infected deer were significantly more closely related than non-infected deer suggesting that contact within family groups was a significant mechanism of disease transmission. Results demonstrate that epidemiological models should incorporate aspects of host ecology likely to affect the probability of disease transmission.
Keywords: bovine tuberculosis, ecology, microsatellites, wildlife, white-tailed deer, zoonoses
1. Introduction
Emerging infectious diseases present a challenge to the scientific community, management agencies, policy makers and the public. Diseases including severe acute respiratory syndrome, west nile virus encephalitis and avian influenza have focused public attention on interrelationships between wildlife, domestic animal and human health. Understanding factors responsible for disease transmission and distribution is critical to controlling disease. Traditionally, disease transmission dynamics have been assumed to be random and to increase proportionally with population size. However, complex social behaviours can result in contacts among infected and susceptible individuals that are not solely explained by density (Altizer et al. 2004). Therefore, social organization should influence contacts among individuals, and the distribution and spread of disease. Molecular genetic markers are ideal tools to elucidate the role of genealogical relationships in disease transmission in wildlife populations without undertaking detailed observational studies.
The emergence of bovine tuberculosis (TB, Mycobacterium bovis) in free-ranging white-tailed deer (Odocoileus virginianus) in the northeast lower peninsula of Michigan also threatens domestic animals and humans, and has resulted in multi-million dollar losses and intensive efforts to eliminate the disease from deer (Schmitt et al. 1997; Bucholz 2005). Bovine TB is spread through direct contact among animals (Thoen & Himes 1981) and indirect contact with contaminated food (Palmer et al. 2004). Until 1999, feeding resulted in large aggregations of deer leading to high rates of animal contact probably facilitating disease transmission. Cessation of feeding and increased harvest was enacted to reduce deer density and contacts among animals, common management strategies for controlling disease. These actions will not be sufficient to eliminate disease, however, if host behaviours result in non-random contacts and thus non-random disease transmission (McCallum et al. 2001).
Female white-tailed deer live in related groups (matrilines) and contact relatives more frequently than non-relatives (Hawkins & Klimstra 1970; Kie & Bowyer 1999). Owing to group fidelity and high contact rates within groups, TB transmission should occur more frequently among relatives. We used molecular genetic markers to estimate relatedness among individuals to test the hypothesis that TB-infected deer would be more closely related than non-infected deer.
2. Material and methods
We collected skeletal tissue from deer harvested from September to December and 1998 and 2000. We estimated genealogical relationships by genotyping all TB-infected deer harvested in a five-county region of northeast Michigan whose harvest location was identified to 2.6 km2 grids (67 deer in 1998; 35 deer in 2000). In order to more accurately characterize population allele frequencies, we also genotyped numerous non-infected deer (574 deer in 1998; 253 deer in 2000).
We extracted DNA using the QIAGEN DNEasy protocol and genotyped samples at 11 microsatellite loci (BL42, BM4107, ETH152, (Talbot et al. 1996); Cervid-1, Cervid-2 (DeWoody et al. 1995); OarFCB193 (Buchanan & Crawford 1993); RT-20, RT-23, RT-24, RT-27 (Wilson et al. 1997); SRCRSP-10 (Bhebhe et al. 1994)) using the polymerase chain reaction. Products were separated using gel electrophoresis on 6% denaturing polyacrylamide gels and visualized using a Hitachi FMBIO-II laser scanner. We assigned genotypes with reference to base pair standards and individuals of known genotype. We used exact tests in Genepop (Raymond & Rousset 1995) to confirm that genotype frequencies were consistent with Hardy–Weinberg and linkage equilibrium.
We calculated relatedness between all pairs of TB-infected deer and an equal number of randomly chosen non-infected deer whose spatial distribution was the same as that of the TB-infected deer. Queller & Goodnight's (1989) coefficient of relatedness (rxy) ranges from −1 to +1 with a value of 0 equal to a pair that is no more or less related than average relatedness in the population. We computed the difference between mean rxy for infected and non-infected deer using non-parametric permutation tests (Wayne et al. 1991; A. Saxton, University of Tennessee, unpublished SAS-based software). Data were pooled and randomly subsampled 500 times. For each subsampling, we calculated the difference between observed and theoretical mean relatedness, and tested the one-tailed probability that average relatedness was higher among TB-infected than non-infected deer.
