Abstract
In an attempt to further elucidate the pathomechanisms in oral squamous cell carcinoma (OSCC), gene expression profiling was performed using a whole-transcriptome chip that contains 35,035 gene-specific 70mere oligonucleotides (Human OligoSet 4.0; Operon, Cologne, Germany) to a set of 35 primary OSCCs. Altogether, 7390 genes were found differentially expressed between OSCC tumor samples and oral mucosa. To characterize the major biologic processes in this tumor collection, MAPPFinder, a component of GenMAPP version 2.1, was applied to this data set to generate a statistically ranked list of molecular signaling pathways. Among others, cancer-related pathways, such as mitogen-activated protein (MAP) kinase signaling (z score = 4.6, P < .001), transforming growth factor-beta signaling (z score = 3.0, P = .015), and signaling pathways involved in apoptosis (z score = 2.1, P = .037), were found deregulated in the OSCC collection analyzed. Focusing on the MAP kinase signaling pathway, subsequent tissue microarray analyses by immunohistochemistry revealed an increase in protein expression of MAP kinase-related proteins ERK1 in 22.8% (48 of 209) and ERK5 in 27.4% (76 of 277), respectively. An association of high ERK5 but not of high ERK1 expression with advanced tumor stage and the presence of lymph node metastases was found (P = .008 and P = .016, respectively). Our analysis demonstrates the reliability of the combined approach of gene expression profiling, signaling pathway analyses, and tissue microarray analysis to detect novel distinct molecular aberrations in OSCC.
Introduction
Oral squamous cell carcinoma (OSCC) belongs to the 10 most common human malignancies worldwide, affecting more than 500,000 individuals per year. The 5-year survival rate for OSCC does not exceed 55% [1,2], which is mainly caused by locally aggressive tumor phenotypes [3]. Furthermore, clinicopathological parameters such as the TNM system, which are generally used as basis for therapeutic decisions, frequently fail to predict the biologic behavior of the tumors or the patients' outcome. To improve clinical management of individual patients, there is a strong requirement for a better understanding of molecular events involved in OSCC pathophysiology. Attempts to find biomarkers that identify cancerous lesions had identified several candidate genes associated with OSCC tumor progression, including p53, p16, MYC, CCND1, EGFR, and CCNL1 [4–7]. Up to now, however, no single gene was shown to have sufficient diagnostic use to predict the biologic behavior of the tumors. Although those studies contributed greatly to our current understanding, they did not explain the complexity of this malignancy.
High-throughput gene expression profiling techniques offer a unique mechanism for interrogating transcriptome-wide levels of thousands of genes expression and have proven value in defining gene expression signatures dividable in important subsets of patients, who would otherwise be undetected by conventional prognostication schemes [8,9]. Over the last few years, gene expression profiling using microarray hybridization has provided new insights in carcinogenesis and tumor cell dissemination. More than just focusing on the expression of a few genes, genomic-scale expression profiles allow the investigator to look at genetic expression variability in the context of broader genetic themes and pathways. Likewise, the expression profiles of cancers may provide the identification of specific biochemical pathways that might be targeted by new therapeutic agents.
In this study, we applied gene expression microarray technology to a collection of 35 OSCC specimens and compared the gene expression with a pool of normal oral mucosal biopsies from healthy patients. To identify pathways, which are recurrently affected by differential mRNA expression, the software package MAPPFinder was used [10–12], which allows the integration and procession of microarray data sets into the biologic context of molecular functions. To validate individual candidate genes, which were found upregulated in gene expression profiling analyses, a tissue microarray (TMA) analysis was subsequently performed in a representative collection of 306 clinically well-defined primary OSCC specimens.
Materials and Methods
RNA Expression Profiling
OSCC specimen and control samples
Thirty-five frozen tissue tumor samples were collected from patients with histologic confirmed OSCC after approval by the institutional review board of the Universitätsklinikum Heidelberg and after obtaining informed consent. Tumor samples were cut and stained by hematoxylin-eosin. Only samples containing at least 80% tumor cells were used for further analyses. Six oral mucosa samples collected from healthy donors served as control tissue. Nucleic acid extraction of high-molecular weight RNA from frozen tumor tissue was carried out by ultracentrifugation as described elsewhere in detail [13].
Transcriptome amplification and labeling
Two micrograms of total RNA from each patient and Human Universal Reference RNA (Stratagene, La Jolla, CA) were used in a T7-polymerase-based transcriptome amplification method, which was described elsewhere in detail [14].
70mere oligonucleotide microarrays
A set of 35,035 gene-specific 70mere oligonucleotide probes (Human OligoSet 4.0; Operon, Cologne, Germany) was printed on glass slides coated with epoxy-silane (Schott Nexterion, Jena, Germany). The microarray chip represents approximately 25,100 unique genes and 39,600 transcripts excluding control oligos. A variety of data sources was used to cover the genes from human mitochondrial genome, RNA genes, micro-RNA genes, the endogenous human viral genes, and the exogenous reporter genes (Operon). Microarray production, prehybridization treatment, hybridization, and posthybridization washes, and data acquisition were performed as described previously [15]. After hybridization and stringent washing, fluorescence intensity images were acquired using a dual-laser scanner (G2505 B; Agilent Technologies, Santa Clara, CA) and were analyzed with the GenePix Pro 6.0 imaging software (Molecular Devices, Union City, CA). All hybridization experiments were repeated with inversely labeled sample and reference.
Data preprocessing
Result files containing all relevant raw data were processed using the in-house-developed ChipYard microarray analysis software (http://www.dkfz.de/genetics/ChipYard/), the statistical programming language R, and packages of the Bioconductor project. Raw fluorescence intensity values were normalized applying variance stabilization. We assessed the quality of all hybridizations by generating scatter plots and gradient plots using the Bioconductor package “limma” (www.bioconductor.org). The raw and normalized data are deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/; Accession No. GSE10121).
Statistical analysis
After normalization, all gene expression data were analyzed for differences between normal and tumor samples using analysis of variance with Bonferroni multiple testing correction. All genes with a log-value between -1 and 1 were excluded from further analyses. To identify pathways, which were likely to be affected by differentially expression, MAPPFinder, a component of Gen- MAPP software package version 2.1, was used [10–12]. MAPPFinder produces a statistically ranked list of Gene Ontology (GO) biologic categories associated with each cluster, from which the most significant nonsynonymous groups are listed. MAPPFinder analysis was performed on a set of 96 MAPPs, a representation of a biologic relationship between genes or gene products, calculating the percentage of genes meeting the criterion for each MAPP. A positive z score indicates that there are more genes meeting the criterion in a MAPP than would be expected by random chance. A negative z score indicates that there are fewer genes meeting the criterion than would be expected by random chance.
