Abstract
DNA studies are revealing the extent of hidden, or cryptic, biodiversity. Two new studies challenge paradigms about cryptic biodiversity and highlight the importance of adding a historical and biogeographic dimension to biodiversity research.
Biodiversity, the variety of life, is one of nature's most exuberant manifestations. Scientists have long struggled to understand the evolutionary and ecological processes underlying the origin, distribution and maintenance of biodiversity. This dilemma is faced not only by researchers working in undersampled regions such as tropical rainforests and marine habitats, but also by those studying densely sampled and well characterized temperate systems. The problem is partly generated by the difficulty of detecting and measuring biodiversity solely on the basis of morphological information. Despite the central and unrivalled position of morphology-based taxonomy in biodiversity research, human visual perception will probably never quite suffice to capture natural complexity. A good example of this is the escalating number of DNA-based studies reporting cryptic species [1,2]. Cryptic, or sibling, species are discrete species that are difficult, or sometimes impossible, to distinguish morphologically and thus have been incorrectly classified as a single taxon. Cryptic species are found from the poles to the Equator and in all major terrestrial and aquatic taxonomic groups [2,3]. For example, a recent meta-analysis yielded 2,207 articles reporting cryptic species in all metazoan phyla and classes, including 996 new species in insects, 267 in mammals, 151 in fishes and 94 in birds [2]. Similarly, a recent report shows that global biodiversity in protozoa is often cryptic and significantly higher than previously considered [4].
Analysis of the genetic diversity distributed within 'species' provides a powerful framework for recognizing cryptic species. In this context, historical considerations are important, as the current genetic architectures of many species have been shaped by global climatic fluctuations, environmental gradients and the separation of populations by geographic barriers during the past 3 million years and, to a lesser extent, by more ancient physical processes [5,6].
Phylogeography and the identification of cryptic biodiversity
Here we discuss two recent studies on cryptic species that take a historical biogeographical perspective on the distribution of genetic diversity in populations, and discuss how such perspectives can inform our knowledge of cryptic biodiversity. In one, published in BMC Biology, a team led by Robert Wayne [7] details the discovery of at least six cryptic species in the giraffe (Giraffa camelopardalis) based on the geographic distribution of genetic diversity in the giraffe population today, and discusses how conditions in the mid to late Pleistocene might have led to such speciation. In the other, in BMC Evolutionary Biology, Elmer et al. [8] report previously unsuspected cryptic species within the upper Amazonian leaflitter frog (Eleutherodactylus ockendeni).
Any biogeographic scenario, recent or historical, begins with the process of population differentiation and speciation. Phylogeography, the study of the geographic distribution of genealogical lineages [6], adds an essential component – time – to the understanding of population structure, reproductive isolation and speciation. Changes over time in the physical and biotic environment of a population lead to demographic variations that are correlated with the structure of population genealogies [6]. As a consequence, phylogeographic studies have the potential for describing the chronology of demographic variation and the reproductive isolation of population units. This is particularly true for surveys that also incorporate intraspecific analyses of migration using sophisticated analytical developments based on the coalescent [9,10], a theory on the evolutionary history of alleles at genetic loci that allows inferences to be made about the timing and demographic events linked to genealogical processes. This combination of phylogeographic and population-genetic approaches offers an important framework for delineating morphologically cryptic species and for appreciating the processes that have shaped speciation. A valuable extension of this framework is to compare phylogeographic data for multiple co-distributed species to test for historical contingencies and processes that have shaped the diversification of entire biotas. These comparative assessments of regional evolutionary subdivision are important in informing biodiversity discovery and management as they can potentially identify hotspots of biodiversity – regions within which entire communities have been affected by events in Earth's history [6,11,12].