In order to evaluate the importance of geographical proximity (i.e. social group contact) to relatedness among TB-infected versus non-infected deer, we calculated relatedness among deer harvested in the same location and among deer separated by increasing distances. Mean relatedness was compared among TB-infected and non-infected deer grouped into distance classes (range 0–40 km). Because there were more non-infected deer than TB-infected deer, the differences in relatedness between TB-infected and non-infected deer were evaluated by bootstrap re-sampling (Manly 1997). For each distance class, we calculated mean relatedness among a randomly selected number of non-infected deer equal to the number of TB-infected deer. This process was repeated 5000 times. We estimated the probability that non-infected deer were more closely related than TB-infected deer as the number of times mean relatedness among non-infected deer was higher than among TB-infected deer divided by 5000 (Manly 1997). We evaluated the difference in relatedness from 1998 to 2000 among TB-infected deer harvested in the same location as the number of times relatedness between pairs of TB-infected deer in 1998 was greater than between pairs in 2000 divided by the total number of pairs.
3. Results
TB prevalence was similar in 1998 (0.95%) and 2000 (0.84%). In both years, we found that the mean relatedness among TB-infected deer was significantly higher than non-infected deer (1998: rxy TB-infected=0.028 (±0.010), rxy non-infected=−0.015 (±0.009), p<0.001; 2000: rxy TB-infected=0.014 (±0.025), rxy non-infected=−0.009 (±0.018), p<0.001). The lower relatedness among TB-infected deer in 2000 was not significant (p=0.758) and was probably due to the broader spatial distribution over which TB-infected individuals were harvested in 2000 (6931 km2) relative to 1998 (3364 km2).
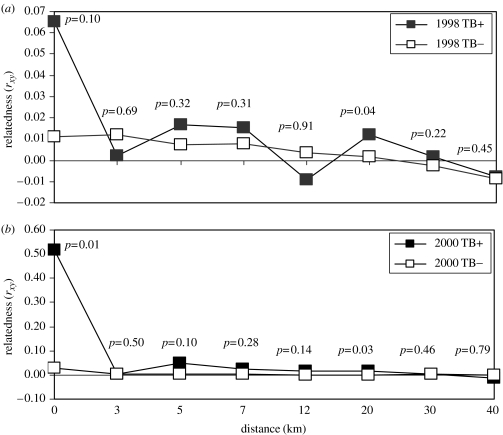
We found that both prior to (1998) and following (2000) the feeding ban and increased harvest, mean relatedness was higher among TB-infected deer than non-infected deer harvested in the same location (within 2.6 km2; figure 1). In addition, mean relatedness among TB-infected deer harvested in the same location was higher in 2000 than in 1998 (p=0.09). For animals harvested at distances exceeding 2.6 km apart, relatedness among non-infected and TB-infected animals was similar with the exception of the 20 km distance class in which the difference was small but significant (figure 1).
Figure 1.
Mean relatedness (rxy; Queller & Goodnight 1989) among TB-infected (filled squares) and non-infected (open squares) white-tailed deer separated by increasing distance prior to (a) (1998) and following (b) (2000) efforts to reduce deer density and ban feeding (note scales on y-axes). p-values indicate the proportion of times mean relatedness among 5000 random samples of non-infected deer was higher than among TB-infected deer.
4. Discussion
Our results suggest that contacts among related deer within social groups contributed to TB transmission. The importance of contacts among relatives was evidenced by the higher relatedness among TB-infected deer in close proximity in 2000 relative to 1998 (figure 1). The largest difference in relatedness among TB-infected animals and non-infected animals occurred for animals that were in close spatial proximity (i.e. the scale of the social group). Relatedness among TB-infected animals harvested from separate spatial locations, and probably from different social groups, was similar to relatedness among non-infected animals. This conclusion is supported by the documentation of significant spatial genetic structuring among groups of deer following the feeding ban (Blanchong et al. 2006), resulting from increased spatial segregation of social groups typical of deer populations (Mathews & Porter 1993).