Quantitative real-time reverse transcriptase-polymerase chain reaction
To validate expression profiling data, mRNA levels of selected candidate genes TRIO, TNFRSF18, EGFR, MAPK3, and MAPK7 were tested by quantitative real-time reverse transcriptase- polymerase chain reaction (RT-PCR). For normalization purposes, mean cDNA expression levels of the housekeeping genes PGK1 and LamininB1 were measured. Details of the experimental setup are described elsewhere [16,17]. Primer sequences used in these experiments are listed in Table W1.
Tissue Microarray Analysis
Tumor material and patients' characteristics
Paraffin-embedded tumor specimen of primary OSCC were obtained from the archives of the Institute of Pathology of the University Hospital Heidelberg after approval by the local institutional review board. For all tumor samples, clinical and follow-up data of the patients were available from the Department of Oral and Craniomaxillofacial Surgery of the University Hospital Heidelberg. Mean age of the patients was 61 years at the time of diagnosis. Tumors were staged according to the TNM system of the International Union against Cancer (UICC). Of 306 tumor specimens available for TMA experiments, 143 tumors were T1/2 and 163 were T3/4 tumors. One hundred twenty-five tumors were graded according to UICC stage I to III and 207 were graded according to stage IV, respectively. One hundred twenty-three tumors presented no regional lymph node metastases at the time of diagnosis, whereas 183 exhibited regional lymph node metastases.
Tissue microarray generation
The OSCC-TMA was generated as previously described [18]. Briefly, hematoxylin-eosin.stained sections were cut from each donor block to define representative tumor regions. Small tissue cylinders with a diameter of 0.6 mm were taken from selected areas of each donor block using a tissue chip microarrayer (Beecher Instruments, Silver Spring, MD) and were transferred to a recipient paraffin block. The recipient paraffin block was cut in 5-µm paraffin sections using standard techniques. Five oral mucosa biopsies from healthy donors were incorporated in the recipient block as control specimen.
Immunohistochemistry
The immunohistochemistry (IHC) was performed using the DAKO Real Kit (DAKO; Hamburg, Germany) according to the manufacturer's protocol. Monoclonal mouse antibodies against the phosphorylated forms of ERK1 and ERK5 (Cell Signaling Technology, Beverly, MA) were used at a dilution of 1:1000 for IHC experiments. The primary antibody for phosphorylated ERK1 was the rabbit monoclonal phosphor-p44/42 mitogen-activated protein (MAP) kinase (Thr202/Tyr204) antibody that detects endogenous levels of human p42 and p44 MAP kinase (ERK1 and ERK2) only when they are phosphorylated at Thr202 and Tyr204, respectively. The primary antibody for phosphorylated ERK5 was rabbit phospho-Erk5 (Thr218/Tyr220) antibody that detects endogenous levels of human ERK5 MAP kinase only when they are phosphorylated at Thr218 and Tyr220, respectively. Evaluation of IHC experiments was based on the percentage of cells, which showed distinct nuclear and cytoplasmic staining. Oral mucosa control specimen showed slight nuclear and cytoplasmic staining, which was set as baseline immunoreactivity (0/None) in the following arbitrary score: none, <5% cytoplasmic and nuclear staining; weak, 5% to 30% cytoplasmic and nuclear staining; moderate, 31% to 60% cytoplasmic and nuclear staining; and strong, >60% cytoplasmic and nuclear staining. For statistical analyses, none and weak stainings were combined and counted as low expression, whereas moderate and strong stainings were grouped together and scored as high expression.
Statistical analysis
Nonparametric univariate analysis using chisquare test was performed to compare the prevalence of high ERK1 and ERK5 expression with T-stadium, UICC stage, and the presence of lymph node metastases of the primary tumors. For overall survival, Kaplan-Meier curves of tumors with high versus low ERK1 and ERK5 expression were analyzed by log-rank test. P ≤ .05 was considered significant. All statistical analyses were performed using R for windows version 2.4.1.
Results
70mere Microarray Expression Profiling
In the present study, n = 35 tumor specimens of primary OSCCs were analyzed for global gene expression using 70mere oligonucleotide microarrays containing 35,035 gene-specific 70mere oligonucleotides. After quality control, an analysis of variance and a following Bonferroni multitest were performed for the identification of differential gene expression between normal tissue and tumor samples. Gene expression was considered as significantly changed, if exceeding the multiple testing cutoff, computed in this case with -log10(P) of 5.65 according to the Bonferroni criterion. Using this approach, we identified 7390 genes significantly and differentially expressed between OSCC and normal oral mucosa (Table W2).
Pathway Analysis
To obtain a comprehensive view of global gene expression in OSCC, pathway analysis was applied to this data set of 7390 genes to identify molecular pathways that contained a number of altered transcript levels in OSCC compared to healthy mucosa. MAPPFinder, a component of the software package GenMAPP version 2.1 containing 96 MAPPs, was used. We ran the MAPPFinder analysis on this data set using two criteria, either an increase (fold change > 1 and -log10(P) > 5.65) or a decrease (fold change < -1 and -log10(P) > 5.65) in gene expression to obtain pathways with mostly upregulated genes and pathways with mostly downregulated genes. A pathway was defined as significantly affected by differentially expressed genes, if P ≤ .05.
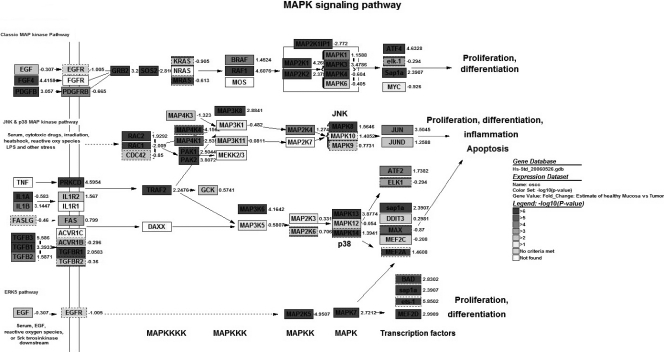
In this respect, several significantly altered pathways were revealed with this approach. We found an amount of twenty-three deregulated pathways with MAPPFinder (Table 1). Of those pathways, which were upregulated in the tumor samples analyzed, several were related to inflammatory response system [B cell receptor, P < .001; interleukin (IL) 5, P < .001; IL-7, P < .001; IL-2, P < .001; tumor necrosis factor alpha (TNFα)-nuclear factor kappa B (NF-κB), P < .001; IL-6, P = .004; IL-4, P = .019; T cell receptor, P = .035]. Other pathways with a high number of differentially expressed genes were cancer-related like MAP kinase signaling (P < .001; Figure 1), transforming growth factor-beta (TGF-β) signaling (P = .015), and signaling pathways involved in apoptosis (P = .037). Pathways with significantly downregulated genes were the ribosomal protein pathway (P < .001) and the pathway of members of the electron transport chain (P < .001). The MAP kinase signaling pathway exhibited the highest number of altered genes. The altered expressed genes of this pathway are listed in Table 2.
