Abstract
Synapsin I is a synaptic vesicle-associated phosphoprotein that has been implicated in the formation of presynaptic specializations and in the regulation of neurotransmitter release. The nonreceptor tyrosine kinase c-Src is enriched on synaptic vesicles, where it accounts for most of the vesicle-associated tyrosine kinase activity. Using overlay, affinity chromatography, and coprecipitation assays, we have now shown that synapsin I is the major binding protein for the Src homology 3 (SH3) domain of c-Src in highly purified synaptic vesicle preparations. The interaction was mediated by the proline-rich domain D of synapsin I and was not significantly affected by stoichiometric phosphorylation of synapsin I at any of the known regulatory sites. The interaction of purified c-Src and synapsin I resulted in a severalfold stimulation of tyrosine kinase activity and was antagonized by the purified c-Src-SH3 domain. Depletion of synapsin I from purified synaptic vesicles resulted in a decrease of endogenous tyrosine kinase activity. Portions of the total cellular pools of synapsin I and Src were coprecipitated from detergent extracts of rat brain synaptosomal fractions using antibodies to either protein species. The interaction between synapsin I and c-Src, as well as the synapsin I-induced stimulation of tyrosine kinase activity, may be physiologically important in signal transduction and in the modulation of the function of axon terminals, both during synaptogenesis and at mature synapses.
Keywords: synaptic vesicles, SH3 domains, synapses
Synapsin I is the major synaptic vesicle protein that binds to the Src homology 3 (SH3) domains of the adapter protein Grb2 in vitro (1). This interaction is mediated by domain D of synapsin I, a 23-kDa proline-rich, strongly basic domain located in the COOH-terminal portion of synapsin I (2). It seemed possible that domain D or other proline-rich regions in synapsin I might interact with other SH3 domain-containing proteins within the nerve terminal and that these interactions might have a physiological role in presynaptic function. One such candidate, the SH3 domain-containing nonreceptor tyrosine kinase c-Src, is expressed at high levels in postmitotic neurons and is enriched on synaptic vesicles, where it accounts for most of the vesicle-associated tyrosine kinase activity (3–6). Using purified components in vitro, we now report that a domain D-mediated interaction of synapsin I with the SH3 domain of c-Src results in a 5-fold activation of the catalytic activity of c-Src. We also present studies employing a variety of biochemical techniques that serve to further characterize the specificity, subcellular location, regulation, and potential functional consequences of the c-Src/synapsin I interaction.
MATERIALS AND METHODS
Materials.
[γ-32P]ATP (>3,000 Ci/mmol; 1 Ci = 37 GBq) was obtained from Amersham. Antibodies were obtained from the following sources: anti-Src clone 327 monoclonal antibody (mAb), Calbiochem–Oncogene Research Products (Cambridge, MA); peroxidase-conjugated secondary antibodies, Bio-Rad; and 125I-labeled anti-mouse and anti-rabbit IgGs (Fab fragments), Amersham. Antibodies against synapsin I, dynamin, synaptojanin, and glutathione S-transferase (GST) have been described (1, 7). The SH3 domain fusion protein constructs were generously provided by the following individuals: pGEX-c-Src-SH3 domain (8), I. Gout (Ludwig Institute, London); pGEX-n-Src-SH3 domain (9), J. S. Brugge (Ariad Pharmaceuticals, Cambridge, MA); pGEX-Grb2 (10), J. Schlessinger (New York University Medical Center, New York). Purified recombinant human c-Src (900,000 units/mg) was purchased from Upstate Biotechnology (Lake Placid, NY). Protein G–Sepharose, glutathione–Sepharose, and pGEX-2T were obtained from Pharmacia. The tyrosine kinase substrate, poly(Glu-80,Tyr-20), and 2-nitro-5-thiocyanobenzoic acid (NTCB) were obtained from Sigma.
Protein Purification and Subcellular Fractionation.
Bacterial cells were transformed to ampicillin resistance with pGEX-2T alone or pGEX-2T-fusion protein constructs containing SH3 domains by electroporation. Large-scale cultures of Luria–Bertani medium containing ampicillin (100 μg/ml) were inoculated with small overnight cultures grown at 37°C and induced with isopropyl β-d-thiogalactopyranoside (100 μM) for 3.5 hr. GST and GST-fusion proteins were extracted from bacterial lysates and purified to homogeneity by affinity chromatography on glutathione–Sepharose as described (11). Synapsin I was purified from bovine brain as described (12). Purified synapsin I was stoichiometrically phosphorylated in vitro using purified protein kinases (13–15) or was subjected to cysteine-specific chemical cleavage with NTCB as described (15). Subcellular fractions were prepared from rat forebrain, and synaptic vesicles were purified through the step of controlled-pore glass chromatography as described (16). Aliquots of purified synaptic vesicles were exposed to 200 mM NaCl treatment, which results in quantitative removal of endogenous synapsin I (16) but only partial depletion (≈50%) of synapsin II (17). Both untreated and salt-treated synaptic vesicles (USV and STSV, respectively) were resuspended in 0.3 M glycine/5 mM Hepes⋅NaOH, pH 7.4, at a protein concentration of 1.5–2 mg/ml.
SH3 Domain Overlay Assays.