We cannot exclude the possibility that higher relatedness among TB-infected deer may be related to genetic resistance. Mackintosh et al. (2000) demonstrated that genetic resistance to experimental inoculation with TB in red deer is heritable. The role of genetic resistance to infection risk in the deer population warrants further exploration. However, contact among animals appears to be the mechanism by which susceptible individuals become infected. Our findings suggest that the contacts that occur within social groups (of related individuals) are an important mechanism of disease transmission.
Complex behaviours and social systems play important roles in pathogen transmission (Altizer et al. 2003). Failure to account for life-history characteristics related to transmission can impede control efforts (Hutchings & White 2000; Woodroffe et al. 2006). For example, badger (Meles meles) culling in the UK aimed at reducing TB disrupted badger social structure and increased TB in cattle (Donnelly et al. 2003). Understanding factors responsible for TB transmission among white-tailed deer in Michigan is a high priority in order to control the disease, protect human and domestic animal health and alleviate economic loss. Standard observational or telemetry methods were not sufficient to resolve relationships among large numbers of individuals in this population and assess the role of genealogical relationship to disease risk. Molecular genetic markers can be used to identify the mechanisms of disease transmission without having to undertake detailed, individual-level fieldwork. Our results demonstrate that epidemiological models should incorporate aspects of host social organization likely to affect disease transmission (Wobeser 2002).
Acknowledgments
The Wildlife Division of the Michigan Department of Natural Resources through the Federal Aid in Wildlife Restoration Act under Pittman-Robertson project W-129-R-17 and the United States Department of Agriculture provided financial support. We thank Wildlife Division personnel for database assistance, the Michigan Animal Health Diagnostic Laboratory, particularly Dr S. Fitzgerald, for use of facilities during sample collection, and numerous Michigan State University undergraduates that collected tissues. We acknowledge the technical assistance of S. Libants and J. Warrillow. Comments by B. Epperson, D. Joly, J. Liu, J. Tsao and N. Walker, and two anonymous referees who improved the manuscript.
References
- Altizer S, et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst. 2003;34:517–547. doi:10.1146/annurev.ecolsys.34.030102.151725 [Google Scholar]
- Altizer S, Davis A.K, Cook K.C, Cherry J.J. Age, sex, and season affect the risk of mycoplasmal conjunctivitis in a southeastern house finch population. Can. J. Zool. 2004;82:755–763. doi:10.1139/z04-050 [Google Scholar]
- Bhebhe E, Kogi J, Holder D, Arevalo E, Derr J.N, Linn R.A, Ruvuna F, Davis S.K, Taylor J.F. Caprine microsatellite dinucleotide repeat polymorphisms at the SR-CRSP-6, SR-CRSP-7, SR-CRSP-8, SR-CRSP-9 and SR-CRSP-10 loci. Anim. Gen. 1994;25:203. doi: 10.1111/j.1365-2052.1994.tb00125.x. [DOI] [PubMed] [Google Scholar]
- Blanchong J.A, Scribner K.T, Epperson B.K, Winterstein S.R. Changes in artificial feeding regulations impact white-tailed deer fine-scale spatial genetic structure. J. Wildl. Manag. 2006;70:1037–1043. doi:10.2193/0022-541X(2006)70[1037:CIAFRI]2.0.CO;2 [Google Scholar]
- Buchanan F.C, Crawford A.M. Ovine microsatellites at the OarFCB11, OarFCB128, OarFCB193, OarFCB266 and OarFCB304 loci. Anim. Gen. 1993;24:145. doi: 10.1111/j.1365-2052.1993.tb00269.x. [DOI] [PubMed] [Google Scholar]
- Bucholz T.J. Michigan Department of Community Health; Lansing, MI: 2005. Bovine TB strain confirmed in Michigan hunter. [Google Scholar]