Cryptic species in the giraffe and in an Amazonian frog
The two articles by Brown et al. [7] and Elmer et al. [8] highlight important DNA-based discoveries of multiple evolutionary diversifications that challenge paradigms about cryptic biodiversity. The first paradigm is that cryptic species are expected to be rare in megafauna, such as large mammals. This is because many large-bodied mammals can disperse over large distances, a life-history attribute expected to prevent local genetic differentiation and reproductive isolation. In an exemplary study, Brown et al. present a phylogeographic and population genetic analysis in one such mammal, the giraffe [7]. Giraffes are capable of long-distance dispersal and have an extensive range in sub-Saharan Africa. Based on adequate sampling and mitochondrial DNA (mtDNA) and nuclear microsatellite DNA data, Brown et al. [7] convincingly show that giraffes are composed of at least six distinct lineages. These lineages show levels of evolutionary and genetic distinctiveness consistent with speciation events during the Pleistocene (divergences estimated at between 1.6 million years and 113,000 years ago). In addition, marked genetic subdivision is also apparent within five of the six lineages, yielding a minimum of 11 independent biological units. The authors propose that a combination of increasing aridity, periodic oscillations in wet and dry conditions and regional changes in habitat (for example, the expansion of the 'Mega Kalahari' desert, an area much larger than the present-day Kalahari desert) may have caused fragmentation of giraffe populations during the Pleistocene and divergence within habitat refugia. This agrees with patterns of phylogeographic structure observed in other large African mammals (for example, hartebeest [13] and zebra [14]) and illustrates the influence of large-scale climatic fluctuations in the diversification of Africa's biodiversity.
Evidence for contemporary reproductive isolation in the wild comes from comparisons between adjacently distributed giraffe lineages, which share essentially no gene flow despite the absence of dispersal barriers. It is suggested that reproductive isolation might be maintained by climatically driven differences in reproductive timing or by sexually imprinted assortative mating due to differences in coat patterns [7]. According to current taxonomy, giraffes are considered to represent a single species and as such are listed as "Lower Risk" on the IUCN Red List (downloaded 22 October 2007) [15]. Given the endangered status of several of the lineages reported by Brown et al. [7], giraffes represent another unfortunate example of the negative consequences of neglected taxonomy on conservation management [16,17].
The other paradigm often mentioned in the literature is that most cryptic species are the product of recent speciation events. Hence, recent speciation would account for the apparent morphological stasis observed in many cryptic species. In a phylogeographic study across eastern Ecuador, Elmer et al. [8] report previously unsuspected cryptic species within the upper Amazonian leaflitter frog (Eleutherodactylus ockendeni). They used comparison of mtDNA to uncover three highly divergent clades and non-overlapping microsatellite allele sizes as further evidence for reproductive isolation among clades. These clades occur together in some geographic regions without interbreeding, providing strong support that they represent distinct species. Elmer et al. [8] estimate divergence times between the three clades that date back to late Oligocene and late Miocene (around 24–9 million years ago). These estimates are inconsistent with the idea that climatic cycles of the Quaternary and associated isolation in refugia promoted speciation in this Amazonian frog. In fact, the ancient events of diversification coincide with periods of major and complex geotectonics in the northern Andes during the Miocene [8]. The reports of ancient species in this frog and in other tropical species (for example [18,19]) imply that species richness in tropical regions has been grossly underdocumented by inventories based on morphology. Notwithstanding a recent suggestion that the proportion of cryptic species in nature is similar across different biogeographic regions [2], efforts to increase systematic population sampling in tropical rainforests, especially in developing countries, are urgently needed to better document species richness.
A unification of historical disciplines to better document biodiversity
Despite its usefulness, the phylogeographic method has serious shortcomings as seems to be the case for any discipline with a historical dimension. Generally, direct genetic evidence about phylogeographic divergence can be gathered only where populations currently exist. Even then, the evidence is temporally fragmented as the result of past population extinctions. These factors can obscure inferences about the prevalence and the spatial scale of cryptic speciation. Obtaining genealogical signal from genetic markers is also challenging if speciation occurred very rapidly, as is often the case in Quaternary biological radiations. Partial solutions to these shortcomings exist, but their effectiveness is dictated by the peculiarities of each biogeographic scenario. Solutions include combining traditional tree-based phylogenetic methods with estimates of demographic parameters that take into account uncertainties in phylogeographic inference [9,10,20-22], adding data from extinct populations [23], adding temporal samples from the same populations [24-27] and adding data from a large number of individuals, localities and fast-evolving genetic markers [21,26-28].