Proteins of subcellular fractions of rat brain, purified synapsin I, and NTCB cleavage fragments were separated by SDS/PAGE and transferred to nitrocellulose membranes. Membranes were incubated for 1 hr at room temperature in blocking buffer [150 mM NaCl/25 mM Tris⋅Cl, pH 7.4/5% (wt/vol) nonfat dry milk], rinsed with Tris-buffered saline (150 mM NaCl/25 mM Tris⋅Cl, pH 7.4), and incubated overnight at 4°C in overlay buffer [3% (wt/vol) BSA/20 mM Tris⋅Cl, pH 7.4/1 mM DTT/0.1% (vol/vol) Tween 20] containing 5 μg/ml of GST or GST-SH3 domain fusion protein. The membranes were washed, incubated for 2 hr with an anti-GST polyclonal antibody, washed, incubated for 1 hr with 125I-labeled anti-rabbit IgG secondary antibodies, washed, and subjected to autoradiography.
SH3 Domain-Binding Assays and Affinity Chromatography.
The binding of GST or GST-SH3 domains to purified synapsin I was assessed by coprecipitation experiments using glutathione–Sepharose essentially as described (1). Affinity resins for the isolation of SH3 domain-binding proteins from brain extracts were prepared by immobilizing either GST or GST-SH3 domain fusion proteins (300 μg protein) on glutathione–Sepharose (100 μl settled beads) by an overnight incubation at 4°C in binding buffer under gentle rotation in small columns. Columns were extensively washed (20-bed volumes) with binding buffer (1) and loaded with a 1% (vol/vol) Triton X-100 extract from an 8-mg sample of crude synaptosomal fraction (P2) of rat cerebral cortex. Loading was performed for 2 hr at 4°C under gentle rotation and was followed by extensive washings with binding buffer and then with detergent-free binding buffer. Elution of the bound proteins was performed with Laemmli sample buffer. Samples were then separated by SDS/PAGE and analyzed by Coomassie blue staining of the gels or by immunoblot assay.
Src Kinase Phosphorylation Assays.
Purified c-Src (3 units per sample) was incubated for 15 min at 30°C in phosphorylation buffer [50 mM Tris⋅Cl, pH 7.4/10 mM MnCl2/1 mM EGTA/0.2 mM Na3VO4/40 mM Mg acetate/1 mM DTT/50 μM [γ-32P]ATP (1.5 μCi per sample)] with 10 μg per sample of poly(Glu-80,Tyr-20) in the absence or presence of increasing concentrations of synapsin I (20–400 nM). In some samples, GST-c-Src-SH3 domain fusion protein (0.2–4 μM) was also present. 32P incorporation into poly(Glu-80,Tyr-20) was quantitated by Cerenkov counting of individual lanes excised from SDS/PAGE gels. Endogenous vesicle-associated tyrosine kinase activity was assessed under similar conditions by incubating either USV or STSV (20 μg protein per sample) in the absence or presence of 10 μg per sample of poly(Glu-80,Tyr-20) and/or purified c-Src (2 units per sample). Samples were separated by SDS/PAGE, and gels were subjected to alkali treatment (1 M KOH for 2 hr at 60°C) as described (4), followed by autoradiography.
Immunoprecipitation.
Nonidet P-40 (2% vol/vol) extracts (400 μg) of crude synaptosomal fraction (P2) obtained from rat cerebral cortex were incubated for 3 hr at 4°C with either anti-synapsin 19.11 mAb, anti-Src 327 mAb, or porcine anti-mouse IgG (10 μg per sample). Immunoprecipitation was performed as described (18) using protein G–Sepharose (25 μl settled prewashed beads) to precipitate the immunocomplexes. After extensive washing of the beads, immunocomplexes were eluted by adding 2-mercaptoethanol-free Laemmli stop buffer and boiling for 5 min and then subjected to SDS/PAGE under nonreducing conditions and immunoblot assay.
Miscellaneous Procedures.
SDS/PAGE was performed according to Laemmli (19). Electrophoretic transfer of proteins to nitrocellulose membranes was performed as described (20). Immunoblots were developed using either 125I-labeled secondary antibodies (Fab fragments) or peroxidase-conjugated secondary antibodies coupled to a chemiluminescence detection system.
RESULTS
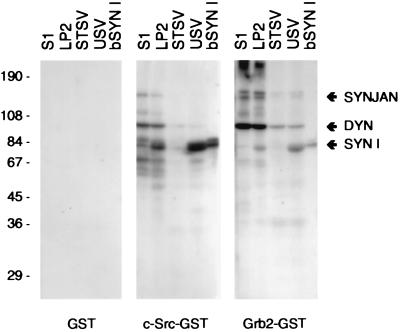
Proteins in various subcellular fractions obtained from rat forebrain, as well as purified bovine synapsin I, were analyzed by overlay assay for their ability to bind to the SH3 domain of c-Src expressed as a fusion protein with GST (Fig. 1). The c-Src-SH3 domain strongly labeled three protein bands, which were identified as dynamin (8), synaptojanin (7), and synapsin I by immunodetection of blots run in parallel (not shown). In agreement with previous data (21), the binding to dynamin and synaptojanin was high in the postnuclear supernatant (S1) and in crude synaptic vesicle fractions (LP2), and decreased in highly purified USV. In contrast, the binding of the c-Src-SH3 domain to synapsin I greatly increased from LP2 to USV. Quantitative depletion of synapsin I from synaptic vesicles by salt treatment virtually abolished the c-Src-SH3 domain-binding activity in this fraction. Overlay assays with GST-Grb2 exhibited a binding pattern that was qualitatively similar to that obtained with the GST-c-Src-SH3 domain (Fig. 1), although several differences in selectivity were also noted.