Table 1.
Identification of Molecular Pathways with Significantly Upregulated (Above) and Downregulated Genes (Below) in OSCC Compared to Healthy Mucosa By MAPPFinder Analysis.
| MAPP Name | n (Changed) | n (Measured) | n (on MAPP) | % (Changed) | % (Present) | z | P |
| MAPK signaling pathway_ | 72 | 151 | 162 | 47.7 | 93.2 | 4.675 | 0 |
| TNFα-NF-κB | 80 | 181 | 187 | 44.2 | 96.8 | 4.096 | 0 |
| IL-2 NetPath 14 | 38 | 75 | 76 | 50.7 | 98.7 | 3.817 | 0 |
| IL-7 NetPath 19 | 25 | 44 | 44 | 56.8 | 100.0 | 3.799 | 0 |
| IL-5 NetPath 17 | 34 | 66 | 69 | 51.5 | 95.7 | 3.726 | 0 |
| B cell receptor NetPath 12 | 68 | 155 | 158 | 43.9 | 98.1 | 3.681 | 0 |
| GPCRDB_Other | 11 | 83 | 119 | 13.3 | 69.7 | .3.484 | 0 |
| GPCRDB class rhodopsin-like | 27 | 212 | 262 | 12.7 | 80.9 | .5.873 | 0 |
| Insulin signaling | 65 | 155 | 159 | 41.9 | 97.5 | 3.144 | .002 |
| IL-6 NetPath 18 | 42 | 96 | 100 | 43.8 | 96.0 | 2.839 | .004 |
| p38 MAPK signaling pathway | 17 | 32 | 34 | 53.1 | 94.1 | 2.777 | .006 |
| IL-3 NetPath 15 | 43 | 99 | 101 | 43.4 | 98.0 | 2.816 | .007 |
| TGF-β receptor | 57 | 145 | 151 | 39.3 | 96.0 | 2.331 | .015 |
| Proteasome degradation | 23 | 50 | 61 | 46.0 | 82.0 | 2.38 | .017 |
| IL-4 NetPath 16 | 27 | 62 | 62 | 43.5 | 100.0 | 2.233 | .019 |
| Peptide GPCRs | 10 | 60 | 73 | 16.7 | 82.2 | .2.37 | .026 |
| IL-9 NetPath 20 | 12 | 23 | 24 | 52.2 | 95.8 | 2.252 | .032 |
| T cell receptor NetPath 11 | 50 | 129 | 135 | 38.8 | 95.6 | 2.053 | .035 |
| mRNA processing reactome | 48 | 121 | 127 | 39.7 | 95.3 | 2.207 | .036 |
| Apoptosis | 33 | 80 | 82 | 41.3 | 97.6 | 2.092 | .037 |
| Fas pathway and stress induction | 17 | 36 | 38 | 47.2 | 94.7 | 2.175 | .041 |
| Fatty acid beta oxidation 3 | 5 | 8 | 8 | 62.5 | 100.0 | 1.959 | .046 |
| Ribosomal_Proteins | 35 | 84 | 88 | 41.7 | 95.5 | 15.668 | 0 |
| Electron transport chain | 21 | 103 | 105 | 20.4 | 98.1 | 7.309 | 0 |
Figure 1.
Overview of differentially expressed genes in 35 OSCC compared to six healthy mucosa specimen involved in MAP kinase signaling (modified from the KEGG MAPK pathway). Dark gray color of gene symbols represents the significance of difference [-log10 (P)]; the corresponding numbers are the average fold change (log fold).
Table 2.
Increased Expression of Genes Involved in MAP-Kinase Signaling Pathway in 35 OSCC Samples Compared to Six Healthy Mucosa Specimens.
| Ensembl ID | Gene Symbol | Gene Name | -log10(P) | log (Fold Change) |
| ENSG00000100311 | PDGFB | Platelet-derived growth factor B chain precursor | 15.467 | 3.05696878 |
| ENSG00000180370 | PAK2 | Serine/threonine-protein kinase PAK 2 | 14.294 | 3.807238826 |
| ENSG00000107566 | CHUK | SPFH domain-containing protein 1 precursor | 14.274 | 4.832758228 |
| ENSG00000114738 | MAPKAPK3 | MAP kinase-activated protein kinase 3 | 14.104 | 4.067114259 |
| ENSG00000089022 | MAPKAPK5 | MAP kinase-activated protein kinase 5 | 12.030 | 2.129482743 |
| ENSG00000137764 | MAP2K5 | Dual-specificity mitogen-activated protein kinase kinase 5 | 11.859 | 4.950684923 |
| ENSG00000105221 | AKT2 | RAC-beta serine/threonine-protein kinase | 11.838 | 5.375739428 |
| ENSG00000154229 | PRKCA | Protein kinase C alpha type | 11.549 | 2.137398935 |
| ENSG00000119699 | TGFB3 | Transforming growth factor beta-3 precursor | 11.317 | 5.585986774 |
| ENSG00000166484 | MAPK7 | Mitogen-activated protein kinase 7 | 11.211 | 2.721242004 |
| ENSG00000105550 | FGF21 | Fibroblast growth factor 21 precursor (FGF-21) | 11.099 | 1.828757871 |
| ENSG00000167193 | CRK | Protooncogene C-crk | 10.696 | 4.945734007 |
| ENSG00000142208 | AKT1 | RAC-alpha serine/threonine-protein | 10.577 | 4.734240647 |
| ENSG00000108861 | DUSP3 | Dual-specificity protein phosphatase 3 | 10.476 | 3.187181556 |
| ENSG00000115953 | PPP3R1 | Calcineurin subunit B isoform 1 | 10.403 | 1.687353652 |
| ENSG00000156711 | MAPK13 | Mitogen-activated protein kinase 13 | 9.925 | 3.877349348 |
| ENSG00000085276 | EVI1 | Ecotropic virus integration 1 site protein | 9.862 | 1.102867786 |
| ENSG00000099725 | PRKY | Serine/threonine-protein kinase PRKY | 9.857 | 4.186894009 |
| ENSG00000126458 | RRAS | Ras-related protein R-Ras | 9.828 | 3.738753854 |
| ENSG00000142733 | MAP3K6 | Mitogen-activated protein kinase kinase kinase 6 | 9.684 | 4.164230763 |
| ENSG00000112658 | SRF | Serum response factor | 9.667 | 2.390663287 |
| ENSG00000169032 | MAP2K1 | Dual-specificity mitogen-activated protein kinase kinase 1 | 9.664 | 4.265539167 |
| ENSG00000112062 | MAPK14 | Mitogen-activated protein kinase 14 | 9.647 | 1.394125597 |
| ENSG00000138794 | CASP6 | Caspase-6 precursor | 9.462 | 2.734601578 |
| ENSG00000149269 | PAK1 | Serine/threonine-protein kinase PAK 1 | 9.420 | 2.504398448 |
| ENSG00000106799 | TGFBR1 | TGF-beta receptor type I precursor | 9.414 | 2.058273508 |
| ENSG00000174775 | HRAS | GTPase HRas precursor | 9.398 | 5.345430439 |
| ENSG00000104365 | IKBKB | Inhibitor of nuclear factor kappa B kinase beta subunit | 9.303 | 2.735869467 |
