Abstract
Motivation: The ability of human immunodeficiency virus-1 (HIV-1) protease to develop mutations that confer multi-drug resistance (MDR) has been a major obstacle in designing rational therapies against HIV. Resistance is usually imparted by a cooperative mechanism that can be elucidated by a covariance analysis of sequence data. Identification of such correlated substitutions of amino acids may be obscured by evolutionary noise.
Results: HIV-1 protease sequences from patients subjected to different specific treatments (set 1), and from untreated patients (set 2) were subjected to sequence covariance analysis by evaluating the mutual information (MI) between all residue pairs. Spectral clustering of the resulting covariance matrices disclosed two distinctive clusters of correlated residues: the first, observed in set 1 but absent in set 2, contained residues involved in MDR acquisition; and the second, included those residues differentiated in the various HIV-1 protease subtypes, shortly referred to as the phylogenetic cluster. The MDR cluster occupies sites close to the central symmetry axis of the enzyme, which overlap with the global hinge region identified from coarse-grained normal-mode analysis of the enzyme structure. The phylogenetic cluster, on the other hand, occupies solvent-exposed and highly mobile regions. This study demonstrates (i) the possibility of distinguishing between the correlated substitutions resulting from neutral mutations and those induced by MDR upon appropriate clustering analysis of sequence covariance data and (ii) a connection between global dynamics and functional substitution of amino acids.
Contact: bahar@ccbb.pitt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
HIV-1 protease plays an important role in the late stage of viral replication by cleavage of premature viral polypeptides to peptides that fold into mature virus proteins. The ability of HIV-1 protease to rapidly acquire a variety of mutants in response to various protease inhibitors (PI) confers the enzyme with high resistance to anti-AIDS treatments. A high cooperativity has been documented among drug-resistant mutations observed in HIV-1 protease (Ohtaka et al., 2003). The sequence data retrieved from treated patients is likely to include mutations that reflect cooperative effects originating from late functional constraints, rather than stochastic evolutionary noise (Atchley et al., 2000). Extensive studies have been made on this protein structure and dynamics (Cecconi et al., 2001; Hornak et al., 2006; Perryman et al., 2004; Zoete et al., 2002) although the molecular mechanisms of multi-drug resistance (MDR) is yet to be elucidated.
HIV-1 protease is particularly suitable for covariance analysis because of the large sets of sequences available, and the observed fast rate of mutations in response to treatments. Sequence covariance analysis is a method widely used for identifying correlated sites in proteins. Such correlations are usually inferred from the statistical analysis of pairwise amino-acid substitutions among the members of the examined family of proteins. Because correlated substitutions are expected to occur between residue pairs directly interacting in the 3-dimensional (3D) structure, sequence covariance analysis, also referred to as correlated mutation analysis (CMA), has long been used for detecting inter-residue contacts within proteins (Eyal et al., 2007a, b; Gobel et al., 1994; Olmea et al., 1999; Shindyalov et al., 1994; Thomas et al., 1996). More recently, the same approach proved useful in identifying communication pathways in allosteric proteins (Hatley et al., 2003; Kass and Horovitz, 2002; Lockless and Ranganathan, 1999; Shulman et al., 2004; Süel et al., 2003), and in studying drug-induced mutations using clinical data (Hoffman et al., 2003; Wu et al., 2003).
The CMA procedure consists of three steps, in general: (i) generation of multiple sequence alignment (MSA) using homologous protein sequences; (ii) quantifying the covariance between different columns in MSA and (iii) identifying groups of highly covariant positions, also called clustering. The underlying assumption is that co-varying residues reflect essential structural/functional inter-residue couplings.
These techniques have some major limitations. The purpose of the method is to identify inter-residue couplings that are directly relevant to protein structure or function. However, the observed signals may not solely arise from such couplings. In fact sequence data are known to be noisy. A strong covariance may be detected among columns due to evolutionary signals that originate from early random mutation events. Noivirt et al. (2005) have shown that the signal due to inter-residue interactions is comparable in magnitude to the noise caused by other stochastic evolutionary events.