- DeWoody J.A, Honeycutt R.L, Skow L.C. Microsatellite markers in white-tailed deer. J. Hered. 1995;86:317–319. doi: 10.1093/oxfordjournals.jhered.a111593. [DOI] [PubMed] [Google Scholar]
- Donnelly C.A, Woodroffe R, Cox D.R, Bourne J, Gettinby G, LeFever A.M, McInerney J.P, Morrison W.I. Impact of localized badger culling on tuberculosis incidence in British cattle. Nature. 2003;426:834–837. doi: 10.1038/nature02192. doi:10.1038/nature02192 [DOI] [PubMed] [Google Scholar]
- Hawkins R.E, Klimstra W.D. A preliminary study of the social organization of white-tailed deer. J. Wildl. Manag. 1970;34:407–419. [Google Scholar]
- Hutchings M.R, White P.C.L. Mustelid scent-marking in managed ecosystems: implications for population management. Mamm. Rev. 2000;30:157–169. doi:10.1046/j.1365-2907.2000.00065.x [Google Scholar]
- Kie J.G, Bowyer R.T. Sexual segregation in white-tailed deer: density-dependent changes in use of space, habitat selection, and dietary niche. J. Mammol. 1999;80:1004–1020. doi:10.2307/1383271 [Google Scholar]
- Mackintosh C.G, Qureshi T, Waldrup K, Labes R.E, Dodds K.G, Griffin J.F.T. Genetic resistance to experimental infection with Mycobacterium bovis in red deer (Cervus elaphus) Infect. Immun. 2000;68:1620–1625. doi: 10.1128/iai.68.3.1620-1625.2000. doi:10.1128/IAI.68.3.1620-1625.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly B.F.J. Chapman and Hall; London, UK: 1997. Randomization, bootstrap and Monte Carlo methods in biology. [Google Scholar]
- Mathews N.E, Porter W.F. Effect of social structure on genetic structure of free-ranging white-tailed deer in the Adirondack mountains. J. Mammol. 1993;74:33–43. doi:10.2307/1381903 [Google Scholar]
- McCallum H, Barlow N, Hone J. How should pathogen transmission be modelled? Trends Ecol. Evol. 2001;16:295–300. doi: 10.1016/s0169-5347(01)02144-9. doi:10.1016/S0169-5347(01)02144-9 [DOI] [PubMed] [Google Scholar]
- Palmer M.V, Waters W.R, Whipple D.L. Shared feed as a means of deer-to-deer transmission of Mycobacterium bovis. J. Wildl. Dis. 2004;40:87–91. doi: 10.7589/0090-3558-40.1.87. [DOI] [PubMed] [Google Scholar]
- Queller D.C, Goodnight K.F. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. doi:10.2307/2409206 [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2). Population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Schmitt S.M, et al. Bovine tuberculosis in free-ranging white-tailed deer from Michigan. J. Wildl. Dis. 1997;33:749–758. doi: 10.7589/0090-3558-33.4.749. [DOI] [PubMed] [Google Scholar]
- Talbot J, Haigh J, Plante Y. A parentage evaluation test in North American elk (wapiti) using microsatellites of ovine and bovine origin. Anim. Gen. 1996;27:117–119. doi: 10.1111/j.1365-2052.1996.tb00480.x. [DOI] [PubMed] [Google Scholar]
- Thoen C.O, Himes E.M. Tuberculosis. In: Davis J.W, editor. Infectious diseases of wild mammals. Iowa State University Press; Ames, IA: 1981. pp. 263–274. [Google Scholar]
- Wayne R.K, et al. Conservation genetics of the endangered Isle Royale gray wolf. Conserv. Biol. 1991;5:41–51. doi:10.1111/j.1523-1739.1991.tb00386.x [Google Scholar]
- Wilson G.A, Strobeck C, Wu L, Coffin J.W. Characterization of microsatellite loci in caribou, Rangifer tarandus, and their use in other artiodactyls. Mol. Ecol. 1997;6:697–699. doi: 10.1046/j.1365-294x.1997.00237.x. doi:10.1046/j.1365-294X.1997.00237.x [DOI] [PubMed] [Google Scholar]
- Wobeser G. Disease management strategies for wildlife. Rev. Sci. Tech. Off. Init. Epiz. 2002;21:159–178. doi: 10.20506/rst.21.1.1326. [DOI] [PubMed] [Google Scholar]
- Woodroffe R, Donnelly C.A, Cox D.R, Bourne F.J, Cheeseman C.L, Delahay R.J, Gettinby G, McInery J.P, Morrison W.I. Effects of culling on badger Meles meles spatial organization: implications for the control of bovine tuberculosis. J. Appl. Ecol. 2006;43:1–10. [Google Scholar]