Figure 1.
Binding of SH3 domains of c-Src and Grb2 to proteins in subcellular fractions from rat forebrain. Overlay assays with the respective SH3 domain–GST fusion proteins or with GST alone were performed as described in Materials and Methods using the indicated amount of protein. The major proteins labeled by the procedure were synaptojanin (SYNJAN), dynamin (DYN), and synapsin I (SYN I). S1, postnuclear supernatant (60 μg); LP2, crude synaptic vesicle pellet (60 μg); STSV, salt-treated synaptic vesicles (60 μg); USV, untreated synaptic vesicles (60 μg); bSYN I, purified bovine synapsin I (1.5 μg).
In addition to c-Src, neurons also express high levels of n-Src, a neuron-specific splice variant that differs by only a 6-aa insert within the SH3 domain (9, 23). In contrast to the results obtained with the GST-c-Src-SH3 domain, overlay assay failed to detect any specific interactions of the GST-n-Src-SH3 domain with nerve terminal proteins (not shown).
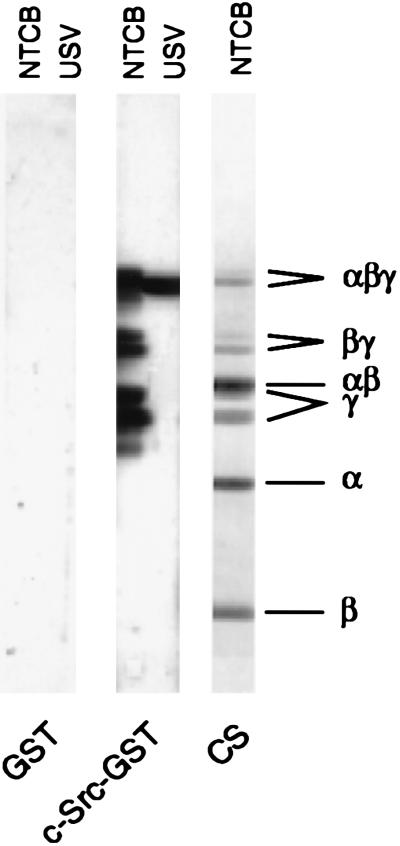
Proline-rich sequences located in both the NH2-terminal portion (domains A and B) and COOH-terminal portion (domains D and E; synapsin Ia) of synapsin I (2) represent potential sites of interaction with SH3 domains. To identify which region of synapsin I was involved in binding the c-Src-SH3 domain, synapsin I was cleaved at cysteine residues with NTCB, and the resultant fragments were analyzed by overlay assay for their ability to interact with the GST-c-Src-SH3 domain. Only those fragments containing the COOH-terminal portion of synapsin I, namely the COOH-terminal fragments (peptide γ doublet), the middle/COOH-terminal fragments (peptide βγ doublet), and uncleaved synapsin Ia/b (peptide αβγ doublet) were labeled by the GST-c-Src-SH3 domain (Fig. 2).
Figure 2.
Binding to the SH3 domain of c-Src involves the COOH-terminal portion of synapsin I. Synapsin I was cleaved at Cys-223, Cys-360, and/or Cys-370 with NTCB as described (15). Aliquots (5 μg) of the resulting fragments (NTCB), together with a sample (30 μg) of untreated synaptic vesicles (USV), were separated by SDS/PAGE on 7–15% polyacrylamide gradient gels and transferred to nitrocellulose membranes, and overlay assays were performed as described in Materials and Methods. Coomassie blue staining (CS) of a gel (30 μg NTCB fragments) run in parallel was used to identify the major cleavage fragments, which together span the entire molecule: peptide α (the 27-kDa NH2-terminal fragment, corresponding to residues 1–222), peptide β (the 15-kDa “middle” fragment, corresponding to residues 223–359/369), and a peptide γ doublet (the 40- and 36-kDa COOH-terminal fragments, corresponding to residues 360/370–706 and 360/370–670 for bovine synapsins Ia and Ib, respectively). Larger fragments resulting from partial cleavage (αβ and βγ doublet), and uncleaved synapsin Ia/b (αβγ doublet) were also visible.
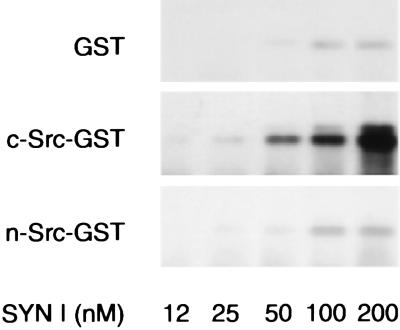
The characteristics of the binding of synapsin I to the SH3 domains of c-Src and n-Src and the potential regulation of this interaction by site-specific seryl phosphorylation of synapsin I were investigated through the use of a solid-phase binding assay. Synapsin I bound to the GST-c-Src-SH3 domain in a dose-dependent fashion, whereas binding to the GST-n-Src-SH3 domain was negligible (Fig. 3). Stoichiometric phosphorylation of bovine synapsin I by either protein kinase A (at site 1; Ser-9), Ca2+/calmodulin-dependent protein kinase II (at sites 2 and 3; Ser-568 and Ser-605, respectively), or mitogen-activated protein kinase (at sites 4, 5, and 6; Ser-62, Ser-67, and Ser-551, respectively) did not significantly alter the amount of synapsin I bound to the GST-c-Src-SH3 domain (not shown).