Several metrics have been used to quantify sequence covariance in proteins. A comparative analysis of some commonly used methods can be found in the studies of Fodor and Aldrich (2004) and Halperin et al. (2006). Yet, not enough attention has been given to date, to the clustering step. This step is important due to various reasons. First, although the CMA is performed in a pairwise manner (mainly due to technical and statistical reasons), it is clear that in nature larger sets of residues are expected to co-evolve to meet particular structural/functional requirements. Second, the clustering procedure is expected to help in distinguishing the real correlations from the background noise. The choice of clustering technique may also depend on the adopted CMA. When an asymmetric metric like the statistical coupling analysis (SCA) introduced by Ranganathan and coworkers (Lockless and Ranganathan, 1999) is used in step 2, a hierarchical clustering is conveniently applied (Chen et al., 2006; Hatley et al., 2003; Shulman et al., 2004; Süel et al., 2003). For symmetric metrics such as Pearson correlation coefficient and MI, on the other hand, a common procedure is to perform a principal component analysis (Wold et al., 1987; Fleishman et al., 2001).
We adopt the MI content as a measure of the correlation between residue substitutions (Atchley et al., 2000; Clarke, 1995; Hoffman et al., 2003; Martin et al., 2005). Accordingly, each of the N columns in the MSA generated for a protein of N residues is considered as a discrete random variable Xi (1 ≤ i ≤ N) that takes on one of the 20 amino-acid types with some probability. The MI (Cover and Thomas, 1991) associated with the random variables Xi and Xj corresponding to the ith and jth columns is defined as
| (1) |
Here P(Xi = xi, Xj = xj) is the joint probability of occurrence of amino-acid types xi and xj at the ith and jth positions, respectively, P(Xi = xi) and P(Xj = xj) are the corresponding singlet probabilities. I(Xi, Xj) is the ijth element of the N × N MI matrix I corresponding to the examined MSA.
In the present study, we introduce the use of spectral partitioning methods for efficient analysis of the MI matrices derived for HIV-1 protease sequences retrieved from the Stanford HIV Drug Resistance database (DB) (http://hivdb.stanford.edu; Rhee et al., 2003) (Table 1). This DB includes sequences obtained from isolates along with information on the type of PIs given to the patients (accessible via the ‘Detailed Treatment Queries’ interface of the DB). The goal is to examine sequence co-variance and distinguish between correlations of different origin. Spectral clustering was originally proposed for partitioning the nodes in an undirected weighted graph G = (V, E). The weight wij of each edge eij is defined as a measure of similarity between nodes vi and vj. This weight matrix W is replaced in our work by the MI matrix. Our objective will be to partition all the nodes/residues into groups, such that the similarity is high among the nodes within a group and low across different groups. This goal will be achieved by minimizing the normalized cut (Shi and Malik, 2000) between groups (see Materials and Methods).
Table 1.
Summary of Data
| Dataset | Treatment | Number of sequences |
|---|---|---|
| 1 | Treated | 7758 |
| 2 | Untreated | 8761 |
| 3 | IDV only | 1112 |
| 4 | IDV + | 2569 |
| 5 | NFV only | 885 |
| 6 | NFV + | 2131 |
In the ‘Treatment’ column, ‘treated’ means at least one PI is used in the treatment. ‘IDV +’ and ‘NFV +’ means that at least one of the other PIs has been used in combination with the one before the ‘+’ sign. IDV and NFV are the respective PI drugs indinavir and nelfinavir.
We show that the method successfully identifies the residues cooperatively involved in MDR, as well as the mutational patterns arising from different drug treatments. The results suggest that spectral partitioning of the covariance data can help in detecting cooperative functional relations and discriminating to a certain degree between the covariance patterns originating from functional constraints and those associated with neutral/stochastic mutation events that occur early in the evolution of the species/family.
2 METHODS
2.1 Mutual information
Mutual information [MI; Equation (1)] describes the mutual dependence of the two random variables Xi, Xj, it can alternatively be expressed as
| (2) |
where
| (3) |
is the entropy of Xi,
| (4) |
is the conditional entropy of Xi given Xj, S(Xi, Xj) is the joint entropy of Xi and Xj. Equation (2) implies that MI is non-negative. The non-negativity of MI permits us to use it in the similarity matrix for spectral clustering.