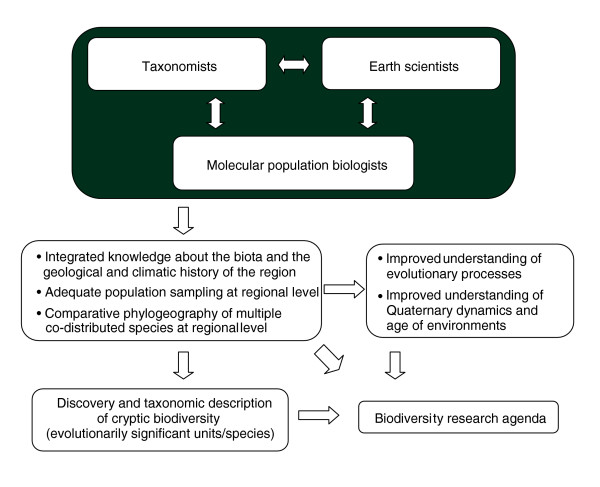
Nevertheless, when combined with data from population genetics and Earth sciences, phylogeographic information can be used to answer key questions concerning past and present aspects of biodiversity and to predict future demographic scenarios. Techniques for studying cryptic diversity using DNA data are becoming cheaper and cheaper, and so finite resources can be reallocated to gather more population samples, in both time and space. Through temporal and spatial sampling a biologist is basically looking at the world as a geologist would. Phylogeographers have two main tools for looking into the past: using sophisticated models of DNA evolution they can infer from present-day data the evolutionary processes that happened in the past [20-22], or they can actually look at the past [23-27], as a geologist would when studying stratigraphic series. However, phylogeographers generally have no formal training on how to explore and interpret physical data about Earth's history. As a result, they have often inefficiently, and sometimes incorrectly, used information from disciplines such as geomorphology, sedimentology, paleoclimatology, volcanology and oceanography. Many of these fields, especially those related to Late Quaternary dynamics, have experienced technological and theoretical advances in recent years that produce data that are probably 'cryptic' to the eyes of many biologists. As a starting point, Earth scientists and phylogeographers should integrate their information to fill in temporal and spatial gaps when reconstructing the history of a particular region and its biota (Figure 1). This can be of mutual benefit to both types of specialists by guiding and rationalizing sampling (both genetic and geological) over the appropriate geographic and temporal landscape. In turn, this can produce a less fragmented picture about the patterns and processes shaping biodiversity.
Figure 1.
An integration among molecular population biologists, Earth scientists and taxonomists to discover, document and understand biodiversity. The diagram exemplifies a comparative phylogeographic study but single-taxon studies are also important. Integrated scientists benefit from the flow of information that occurs from all sections of the diagram (not shown).
Justifiably or not, species as established in the current taxonomy are often used as units in biodiversity research and in conservation policy. Thus, investment towards a better resourced morphology-based taxonomy is urgently needed to implement a modern and integrated system to ensure that newly reported cryptic species will be described following their discovery [29]. Human activity has had a greater impact on biodiversity in the past 50 years than at any time in human history, and the rate of change is predicted to continue or to increase [30]. Some of the key drivers affecting the loss of biodiversity worldwide are habitat alteration, climate change, overexploitation and invasive alien species. By improving the way we discover, document and measure biodiversity, we will move towards understanding the consequences of changes in these drivers for biodiversity. For this to become a reality, biodiversity programs need to bring a spatial and temporal perspective to the forefront of their research agenda. Biologists need to dedicate more time to fieldwork (for example, the giraffe study) and expand their intellectual 'confidence zone' to better address temporal axes of diversification (for example, the frog study). The prevalence of cryptic species, even in charismatic and well studied animals like the giraffe, highlights the importance of combining multidisciplinary approaches in order to capture nature's complexity.
Acknowledgments
Acknowledgements
We thank the participants of the 'Phylogeography and Coalescence Workshop' (Melbourne, 2007) for constructive discussions and David Briscoe, Rob Fleischer and Rus Hoelzel for comments on the manuscript.