Figure 3.
Characterization of the interactions of synapsin I with the SH3 domains of c-Src and n-Src. Increasing concentrations of purified synapsin I were incubated with either GST alone (200 nM), GST-c-Src-SH3 domain fusion protein (200 nM), or GST-n-Src-SH3 domain fusion protein (200 nM), which had been coupled to glutathione–Sepharose. Synapsin I recovered in each pellet was separated by SDS/PAGE and detected by immunoblot assay.
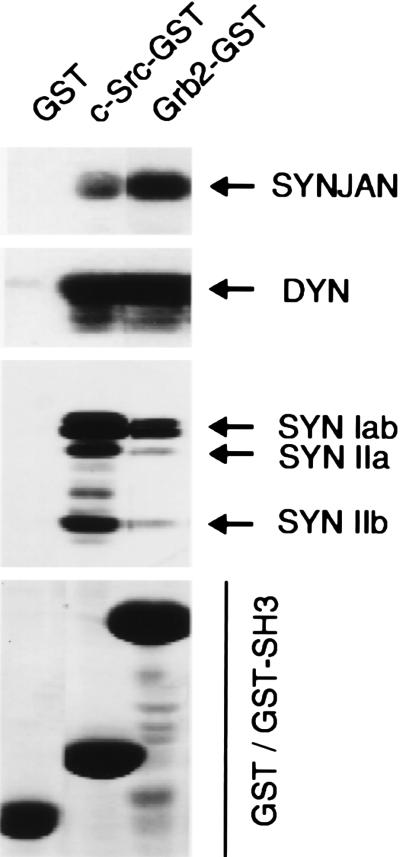
The c-Src-SH3 domain was also used to affinity-purify binding proteins from a detergent extract of a crude synaptosomal fraction (P2) from rat forebrain. In agreement with the results obtained in overlay assays, major proteins eluted from the c-Src-SH3 domain affinity column included synaptojanin, dynamin, and synapsin I (Fig. 4). When the protein composition of the eluent pools from the c-Src- and Grb2-SH3 domain affinity columns were compared, the recovery of dynamin was similar for both columns. Synaptojanin was preferentially recovered by the Grb2-SH3 domain column, whereas synapsin I was preferentially recovered by the c-Src-SH3 domain column. In addition to synapsin I, synapsins IIa and IIb were recovered in eluates from the c-Src-SH3 domain affinity column.
Figure 4.
Affinity purification of SH3 domain-binding proteins from a detergent extract of a crude synaptosomal fraction of rat brain. Affinity resins were prepared by coupling either GST-c-Src-SH3 domain, GST-Grb2, or GST alone to glutathione–Sepharose. Columns were loaded with a 1% (vol/vol) Triton X-100 extract from an 8-mg sample of crude synaptosomal fraction (P2) from rat cerebral cortex. Bound proteins were eluted with Laemmli stop buffer and separated by SDS/PAGE. Recovery of synaptojanin (SYNJAN), dynamin (DYN), synapsin I (SYN Iab), and synapsin II (SYN IIa; SYN IIb) was assessed by immunoblot assay. The relative amounts of GST, GST-c-Src-SH3 domain, and GST-Grb2 eluted from the column resins are shown by Coomassie staining in the bottom panel (GST/GST-SH3).
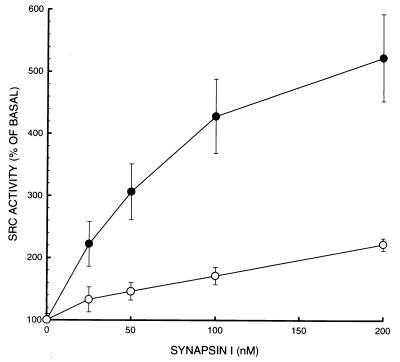
The SH3 domain-mediated interaction between synapsin I and c-Src modulated the catalytic activity (Fig. 5). The ability of purified c-Src to phosphorylate poly(Glu-80,Tyr-20) was stimulated by synapsin I in a dose-dependent fashion (up to approximately 5-fold over basal activity at 200 nM synapsin I). Addition of a relative excess of the GST-c-Src-SH3 domain to the phosphorylation mixture antagonized the synapsin I-induced stimulation of c-Src kinase activity.
Figure 5.
Stimulation of c-Src tyrosine kinase activity by synapsin I. Purified c-Src (3 units per sample) was incubated for 15 min at 30°C in phosphorylation buffer containing poly(Glu-80,Tyr-20) (10 μg per sample) and varying concentrations of synapsin I, in the absence (•) or presence (○) of 4 μM GST-c-Src-SH3 domain. Samples were subjected to SDS/PAGE, and 32P incorporation into the substrate polypeptide was quantitated by Cerenkov counting of individual lanes excised from the gel. Data represent means ± SEM of 5 independent experiments performed on different days, using two different preparations of c-Src and two different preparations of synapsin I.