| ENSG00000162889 | MAPKAPK2 | MAP kinase-activated protein kinase 2 | 9.128 | 3.689152094 |
| ENSG00000109971 | HSPA8 | Heat shock cognate 71 kDa protein | 9.061 | 4.628190782 |
| ENSG00000077782 | FGFR1 | Basic fibroblast growth factor receptor 1 precursor | 9.035 | 4.913382054 |
| ENSG00000075429 | CACNG5 | Voltage-dependent calcium channel gamma-5 subunit | 8.976 | .1.9493298 |
| ENSG00000127191 | TRAF2 | TNF receptor-associated factor 2 | 8.954 | 2.247565686 |
| ENSG00000113013 | HSPA9B | Stress-70 protein, mitochondrial precursor | 8.893 | 5.378858628 |
| ENSG00000177885 | GRB2 | Growth factor receptor-bound protein 2 | 8.777 | 3.249343399 |
| ENSG00000132155 | RAF1 | RAF protooncogene serine/threonine-protein kinase | 8.731 | 4.607548955 |
| ENSG00000011485 | PPP5C | Serine/threonine-protein phosphatase 5 | 8.703 | 2.290087262 |
| ENSG00000071054 | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | 8.438 | 4.156174225 |
| ENSG00000161326 | DUSP14 | Dual-specificity protein phosphatase 14 | 8.327 | 4.528707616 |
| ENSG00000067191 | CACNB1 | Voltage-dependent L-type calcium channel beta-1 subunit | 7.982 | 1.761273845 |
| ENSG00000107643 | MAPK8 | Mitogen-activated protein kinase 8 | 7.942 | 1.56455685 |
| ENSG00000106211 | HSPB1 | Heat shock protein beta-1 (HspB1) | 7.838 | 1.440732073 |
| ENSG00000126934 | MAP2K2 | Dual-specificity mitogen-activated protein kinase kinase 2 | 7.818 | 2.378935599 |
| ENSG00000140285 | FGF7 | Fibroblast growth factor 7 precursor (FGF-7) | 7.738 | .1.472920826 |
| ENSG00000120875 | DUSP4 | Dual-specificity protein phosphatase 4 | 7.623 | 2.488597455 |
| ENSG00000105329 | TGFB1 | Transforming growth factor beta-1 precursor | 7.621 | 3.393246251 |
| ENSG00000134259 | NGFB | Beta-nerve growth factor precursor (Beta-NGF) | 7.586 | 1.28841258 |
| ENSG00000075388 | FGF4 | Fibroblast growth factor 4 precursor (FGF-4) | 7.549 | 4.415819419 |
| ENSG00000128272 | ATF4 | Cyclic AMP-dependent transcription factor ATF-4 | 7.490 | 4.632776685 |
| ENSG00000126583 | PRKCG | Protein kinase C gamma type | 7.399 | 2.201198322 |
| ENSG00000166501 | PRKCB1 | Protein kinase C beta type | 7.396 | 2.359630993 |
| ENSG00000132906 | CASP9 | Caspase-9 precursor | 7.367 | 4.394431415 |
| ENSG00000100485 | SOS2 | Son of sevenless homolog 2 | 7.271 | 2.816563747 |
| ENSG00000128340 | RAC2 | Ras-related C3 Botulinum toxin substrate 2 precursor | 7.256 | 1.929198943 |
| ENSG00000107968 | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 7.202 | 2.884100613 |
| ENSG00000138834 | MAPK8IP3 | C-jun-amino-terminal kinase-interacting protein 3 | 7.163 | 2.707867082 |
| ENSG00000123739 | PLA2G12A | Group XIIA secretory phospholipase A2 precursor | 7.105 | .1.889796803 |
| ENSG00000143851 | PTPN7 | Tyrosine-protein phosphatase nonreceptor type 7 | 6.948 | 1.921973144 |
| ENSG00000102882 | MAPK3 | Mitogen-activated protein kinase 3 | 6.906 | 3.478604152 |
| ENSG00000176697 | BDNF | Brain-derived neurotrophic factor precursor (BDNF) | 6.825 | 1.762730087 |
| ENSG00000141480 | ARRB2 | Beta-arrestin-2 (Arrestin, beta 2) | 6.756 | 2.512394619 |
| ENSG00000085511 | MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | 6.655 | 1.171686631 |
| ENSG00000186895 | FGF3 | Fibroblast growth factor 3 precursor (FGF-3) | 6.651 | 1.401698475 |
| ENSG00000115904 | SOS1 | Son of sevenless homolog 1 | 6.630 | 2.41943162 |
| ENSG00000141510 | TP53 | Tumor suppressor p53-binding protein 1 | 6.479 | 2.802984167 |
| ENSG00000073009 | IKBKG | NF-κB essential modulator (NEMO) | 6.358 | 2.203573687 |
| ENSG00000170458 | CD14 | Monocyte differentiation antigen CD14 precursor | 6.330 | 3.763791737 |
| ENSG00000155903 | RASA2 | Ras GTPase-activating protein 2 | 6.092 | 1.297339857 |
| ENSG00000136238 | RAC1 | Ras-related C3 Botulinum toxin substrate 1 precursor | 6.078 | .2.009429284 |
| ENSG00000120129 | DUSP1 | Dual-specificity protein phosphatase 1 | 6.043 | 2.637357306 |
| ENSG00000138032 | PPM1B | Protein phosphatase 2C isoform beta | 6.006 | 3.908525816 |
| ENSG00000058335 | RASGRF1 | Guanine nucleotide-releasing protein | 5.905 | 2.008095195 |
| ENSG00000134853 | PDGFRA | Alpha platelet-derived growth factor receptor precursor | 5.808 | 1.093708029 |
| ENSG00000113578 | FGF1 | Fibroblast growth factor 1 precursor (FGF-1) | 5.786 | 2.292817534 |
| ENSG00000135090 | TAOK3 | Serine/threonine-protein kinase TAO3 | 5.759 | 2.493439143 |
| ENSG00000072062 | PRKACA | cAMP-dependent protein kinase, alpha-catalytic subunit | 5.661 | 1.435441441 |
| ENSG00000104814 | MAP4K1 | Mitogen-activated protein kinase kinase kinase kinase 1 | 5.626 | 2.539747316 |
| ENSG00000142875 | PRKACB | cAMP-dependent protein kinase, beta-catalytic subunit | 5.604 | 2.081661093 |
For each transcript, the mean value of these six mucosa specimens was subtracted. An increased expression was defined if this value was higher than three standard deviations of these six healthy mucosa specimens.