2.2 Spectral graph partitioning
The normalized cut for two disjoint sets of nodes A and B is defined as
| (5) |
where cut(A, B) is the total weight of edges connecting the nodes in A and B,
and assoc(A, V) is the total weight of connections from A to all nodes in the graph. Shi and Malik (2000) have derived an algorithm to approximately solve the optimization problem of minimizing Ncut(A, B). By adopting a solution for the discrete clustering problem in a continuous space, the problem reduces to solving the generalized eigenvalue problem
| (6) |
where W = {wij} is the matrix of the edge weights, also called similarity or affinity matrix, D is the diagonal matrix with elements, di = Σj wij. λ and y are the generalized eigenvalues and eigenvectors of W, respectively. The difference D−W, also called the Laplacian matrix, is symmetric and positive semi-definite (Chung, 1997). In order to partition a graph of N nodes into k clusters, we utilize the first k eigenvectors y1, …,yk. For the particular case of bi-partitioning the graph (i.e. k = 2), y2 becomes the only eigenvector used as a criterion, since λ1 = 0. In our application, each column in the MSA corresponds to a residue, which in turn is represented as a node in the graph. The MI matrix I replaces W, and the graph (protein) is bi-partitioned based on the elements of y2 (see below).
2.3 k-way clustering
We also performed k-way partitioning of the data using k = 3, 4 and 5. Dataset 1 was chosen for these additional calculations, as the largest dataset that contains data about viruses exposed to PIs. We used the city block distance in k-means clustering. For each k we performed ten runs, and reported the results for the one with the minimum point-to-centroid distance sums.
2.4 Protein dynamics
The Gaussian Network Model (GNM) was applied according to the standard protocol (Yang et al., 2006). We used a cutoff distance of 7.3 Å between Cα atoms to define the Kirchhoff matrix of inter-residue contacts. Details on the methodology may be found in our earlier work (Bahar et al., 1997; Haliloglu et al., 1997) and recent reviews (Rader et al., 2006).
3 RESULTS AND DISCUSSION
3.1 Spectral clustering of CMA results
To investigate the correlation between drug treatment and mutational patterns, we compiled six datasets of sequences, summarized in Table 1. We collected sequences of all subtypes and aligned them against the consensus subtype B sequence (Korber and Myers, 1992). Any sequence shorter than 99 residues was excluded, and all residues with ambiguity were treated as gaps. A MI matrix was generated for each dataset of HIV-1 protease sequences listed in Table 1.
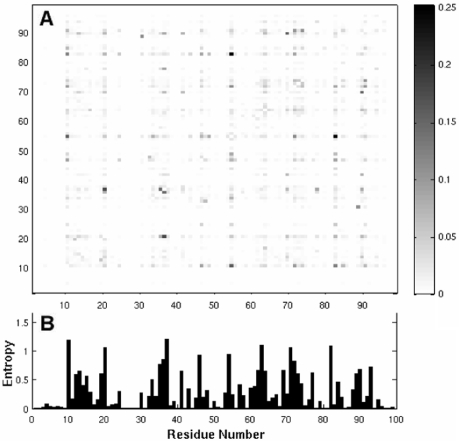
The result for dataset 1 is illustrated in Figure 1. The plot underneath represents the entropy profile, calculated using Equation (3). Peaks are distinguished at positions such as 10, 20, 63 and 82, reflecting the high tendency of these residues to undergo substitutions.
Fig. 1.
MI map (A) and entropy profile (B) for sequences in Dataset 1. The entries in the map are calculated using Equation (1) for the 7758 sequences compiled in Dataset 1 (Table 1). The MI varies in the range 0 < I(X, Y) < 0.25, as indicated by the gray scale on the right. Panel B displays the entropy profile, with the peaks indicating those sites exhibiting the largest variation among the members of this dataset.