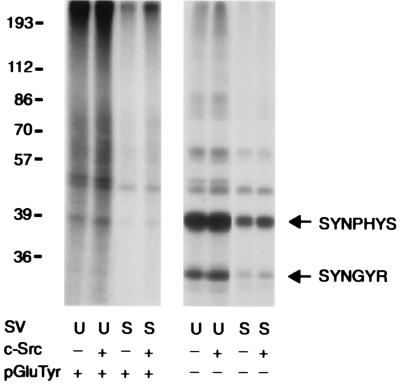
We addressed the possibility that the presence of synapsins on synaptic vesicles might affect the endogenous tyrosine kinase activity associated with vesicles and regulate the phosphorylation state of synaptic vesicle-associated tyrosyl phosphoproteins. The tyrosine kinase activity of USV was higher than that of STSV (Fig. 6), as evaluated by 32P incorporation into either poly(Glu-80,Tyr-20) or endogenous vesicle-associated tyrosine kinase substrates such as synaptophysin and p29/synaptogyrin (4, 5). Phosphorylation of vesicle-associated proteins was not increased further by the addition of exogenous purified c-Src. To ensure that salt treatment did not affect the association of Src with the synaptic vesicle membrane, the level of Src immunoreactivity present in rat brain subcellular fractions was analyzed at various stages of vesicle purification (Fig. 7). A specific enrichment of Src was observed in the synaptic vesicle fraction as well as in some other membrane fractions. Exposure of purified vesicles to 200 mM NaCl did not change their Src content, although it brought about an almost complete dissociation of synapsin I.
Figure 6.
Endogenous synaptic vesicle-associated tyrosine kinase activity is regulated by synapsin I. Untreated synaptic vesicles (U, 20 μg per sample) or vesicles that had been quantitatively depleted of endogenous synapsin I by salt treatment (S, 20 μg per sample) were incubated in phosphorylation buffer for 15 min at 30°C in the presence (Left) or absence (Right) of poly(Glu-80,Tyr-20) (10 μg per sample; pGluTyr). Where indicated, samples also contained purified c-Src (2 units per sample; c-Src). Kinase activity was evaluated by autoradiography of alkali-treated gels after 32P-labeled substrates were separated by SDS/PAGE. Two synaptic vesicle tyrosyl phosphoproteins, synaptophysin (SYNPHYS) and synaptogyrin (SYNGYR), are identified. The diffuse nature of 32P labeling observed in the lanes of the Left panel reflects the size heterogeneity of the poly(Glu-80,Tyr-20) substrate.
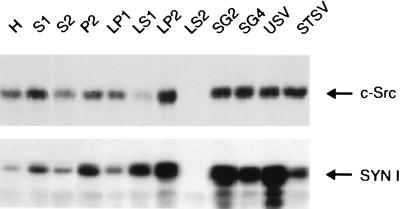
Figure 7.
Distribution of Src and synapsin I immunoreactivities in subcellular fractions of rat forebrain obtained at various stages of vesicle purification. Protein samples (15 μg) of subcellular fractions were separated by SDS/PAGE and analyzed by immunoblot assay. Relative to the homogenate, Src was enriched in highly purified vesicle fractions, and its association with the vesicle membrane was not affected by the salt treatment used to deplete synapsin I. H, homogenate; S1, postnuclear supernatant; S2, supernatant of P2; P2, crude synaptosomes; LP1, crude synaptic plasma membranes; LS1, supernatant of LP1; LP2, crude synaptic vesicles; LS2, synaptosol; SG2, synaptic vesicles; SG4, small synaptic membranes; USV, untreated synaptic vesicles; STSV, salt-treated synaptic vesicles.
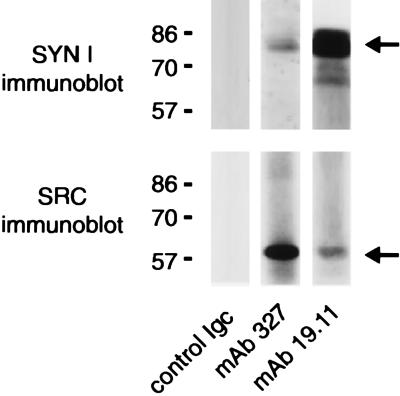
To determine whether synapsin I and c-Src are physiological binding partners in intact brain, we performed immunoprecipitations from detergent extracts of crude (P2) synaptosomal pellets (Fig. 8). Anti-synapsin antibodies coprecipitated synapsin I with a portion of the total pool of Src. Conversely, anti-Src antibodies coprecipitated Src with a portion of the total pool of synapsin I.
Figure 8.
Coimmunoprecipitation of Src with synapsin I from detergent extracts of a crude synaptosomal fraction of rat brain. Nonidet P-40 (2% vol/vol) extracts (400 μg) of rat forebrain crude synaptosomal (P2) fraction were incubated with either a control porcine anti-mouse IgG, the anti-synapsin 19.11 mAb, or the anti-Src 327 mAb (10 μg per sample). Immunocomplexes were sedimented with protein G–Sepharose. The samples were resolved by SDS/PAGE under denaturing, nonreducing conditions and transferred to nitrocellulose membranes. Membranes were probed with either anti-synapsin I G-94 polyclonal antibody (Upper) or the anti-Src 327 mAb (Lower), followed by a peroxidase-conjugated secondary antibody and chemiluminescence detection. Immunoreactive bands corresponding to synapsin I and Src are indicated.