Quantitative Real-Time RT-PCR
To confirm the findings of the microarray analysis, we performed quantitative real-time RT-PCR using primers specific for TRIO, TNFRSF18, EGFR, MAPK3/ERK1, and MAPK7/ERK5. The fold differences in expression between tumor specimen and control mucosa specimen predicted by 70mere microarray expression profiling were compared to those fold differences obtained by RT-PCR (see Figure W1).
Quantitative PCR confirmed the direction of fold change in 62 (84.9%) of 73 analyses. Overall, these results confirm our findings of differential gene expression by microarray analysis.
Tissue Microarray Analyses
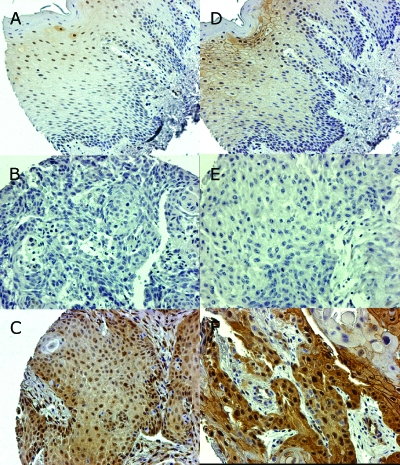
From those pathways found deregulated by cDNA microarray expression profiling in the OSCC tumor samples, further analyses were focused on the MAP kinase signaling pathway, because MAP kinase signaling is supposed to be critically involved in a variety of cancerspecific functions, such as proliferation, dedifferentiation, and evasion from apoptosis. For TMA analysis, antibodies against ERK1 and ERK5, which play a central role in signal transduction in the MAP kinase signaling pathway, were selected. To test whether OSCC also show an increase of ERK1 and ERK5 protein expression, an immunohistochemical analysis was performed on TMA sections (Figure 2). To assess the functional properties of these proteins in the primary tumor tissue, antibodies against the active phosphorylated form of ERK1 and ERK5 were used. The overall frequency of high pERK5 expression was 27.4% (79 of 277). There was a significantly higher prevalence for high pERK5 expression in T1/2 versus T3/4 tumors (P = .015), in Stage I to III versus Stage IV tumors (P = .008), and for tumors with lymph node metastasis (N1-3) versus tumors without lymph node metastasis (N0, P = .016). For high pERK1 expression, the overall frequency was 22.8% (48 of 209) without obtaining any correlation of high pERK1 expression with the clinical parameters analyzed. All data obtained from TMA analyses are shown in Table 3. Kaplan-Meier analysis revealed no difference in the overall survival for tumors with high expression versus low/no expression of the proteins analyzed (P > .05, data not shown).
Figure 2.
Detection of differential pERK1 (left side) and pERK5 (right side) protein expression on TMA sections as detected by IHC. Normal oral mucosa specimens (A and D; original magnification, x20) exhibit a slight pERK1 and pERK5 staining. For OSCC specimen, two examples of absent (B and E; original magnification, x20) and high (C and F; original magnification, x20) ERK1 and ERK5 protein expression are shown.
Table 3.
Frequency of Overexpressed ERK1 and ERK5 in Clinically Defined Tumor Subgroups.
| HNSCC (N = 306) | n | ERK1 | ERK5 |
| T1/2 | 143 | 18.3% (20/109) | 20.3% (27/133) |
| vs | |||
| T3/4 | 163 | 28% (28/100) | 34% (49/144) |
| P | .136 | .015 | |
| N0 | 124 | 18.2% (18/99) | 19.3% (22/114) |
| vs | |||
| N1-3 | 182 | 27.3% (30/110) | 33.1% (54/163) |
| P | .163 | .016 | |
| Stages I–III | 123 | 18.0% (16/89) | 17.9% (19/106) |
| vs | |||
| Stage IV | 183 | 26.7% (32/120) | 33.3% (57/171) |
| P | .19 | .008 |
P values for univariate subgroup analysis are added. P values ≤ .05 are considered as significant.
Discussion
Gene expression profiling to OSCC specimen using different types of cDNA arrays has been performed in several studies to define distinct expression signatures for specific clinical stages [19,20]. To actually understand the process of OSCC progression and metastases, however, it would be essential to know which biologic aberrations are represented by these classifying genes and to what extent they contribute to tumorigenesis. To obtain a comprehensive overview of activated signal transduction pathways in biologic systems, several programs have been developed, which allow the integration and procession of microarray data sets into the context of molecular functions and interrelated dependencies of annotated genes [21–23]. In contrast to those approaches, however, which mainly focus on welldefined metabolic pathways, the program MAPPFinder, which was used in the present analysis, is closely linked to a broader base of pathway information provided by the GO Consortium [11,12]. MAPPFinder dynamically links gene expression data to the GO hierarchy at the level of biologic processes, thereby making it a valuable tool to define novel aberrant pathways in homogenous cohorts of tumor samples [24].