References
- Bickford D, Lohman DJ, Sohdi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Schwenk K. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol Biol. 2007;7:121–126. doi: 10.1186/1471-2148-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton N. Sibling species in the sea. Annu Rev Ecol Syst. 1993;24:189–216. doi: 10.1146/annurev.es.24.110193.001201. [DOI] [Google Scholar]
- Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol Biol. 2007;7:162. doi: 10.1186/1471-2148-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography The History and Formation of Species. Cambridge, London: Harvard University Press; 2000. [Google Scholar]
- Brown DM, Brenneman RA, Georgiadis NJ, Koepfli K, Pollinger JP, Mila B, Louis EL, Jr, Grether GF, Jacobs DK, Wayne RK. Extensive population genetic structure in the giraffe. BMC Biol. 2007;5:57. doi: 10.1186/1741-7007-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, Davila JA, Lougheed SC. Cryptic diversity and deep divergence in an upper Amazonian frog, Eleutherodactylus ockendeni. BMC Evol Biol. 2007;7:247. doi: 10.1186/1471-2148-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LL, Maddison WP. Statistical phylogeography. Mol Ecol. 2002;11:2623–2635. doi: 10.1046/j.1365-294X.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Beerli P. Perspective: Gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Bermingham E, Moritz C. Comparative phylogeography: concepts and applications. Mol Ecol. 1998;7:367–369. doi: 10.1046/j.1365-294x.1998.00424.x. [DOI] [Google Scholar]
- Moritz C, Faith DP. Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol Ecol. 1998;7:419–429. doi: 10.1046/j.1365-294x.1998.00317.x. [DOI] [Google Scholar]
- Flagstad O, Syvertsen PO, Stenseth NC, Jakobsen KS. Environmental change and rates of evolution: the phylogeographic pattern within the hartebeest complex as related to climatic variation. Proc R Soc B. 2001;268:667–677. doi: 10.1098/rspb.2000.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JA, Rohland N, Glaberman S, Fleischer RC, Caccone A, Hofreiter M. A rapid loss of stripes: the evolutionary history of the extinct quagga. Biol Lett. 2005;3:291–295. doi: 10.1098/rsbl.2005.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2007 IUCN Red List of Threatened Species http://www.iucnredlist.org [DOI] [PMC free article] [PubMed]
- Daugherty CH, Cree A, Hay JM, Thompson MB. Neglected taxonomy and continuing extinctions of tuatara (Sphenodon) Nature. 1990;347:177–179. doi: 10.1038/347177a0. [DOI] [Google Scholar]
- Russello M, Glaberman S, Gibbs J, Marquez C, Powell J, Caccone A. A cryptic taxon of Galápagos tortoise in conservation peril. Biol Lett. 2005;1:287–290. doi: 10.1098/rsbl.2005.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: An integrated molecular approach. Ann Rev Ecol Syst. 2000;31:533–563. doi: 10.1146/annurev.ecolsys.31.1.533. [DOI] [Google Scholar]
- Parra-Olea G, Wake DB. Extreme morphological and ecological homoplasy in tropical salamanders. Proc Natl Acad Sci USA. 2001;98:7888–7891. doi: 10.1073/pnas.131203598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa EP, Cook JA, Patton JL. Genetic footprints of demographic expansion in North America, but not Amazonia, during the Late Quaternary. Proc Natl Acad Sci USA. 2003;100:10331–10334. doi: 10.1073/pnas.1730921100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheregaray LB, Ciofi C, Geist D, Gibbs J, Caccone G, Powell JR. Genes record a prehistoric volcano eruption in the Galápagos. Science. 2003;302:75. doi: 10.1126/science.1087486. [DOI] [PubMed] [Google Scholar]
- Sunnucks P, Blacket MJ, Taylor JM, Sands CJ, Ciavaglia SA, Garrick R, Tait NN, Rowell DM, Pavlova A. A tale of two flatties: different responses of two terrestrial flatworms to past environmental climatic fluctuations at Tallaganda in montane south-eastern Australia. Mol Ecol. 2006;14:4513–4531. doi: 10.1111/j.1365-294X.2006.03107.x. [DOI] [PubMed] [Google Scholar]
- Leonard JA, Vilà C, Fox-Dobbs K, Koch PL, Wayne RK, Valkenburgh BV. Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr Biol. 2007;17:1146–1150. doi: 10.1016/j.cub.2007.05.072. [DOI] [PubMed] [Google Scholar]
- Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- Paxinos EE, James HF, Olson SL, Ballou JD, Leonard JA, Fleischer RC. Prehistoric decline of genetic diversity in the Nene. Science. 2002;296:1827. doi: 10.1126/science.296.5574.1827. [DOI] [PubMed] [Google Scholar]
- Bunje PMW, Barluenga M, Meyer A. Sampling genetic diversity in the sympatrically and allopatrically speciating Midas cichlid species complex over a 16 year time series. BMC Evol Biol. 2007;7:25. doi: 10.1186/1471-2148-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA. 2002;99:2445–2449. doi: 10.1073/pnas.032477999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheregaray LB, Sunnucks P, Briscoe DA. A rapid fish radiation associated with the last sea level changes in southern Brazil: the silverside Odontesthes perugiae complex. Proc R Soc B. 2002;269:65–73. doi: 10.1098/rspb.2001.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick-Steiner BC, Seifert B, Stauffer C, Christian E, Crozier RH, Steiner FM. Without morphology cryptic species stay in taxonomic crypsis following discovery. Trends Ecol Evol. 2007;22:391–392. doi: 10.1016/j.tree.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Millennium Ecosystem Assessment Ecosystems and Human Well-being: Biodiversity Synthesis. 2007. http://www.millenniumassessment.org