DISCUSSION
Prominent c-Src-SH3 domain-mediated interactions were observed with three identified proteins, namely dynamin, synaptojanin, and synapsin I, in subcellular fractions of rat brain. Synapsin I was the major protein binding the SH3 domain of c-Src in highly purified synaptic vesicle preparations. The interaction of synapsin I with the c-Src-SH3 domain was mediated by domain D and was not significantly altered by site-specific seryl phosphorylation of synapsin I. Pools of synapsin I and Src were coprecipitated from brain extracts using either anti-synapsin- or anti-Src-specific antibodies. In vitro, c-Src/synapsin I interaction was accompanied by a marked stimulation of catalytic activity, and this activation was antagonized by the addition of purified GST-c-Src-SH3 domain.
A thorough description of the interaction of SH3 domains with their ligands has emerged from a variety of structural studies. Despite the presence of the PXXP core motif in many proteins, protein–protein interactions mediated by SH3 domains exhibit a high degree of specificity (24, 25). Synapsin I contains several PXXP motifs within domains A, B, and E (synapsin Ia) and, with higher frequency, within domain D (2). Among these motifs, the sequences RPQPPPP (residues 27–33, at the domain A-B border) and PAGPTR (residues 597–602, bovine domain D) conform exactly to the minimal consensus sequence described for class I (RXXPXXP) and class II (PXXPXR) ligands, respectively (24, 25). Because fragment analysis has demonstrated that the COOH-terminal portion of synapsin I is involved in binding the SH3 domains of c-Src (this paper) and Grb2 (1), the binding site(s) may correspond to the class II ligand consensus sequence or, alternatively, to an atypical SH3 domain-binding sequence found in domain D. Analysis by screening libraries of phage-displayed or synthetic peptides has shown that the c-Src-SH3 domain exhibits a preference for the class I binding motif, RPLPPLP (26–29), although it can also bind to class II ligands (28, 30). In this respect, class II ligands have been reported to be more promiscuous in their binding to SH3 domains than class I ligands, and they exhibit more flexibility in the distance between the PXXP motif and the neighboring arginine residue, a property that can contribute to a broader specificity of this interaction (28, 31).
Although not readily apparent from overlay assays with the subcellular fractions of rat brain, we observed that synapsin II copurified with synapsin I on the GST-c-Src-SH3 domain affinity column. This observation may reflect a direct interaction of synapsin II with the c-Src-SH3 domain or may result from an association of synapsin II with the resin-bound pool of synapsin I. The former interaction appears to be more likely, because synapsin II contains class I ligand motifs, together with other PXXP core motifs, in domains B, G, and H (synapsin IIa) (2), and GST-c-Src-SH3 domain bound to purified recombinant synapsin IIa in blot overlay assays (F.O. and F.B., unpublished observations).
In addition to c-Src, the expression of two neuron-specific Src isoforms has been detected in mammalian brain (9, 22). These isoforms arise from alternative splicing events that direct the insertion of either one or both of two short exons, termed the NI exon (18 nt, encoding a 6-aa insert) and the NII exon (33 nt, encoding an 11-aa insert), into the region encoding the SH3 domain (22, 23). Structural studies have revealed that the insert of n-Src, the isoform containing the 6-residue insert encoded by exon NI, forms a loop that is oriented toward the ligand-binding face of the SH3 domain in a fashion that could modulate ligand-binding interactions (24, 25). We found that binding of synapsin I to the GST-n-Src-SH3 domain was negligible, a characteristic that is common to many other identified c-Src SH3-binding proteins, which interact poorly, if at all, with n-Src (3). The presence of three additional basic residues within the 6-residue insert of the n-Src-SH3 domain may contribute to its reduced affinity for the strongly basic COOH-terminal region of synapsin I.
Interaction of synapsin I with purified c-Src kinase resulted in a severalfold activation of the enzyme that was antagonized by the presence of an excess of the GST-c-Src-SH3 domain. This stimulatory effect supports the functional importance of interactions involving the SH3 domain in the regulation of Src kinase activity in addition to the intramolecular interaction between the SH2 domain and the COOH-terminal tail phosphorylated on Tyr-527 (see refs. 3 and 32 for review). Specific mutations within the Src-SH3 domain were found to activate Src (33, 34). A p130CAS-related protein that binds the SH3 domain of c-Src was found to increase c-Src activity (35). More recently, the HIV Nef protein, a high-affinity ligand for the SH3 domain of Hck (a member of the Src kinase family), was found to cause a severalfold increase in the catalytic activity of Hck that was mediated by its SH3 domain-binding motif (36). Recent solutions of the crystal structures of “repressed” forms of c-Src (37) and Hck (38) have revealed that an intramolecular interaction between the SH3 domain and the peptide linker connecting the SH2 domain and the catalytic domain serves as an adapter that allows a direct contact between the SH3 domain and the small lobe of the catalytic domain. This contact produces a repositioning of the C-helix that disables the active site and represses catalytic activity (37, 38). According to this model, synapsin I, by interacting with high affinity with the c-Src-SH3 domain, may function as an endogenous activator of c-Src in neurons. Our preliminary results indicating that synapsin II may also interact directly with the SH3 domain of c-Src suggest that synapsin II may also serve to regulate the kinase activity of c-Src.