In the present study, a whole-transcriptome expression analysis was performed for 35 OSCC specimens resulting in 7390 differentially expressed transcripts compared to a pool of healthy mucosa samples. By subsequent pathway analysis using MAPPFinder, 24 aberrantly expressed molecular pathways were detected. Among the cancerrelated pathways in this data set, most distinct aberrations—mostly upregulations—were found in the MAP kinase signaling network. The MAP kinase signaling pathways are involved in various cellular functions, including cell proliferation, differentiation, and migration by the activation of protooncogenes such as JUN, FOS, MYC, and ELK1 (Figure 1). FourMAP kinase signaling pathways have been identified, namely, the ERK1 pathway, the c-jun N-terminal-regulated kinase (JNK) pathway, the p38 pathway, and the ERK5 pathway [25]. Whereas the JNK and the p38 pathway are signaling cascades, which are mainly stress-activated by proinflammatory cytokines, ERK1 and ERK5 are additionally induced by epidermal growth factor receptor (EGFR) activation [26]. Because EGFR activation is known to be decisively involved in head and neck squamous cell carcinoma (HNSCC) initiation and progression [27], subsequent analyses were focused on ERK1 and ERK5 signaling pathways in the present study. High ERK1 expression and an association with tumor progression and adverse clinical outcome had been observed in several tumor entities, e.g. in primary hepatocellular carcinoma [28], cholangiocarcinoma [29], breast cancer [30], and non-small cell lung cancer [31]. For HNSCC, an involvement of ERK1 signaling in angiogenic processes through vascular endothelial growth factor activation was postulated [32]. In our present TMA analysis, high ERK1 protein expression was found in about 20% of primary OSCC tumor samples. Surprisingly, however, an association with clinical parameters such as advanced UICC stage and presence of lymph node metastases was not found for high ERK1 but for high ERK5 protein expression, although ERK1 and ERK5 coexpression was frequently found. Mitogen signal-regulated ERK5 overexpression was shown to be associated with metastatic prostate cancer [33], but for HNSCC or OSCC, a participation of ERK5 expression in tumor progression has not yet been described. Analyses of tumor cell systems, however, suggested that MAP kinase signaling through ERK5 is a distinct molecular pathway, which might regulate cellular functions such as the activation of protooncogenes originally attributed to ERK1 [34]. Furthermore, it could be shown that ERK5 but not ERK1 signaling contributes to Src-mediated disruption of actin cytoskeleton organization [35]. Because the disorganization of the cytoskeleton is known to be one of the initial steps that are required for the development of metastases, this observation is in concordance with our data of a correlation of high ERK5 expression and the presence of lymph node metastases in OSCC. Therefore, one might speculate that ERK5 signaling is more important in the EGFR-mediated metastasizing process in OSCC than in the classic ERK1 signaling pathway.
A further distinct finding in signaling pathway analysis of the expression profiling data obtained from the OSCC collection was frequently upregulated pathways involved in inflammatory response, e.g., B cell receptor signaling, TNFα signaling, IL-2 signaling, IL-5 signaling, and T cell receptor signaling (Table 1). In general, it is a well-investigated phenomenon that inflammation plays an important role in tumor promotion, particularly in HNSCC development [36]. It was recently postulated that tumor cells can modify the surrounding stroma through the production of cytokines and growth factors and that this locally changed host microenvironment influences the proliferative and invasive behavior of tumor cells [37]. Particularly for HNSCC, it has been demonstrated that primary tumors express a variety of proinflammatory cytokines, including IL-1a, IL-6, IL-8, and granulocyte macrophage colony-stimulating factor that may attract immune effector cells to the tumor microenvironment [38]. In this context, one might interpret the results of the pathway analysis, which showed an increased expression of proinflammatory molecules in the tumor biopsies, as a distinct tumor-specific feature involved in the pathogenesis of the infiltrating process. Conversely, however, inflammatory cytokines are also expressed by cells of the immune system themselves during such an inflammatory process. Therefore, the inflammatory cell infiltration at the invasion front of the tumor might be the source of high cytokine expression as well. Although only tumor samples containing at least 80% tumor cell load in the biopsy were used for expression profiling analysis in the present study, a contamination with cells of the immune system cannot be definitely excluded. Nevertheless, with the development of more efficient microdissection techniques and RNA amplification protocols, a whole-transcriptome expression profiling with smaller RNA samples would be possible. Then, a comprehensive signaling pathway analysis of the expression of immunomodulatory molecules might be a promising approach to further define the role of inflammation in OSCC progression.
Furthermore, pathway analyses showed a significant upregulation of apoptosis-related genes. Although evading apoptosis is a hallmark of almost all malignant tumors, little is known about the actual role of apoptosis-related genes in OSCC development. Previous analyses, for example, suggested increased levels of the anti-apoptotic protein bcl-2 in OSCC samples [39–41]. A recent study, however, using an animal model system exhibiting chemically induced OSCC, found a high expression of the proapoptotic protein bax but decreased levels of bcl-2 in early oral cancer specimens [39]. Another study on advanced primary OSCC showed an association of high expression of the antiapoptotic protein survivin, with favorable outcome of the patients [42]. Tissue microarray technology might be helpful to further delineate the precise role of apoptosis-related genes in OSCC initiation and progression.
Whereas most of the aberrant signaling pathways were identified as upregulated, the expression of ribosomal proteins was found as significantly downregulated in the tumor collection analyzed. Furthermore, MAPPFinder analysis defined the highest z value for ribosomal protein signaling of all pathways analyzed, indicating that its downregulation might be a decisive aberration in OSCC specimen compared to oral mucosa specimen. Only a few published studies have evaluated the role of cytoplasmic ribosomal proteins in carcinogenesis. Significant changes in the expression of several ribosomal proteins have been reported to occur in colorectal carcinoma [43]. In ovarian tumor cell lines, ribosomal proteins, such as S8, S24, and L32, are much more abundantly expressed in differentiated tumor cells than in lessdifferentiated ones [44]. For OSCC, a downregulation of some ribosomal proteins were found in a previous cDNA expression profiling study, with the downregulation of ribosomal protein S13 discriminating between metastatic and nonmetastatic OSCC in small tumor collection [45]. Because of the observation that a large number of genes encoding ribosomal protein are downregulated in OSCC in our study, further detailed investigation to prove the significance of such global downregulation is warranted.
Pathway signaling analyses results not only contribute to a better understanding of molecular and cellular functions and the definition of potential biomarkers but also open the gate to novel molecular targets for specific therapeutic approaches. For ERK1, the negative effect on cell proliferation using specific ERK1 inhibitors has been shown in several tumor systems. In metastasizing neuroblastoma, ERK1 inhibition by the bisphosphonate zoledronic results in a decrease in tumor cell proliferation and an increase in tumor cell apoptosis [46]. For papillary thyroid carcinoma carrying an ERK1-activating BRAF mutation, specific ERK1 inhibition resulted in tumor cell growth arrest [47]. Similar effects of tumor cell growth inhibition after blocking ERK1-mediated signaling were found in cholangiocarcinoma [48] and hepatocellular carcinoma [49]. For HNSCC, targeted therapy has been recently established in clinical management by the use of the monoclonal antibody cetuximab that selectively inhibits EGFR. Although cetuximab has proven antitumor activity as a single agent and in combination with radio- and chemotherapy [50], a significant number of patients do not adequately respond to cetuximab treatment. Therefore, it has been postulated that a combination therapy with the additional specific inhibition of downstream mediators of EGFR signaling might be a useful approach to increase cetuximab response [27,51]. In this context, according to the data from the present study, ERK5, as a downstream mediator of EGFR, which is associated with advanced tumor stage, might be a novel molecular target. The use of specific ERK5 inhibitors to block EGFR-induced tumor cell proliferation might be a promising approach to support antitumor activity of cetuximab in HNSCC treatment.