Purified synaptic vesicle preparations possess an endogenous level of tyrosine kinase activity that was markedly decreased by mild salt treatment. This procedure resulted in virtually quantitative depletion of synapsin I from the vesicles but did not alter the level of vesicle-associated Src, the enzyme that accounts for the majority of endogenous synaptic vesicle-associated tyrosine kinase activity (ref. 5; F.V., F.B., and P.G., unpublished observations). Although other proteins may contribute to the observed effects, our results suggest that the higher level of tyrosine kinase activity in untreated vesicles was due to an SH3 domain-mediated interaction of endogenous c-Src and synapsin I, stimulating c-Src activity on the vesicle membrane. This, in turn, raises the possibility that, in intact nerve terminals, c-Src activity may be regulated by the extent of association of synapsin I, a peripheral protein of synaptic vesicles, with the vesicle membrane. Such a possibility is supported by the observation that the binding of synapsin I to the vesicle membrane is regulated by signal transduction processes (39).
Src is implicated in a variety of cellular processes and plays a prominent role in the regulation of the actin-based cortical cell cytoskeleton (3, 40). In PC12 cells, enhanced expression of Src induced nerve-growth-factor-independent neurite outgrowth (41). Conversely, microinjection of anti-Src antibodies inhibited both nerve growth factor- and fibroblast growth factor-dependent neurite elongation (42). Although the function of Src must be at least partially redundant with that of other proteins (for example, Src-deficient mice do not exhibit major defects in synaptic function; see refs. 3 and 43 for review), impaired neurite outgrowth was observed when cerebellar neurons derived from Src-deficient mice were cultured on a substrate of the L1 cell adhesion molecule (44).
Synapsin I (an actin-binding protein; see ref. 39 for review) is concentrated in the distal portion of growing neurites (45), and neurite outgrowth was impaired in primary cultures of embryonic hippocampal neurons derived from synapsin I-deficient mice (46). A wide range of evidence, obtained from in vitro models and in vivo, has demonstrated that synapsin I is essential for the proper assembly and maintenance of presynaptic vesicle clusters (47–51). Interactions between synapsin I and c-Src may be of functional importance both during development and at mature synapses. The important role played by c-Src at other types of intercellular junctions (see refs. 3 and 40 for review) supports the possible involvement of c-Src and the c-Src/synapsin I interaction in the biology of the nerve terminal.
Acknowledgments
We thank I. Gout, J. Schlessinger, and J. Brugge for providing the SH3 domain fusion protein constructs. This work was supported by grants from Progetto Nazionale Sclerosi Multipla, Istituto Superiore di Sanità (N. 31 to F.B.), Telethon (N. 753 to F.B. and N. 581 to F.V.), and the U.S. Public Health Service (MH39327 to P.G. and CA46128 to P.D.C). F.O. is a recipient of a fellowship from the Anna Villa Rusconi Foundation (Varese, Italy).
ABBREVIATIONS
- SH2 and SH3
Src homology 2 and 3
- GST
glutathione S-transferase
- NTCB
2-nitro-5-thiocyanobenzoic acid
- USV
untreated synaptic vesicles
- STSV
salt-treated synaptic vesicles
References
- 1.McPherson P S, Czernik A J, Chilcote T J, Onofri F, Benfenati F, Greengard P, Schlessinger J, De Camilli P. Proc Natl Acad Sci USA. 1994;91:6486–6490. doi: 10.1073/pnas.91.14.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Südhof T C, Czernik A J, Kao H-T, Takei K, Johnston P A, Horiuchi A, Kanazir S D, Wagner M A, Perin M S, De Camilli P, Greengard P. Science. 1989;245:1474–1480. doi: 10.1126/science.2506642. [DOI] [PubMed] [Google Scholar]
- 3.Brown M T, Cooper J A. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 4.Pang D T, Wang J K T, Valtorta F, Benfenati F, Greengard P. Proc Natl Acad Sci USA. 1988;85:762–766. doi: 10.1073/pnas.85.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnekow A, Jahn R, Schartl M. Oncogene. 1990;5:1019–1024. [PubMed] [Google Scholar]
- 6.Linstedt A D, Vetter M L, Bishop J M, Kelly R B. J Cell Biol. 1992;117:1077–1084. doi: 10.1083/jcb.117.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPherson P S, Garcia E P, Slepnev V I, David C, Zhang X, Grabs D, Sossin W S, Bauerfeind R, Nemoto Y, De Camilli P. Nature (London) 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- 8.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, Campbell I D, Waterfield M D. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 9.Brugge J S, Cotton P C, Queral A E, Barrett J N, Nonner D, Keane R W. Nature (London) 1985;316:554–557. doi: 10.1038/316554a0. [DOI] [PubMed] [Google Scholar]
- 10.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ulrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. Cell. 1991;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 11.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 12.Bähler M, Greengard P. Nature (London) 1987;326:704–707. doi: 10.1038/326704a0. [DOI] [PubMed] [Google Scholar]