In conclusion, the combined experimental approach of wholetranscriptome expression profiling, automated signaling pathway analysis and protein expression analyses by tissue microarrays resulted in the rapid definition of molecular components, which are critically involved in tumor progression. It allowed the evaluation of diagnostic and prognostic biomarkers as well as the definition of novel promising therapeutic targets. ERK5-mediated signaling of EGFR activation might be more important for OSCC progression than ERK1-mediated signaling, suggesting ERK5 as a potential interference point in future therapeutic approaches.
Supplementary Material
Acknowledgments
The authors thank Cordula Tschuch, Sebastian Barbus, Daniel Haag, and Frauke Devens for chip printing and technical assistance.
Footnotes
Supported in part by the National Genome Research Network (NGFN2-01GS0460/01GR0418), the Tumorzentrum Heidelberg/Mannheim (FSP1-4), and the Medical Faculty of the University Heidelberg.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figure W1 and are available online at www.neoplasia.com.
Both authors contributed equally to this work.
References
- 1.Funk GF, Karnell LH, Robinson RA, Zhen WK, Trask DK, Hoffman HT. Presentation, treatment, and outcome of oral cavity cancer: a National Cancer Data Base report. Head Neck. 2002;24:165–180. doi: 10.1002/hed.10004. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg MA, Estefan DJ. Assessing oral malignancies. Am Fam Physician. 2002;65:1379–1384. [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 4.Sticht C, Hofele C, Flechtenmacher C, Bosch FX, Freier K, Lichter P, Joos S. Amplification of cyclin L1 is associated with lymph node metastases in head and neck squamous cell carcinoma (HNSCC) Br J Cancer. 2005;92:770–774. doi: 10.1038/sj.bjc.6602400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigo JP, Lazo PS, Ramos S, Alvarez I, Suarez C. MYC amplification in squamous cell carcinomas of the head and neck. Arch Otolaryngol Head Neck Surg. 1996;122:504–507. doi: 10.1001/archotol.1996.01890170038008. [DOI] [PubMed] [Google Scholar]
- 6.Miyaguchi M, Olofsson J, Hellquist HB. Expression of epidermal growth factor receptor in laryngeal dysplasia and carcinoma. Acta Otolaryngol. 1990;110:309–313. doi: 10.3109/00016489009122553. [DOI] [PubMed] [Google Scholar]
- 7.Boyle JO, Hakim J, Koch W, van der Riet P, Hruban RH, Roa RA, Correo R, Eby YJ, Ruppert JM, Sidransky D. The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res. 1993;53:4477–4480. [PubMed] [Google Scholar]
- 8.Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 9.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 10.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163:1033–1043. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlingemann J, Thuerigen O, Ittrich C, Toedt G, Kramer H, Hahn M, Lichter P. Effective transcriptome amplification for expression profiling on sense-oriented oligonucleotide microarrays. Nucleic Acids Res. 2005;33:e29. doi: 10.1093/nar/gni029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tews B, Felsberg J, Hartmann C, Kunitz A, Hahn M, Toedt G, Neben K, Hummerich L, von Deimling A, Reifenberger G, et al. Identification of novel oligodendroglioma-associated candidate tumor suppressor genes in 1p36 and 19q13 using microarray-based expression profiling. Int J Cancer. 2006;119:792–800. doi: 10.1002/ijc.21901. [DOI] [PubMed] [Google Scholar]
- 16.Korz C, Pscherer A, Benner A, Mertens D, Schaffner C, Leupolt E, Dohner H, Stilgenbauer S, Lichter P. Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood. 2002;99:4554–4561. doi: 10.1182/blood.v99.12.4554. [DOI] [PubMed] [Google Scholar]
- 17.Schwaenen C, Nessling M, Wessendorf S, Salvi T, Wrobel G, Radlwimmer B, Kestler HA, Haslinger C, Stilgenbauer S, Dohner H, et al. Automated array-based genomic profiling in chronic lymphocytic leukemia: development of a clinical tool and discovery of recurrent genomic alterations. Proc Natl Acad Sci USA. 2004;101:1039–1044. doi: 10.1073/pnas.0304717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freier K, Bosch FX, Flechtenmacher C, Devens F, Benner A, Lichter P, Joos S, Hofele C. Distinct site-specific oncoprotein overexpression in head and neck squamous cell carcinoma: a tissue microarray analysis. Anticancer Res. 2003;23:3971–3977. [PubMed] [Google Scholar]
- 19.O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24:1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- 20.Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y, Ohnishi M, Ohyama T, Shingaki S, Kaji M, et al. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer. 2003;106:683–689. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- 21.Karp PD, Riley M, Paley SM, Pellegrini-Toole A. The MetaCyc database. Nucleic Acids Res. 2002;30:59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grosu P, Townsend JP, Hartl DL, Cavalieri D. Pathway processor: a tool for integrating whole-genome expression results into metabolic networks. Genome Res. 2002;12:1121–1126. doi: 10.1101/gr.226602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luyf AC, de Gast J, van Kampen AH. Visualizing metabolic activity on a genome-wide scale. Bioinformatics. 2002;18:813–818. doi: 10.1093/bioinformatics/18.6.813. [DOI] [PubMed] [Google Scholar]
- 24.Zucchini C, Bianchini M, Valvassori L, Perdichizzi S, Benini S, Manara MC, Solmi R, Strippoli P, Picci P, Carinci P, et al. Identification of candidate genes involved in the reversal of malignant phenotype of osteosarcoma cells transfected with the liver/bone/kidney alkaline phosphatase gene. Bone. 2004;34:672–679. doi: 10.1016/j.bone.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Garrington TP, Johnson GL. Organization and regulation of mitogenactivated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 26.Kato Y, Tapping RI, Huang S, Watson MH, Ulevitch RJ, Lee JD. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature. 1998;395:713–716. doi: 10.1038/27234. [DOI] [PubMed] [Google Scholar]
- 27.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol. 2006;24:2666–2672. doi: 10.1200/JCO.2005.04.8306. [DOI] [PubMed] [Google Scholar]
- 28.Tsuboi Y, Ichida T, Sugitani S, Genda T, Inayoshi J, Takamura M, Matsuda Y, Nomoto M, Aoyagi Y. Overexpression of extracellular signal-regulated protein kinase and its correlation with proliferation in human hepatocellular carcinoma. Liver Int. 2004;24:432–436. doi: 10.1111/j.1478-3231.2004.0940.x. [DOI] [PubMed] [Google Scholar]
- 29.Jinawath A, Akiyama Y, Yuasa Y, Pairojkul C. Expression of phosphorylated ERK1/2 and homeodomain protein CDX2 in cholangiocarcinoma. J Cancer Res Clin Oncol. 2006;132:805–810. doi: 10.1007/s00432-006-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milde-Langosch K, Bamberger AM, Rieck G, Grund D, Hemminger G, Muller V, Loning T. Expression and prognostic relevance of activated extracellular-regulated kinases (ERK1/2) in breast cancer. Br J Cancer. 2005;92:2206–2215. doi: 10.1038/sj.bjc.6602655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicent S, Lopez-Picazo JM, Toledo G, Lozano MD, Torre W, Garcia-Corchon C, Quero C, Soria JC, Martin-Algarra S, Manzano RG, et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer. 2004;90:1047–1052. doi: 10.1038/sj.bjc.6601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bancroft CC, Chen Z, Dong G, Sunwoo JB, Yeh N, Park C, Van Waes C. Coexpression of proangiogenic factors IL-8 and VEGF by human head and neck squamous cell carcinoma involves coactivation by MEK-MAPK and IKK-NF-kappaB signal pathways. Clin Cancer Res. 2001;7:435–442. [PubMed] [Google Scholar]
- 33.Mehta PB, Jenkins BL, McCarthy L, Thilak L, Robson CN, Neal DE, Leung HY. MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene. 2003;22:1381–1389. doi: 10.1038/sj.onc.1206154. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Tournier C. Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal. 2006;18:753–760. doi: 10.1016/j.cellsig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gleich LL, Salamone FN. Molecular genetics of head and neck cancer. Cancer Control. 2002;9:369–378. doi: 10.1177/107327480200900502. [DOI] [PubMed] [Google Scholar]
- 37.Pupa SM, Menard S, Forti S, Tagliabue E. New insights into the role of extracellular matrix during tumor onset and progression. J Cell Physiol. 2002;192:259–267. doi: 10.1002/jcp.10142. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Malhotra PS, Thomas GR, Ondrey FG, Duffey DC, Smith CW, Enamorado I, Yeh NT, Kroog GS, Rudy S, et al. Expression of proinflammatory and proangiogenic cytokines in patients with head and neck cancer. Clin Cancer Res. 1999;5:1369–1379. [PubMed] [Google Scholar]
- 39.Derka S, Vairaktaris E, Papakosta V, Vassiliou S, Acil Y, Vylliotis A, Spyridonidou S, Lazaris AC, Mourouzis C, Kokkori A, et al. Cell proliferation and apoptosis culminate in early stages of oral oncogenesis. Oral Oncol. 2006;42:540–550. doi: 10.1016/j.oraloncology.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Loro LL, Vintermyr OK, Liavaag PG, Jonsson R, Johannessen AC. Oral squamous cell carcinoma is associated with decreased bcl-2/bax expression ratio and increased apoptosis. Hum Pathol. 1999;30:1097–1105. doi: 10.1016/s0046-8177(99)90229-0. [DOI] [PubMed] [Google Scholar]
- 41.Singh BB, Chandler FW, Jr, Whitaker SB, Forbes-Nelson AE. Immunohistochemical evaluation of bcl-2 oncoprotein in oral dysplasia and carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1998;85:692–698. doi: 10.1016/s1079-2104(98)90037-3. [DOI] [PubMed] [Google Scholar]
- 42.Freier K, Pungs S, Sticht C, Flechtenmacher C, Lichter P, Joos S, Hofele C. High survivin expression is associated with favorable outcome in advanced primary oral squamous cell carcinoma after radiation therapy. Int J Cancer. 2007;120:942–946. doi: 10.1002/ijc.22380. [DOI] [PubMed] [Google Scholar]
- 43.Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA, Nakayama J. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem. 2003;51:567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- 44.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendez E, Cheng C, Farwell DG, Ricks S, Agoff SN, Futran ND, Weymuller EA, Jr, Maronian NC, Zhao LP, Chen C. Transcriptional expression profiles of oral squamous cell carcinomas. Cancer. 2002;95:1482–1494. doi: 10.1002/cncr.10875. [DOI] [PubMed] [Google Scholar]
- 46.Peng H, Sohara Y, Moats RA, Nelson MD, Jr, Groshen SG, Ye W, Reynolds CP, DeClerck YA. The activity of zoledronic acid on neuroblastoma bone metastasis involves inhibition of osteoclasts and tumor cell survival and proliferation. Cancer Res. 2007;67:9346–9355. doi: 10.1158/0008-5472.CAN-06-4508. [DOI] [PubMed] [Google Scholar]
- 47.Henderson YC, Fredrick MJ, Clayman GL. Differential responses of human papillary thyroid cancer cell lines carrying the RET/PTC1 rearrangement or a BRAF mutation to MEK1/2 inhibitors. Arch Otolaryngol Head Neck Surg. 2007;133:810–815. doi: 10.1001/archotol.133.8.810. [DOI] [PubMed] [Google Scholar]
- 48.Jinawath A, Akiyama Y, Sripa B, Yuasa Y. Dual blockade of the Hedgehog and ERK1/2 pathways coordinately decreases proliferation and survival of cholangiocarcinoma cells. J Cancer Res Clin Oncol. 2007;133:271–278. doi: 10.1007/s00432-006-0166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt CM, Wang Y, Wiesenauer C. Novel combination of cyclooxygenase-2 and MEK inhibitors in human hepatocellular carcinoma provides a synergistic increase in apoptosis. J Gastrointest Surg. 2003;7:1024–1033. doi: 10.1016/j.gassur.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Astsaturov I, Cohen RB, Harari P. Targeting epidermal growth factor receptor signaling in the treatment of head and neck cancer. Expert Rev Anticancer Ther. 2006;6:1179–1193. doi: 10.1586/14737140.6.9.1179. [DOI] [PubMed] [Google Scholar]
- 51.Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Res. 2004;64:5355–5362. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.