- 13.Schiebler W, Jahn R, Doucet J-P, Rothlein J, Greengard P. J Biol Chem. 1986;261:8383–8390. [PubMed] [Google Scholar]
- 14.Jovanovic J N, Benfenati F, Siow Y L, Sihra T S, Sanghera J S, Pelech S L, Greengard P, Czernik A J. Proc Natl Acad Sci USA. 1996;93:3679–3683. doi: 10.1073/pnas.93.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bähler M, Benfenati F, Valtorta F, Czernik A J, Greengard P. J Cell Biol. 1989;108:1841–1849. doi: 10.1083/jcb.108.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huttner W B, Schiebler W, Greengard P, De Camilli P. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiel G, Südhof T C, Greengard P. J Biol Chem. 1990;265:16527–16533. [PubMed] [Google Scholar]
- 18.Valtorta F, Villa A, Jahn R, De Camilli P, Greengard P, Ceccarelli B. Neuroscience. 1988;24:593–603. doi: 10.1016/0306-4522(88)90353-3. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Towbin L, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McPherson P S, Takei K, Schmid S L, De Camilli P. J Biol Chem. 1994;269:30132–30139. [PubMed] [Google Scholar]
- 22.Pyper J M, Bolen J B. Mol Cell Biol. 1990;10:2035–2040. doi: 10.1128/mcb.10.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez R, Mathey-Prevot B, Bernards A, Baltimore D. Science. 1987;237:411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- 24.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 25.Mayer B J, Eck M J. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 26.Rickles R J, Botfield M C, Weng Z, Taylor J A, Green O M, Brugge J S, Zoller M J. EMBO J. 1994;13:5598–5604. doi: 10.1002/j.1460-2075.1994.tb06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparks A B, Quilliam L A, Thorn J M, Der C J, Kay B K. J Biol Chem. 1994;269:23853–23856. [PubMed] [Google Scholar]
- 28.Alexandropoulos K, Cheng G, Baltimore D. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quilliam L A, Kay B K. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 31.Ren R, Ye Z, Baltimore D. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 32.Mayer B J. Curr Biol. 1997;7:295–298. doi: 10.1016/s0960-9822(06)00141-2. [DOI] [PubMed] [Google Scholar]
- 33.Kato J-Y, Takeya T, Grandori C, Iba H, Levy J B, Hanafusa H. Mol Cell Biol. 1986;6:4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erpel T A, Superti-Furga G, Courtneidge S A. EMBO J. 1995;14:963–975. doi: 10.1002/j.1460-2075.1995.tb07077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandropoulos K, Baltimore D. Genes Dev. 1996;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 36.Moarefi I, LaFevre Bernt M, Sicheri F, Huse M, Lee C-H, Kuriyan J, Miller W T. Nature (London) 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 37.Xu W, Harrison S C, Eck M J. Nature (London) 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 38.Sicheri F, Moarefi I, Kuriyan J. Nature (London) 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 39.Greengard P, Valtorta F, Czernik A J, Benfenati F. Science. 1993;259:780–785. doi: 10.1126/science.8430330. [DOI] [PubMed] [Google Scholar]
- 40.Erpel T, Courtneidge S. Curr Op Cell Biol. 1995;7:176–182. doi: 10.1016/0955-0674(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 41.Alemà S, Casalbore P, Agostini E, Tatò F. Nature (London) 1985;316:557–559. doi: 10.1038/316557a0. [DOI] [PubMed] [Google Scholar]
- 42.Kremer N E, D’Arcangelo G, Thomas S M, DeMarco M, Brugge J S, Halegoua S. J Cell Biol. 1991;115:809–819. doi: 10.1083/jcb.115.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowell C A, Soriano P. Genes Dev. 1996;10:1845–1857. doi: 10.1101/gad.10.15.1845. [DOI] [PubMed] [Google Scholar]
- 44.Ignelzi M A, Jr, Miller D R, Soriano P, Maness P F. Neuron. 1994;12:873–884. doi: 10.1016/0896-6273(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 45.Fletcher T L, Cameron P, De Camilli P, Banker G. J Neurosci. 1991;11:1617–1626. doi: 10.1523/JNEUROSCI.11-06-01617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chin L-S, Li L, Ferreira A, Kosik K S, Greengard P. Proc Natl Acad Sci USA. 1995;92:9230–9234. doi: 10.1073/pnas.92.20.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valtorta F, Iezzi N, Benfenati F, Lu B, Poo M-m, Greengard P. Eur J Neurosci. 1995;7:261–270. doi: 10.1111/j.1460-9568.1995.tb01062.x. [DOI] [PubMed] [Google Scholar]
- 48.Pieribone V A, Shupliakov O, Brodin L, Hilfiker-Rothenfluh S, Czernik A J, Greengard P. Nature (London) 1995;375:493–497. doi: 10.1038/375493a0. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Chin L-S, Shupliakov O, Brodin L, Sihra T S, Hvalby Ø, Jensen V, Zheng D, McNamara J O, Greengard P, Andersen P. Proc Natl Acad Sci USA. 1995;92:9235–9239. doi: 10.1073/pnas.92.20.9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takei Y, Harada A, Takeda S, Kobayashi K, Terada S, Noda T, Takahashi T, Hirokawa N. J Cell Biol. 1995;131:1789–1800. doi: 10.1083/jcb.131.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan T A, Li L, Chin L-S, Greengard P, Smith S J. J Cell Biol. 1996;134:1219–1227. doi: 10.1083/jcb.134.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]