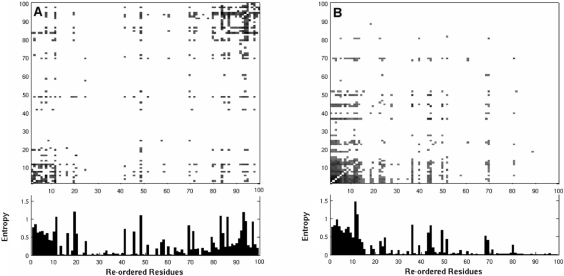
In order to extract more distinctive information, each MI matrix was subjected to spectral graph bi-partitioning as described above, and the elements were re-ordered (i.e. rows/columns were shuffled) according to the rank of residues indicated by the dominant eigenvector y2 (i.e. by sorting the elements of y2 in descending order). Figure 2 displays the MI maps as a function of the re-ordered residues for datasets 1 and 2. The exact labeling of residues following rank ordering can be found in the Supplementary Materials. For visual clarity, the top ranking (highest MI) pairs of amino acids (500 out of a total of 99 × 99 pairs) are displayed. The bar plots refer to the entropy at each site.
Fig. 2.
MI maps with residues re-ordered according to spectral graph bi-clustering (A) Re-organized MI matrix for treated data (dataset 1). Two distinctive types of correlated mutations can be seen at the lower left and upper right portions of the map. (B) Re-organized MI matrix for untreated data (dataset 2). One of the previous clusters is observed (lower left), while the 2nd (top right) is non-existent. The latter is attributed to correlated substitutions induced in the presence of inhibitors, while the former (upper right) refers to evolutionary changes observed between HIV-1 protease subtypes. See Figure 3 for the identity of residues belonging to the two clusters, and the Supplementary Material for the identity of rank-ordered residues for each dataset listed in Table 1. The bar plots refer to the sequence entropy associated with each position. Equivalent figures for the other four datasets can be found in the Supplementary Material.
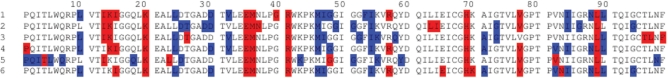
Comparison of panels A and B of Figure 2 reveals that dataset 1 (panel A) contains two distinctive clusters of correlated residues located at the upper right and lower left portions of the map, while dataset 2 does not contain the 2nd cluster (at the upper right) (panel B). The identity of the residues at these two extreme ends of the maps generated for all datasets in Table 1 can be seen in Figure 3. Here we colored in blue and red the first and last 12 residues rank ordered after the spectral bi-partitioning of the MI matrix for each dataset (labeled). Interestingly, all datasets, treated with different regimens or untreated, exhibit similar patterns, with the two groups of residues exhibiting most distinctive correlation behavior clustered at similar sequence positions.
Fig. 3.
Sequence position of two most distinctive clusters of residues. Results are reported for each of the six datasets listed in Table 1. The two clusters include the two extreme subsets of 12 residues rank ordered according to the spectral bi-partitioning of the MI matrix computed for each dataset. The DRC residues are colored blue, the residues belonging to the PhVC are colored red.
3.2 Examination of the two distinctive clusters
Given that the respective datasets 1 and 2 refer to treated and untreated sequences, the cluster at the top right in Figure 2A, which does not exist in panel B, is attributed to the substitutions induced by drug treatment. We will refer to these positions as drug resistance cluster (DRC) sites.
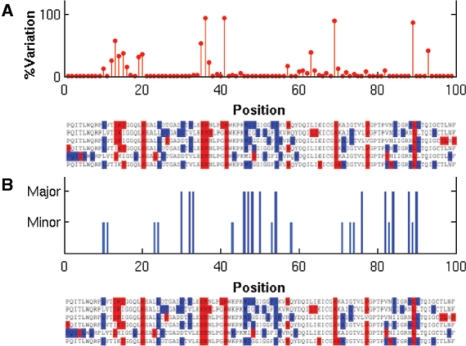
The 2nd cluster of residues, on the other hand, is interestingly found to primarily contain positions reported to exhibit sequence variability between different viral subtype isolates (Gonzales et al., 2001). To verify this feature, we collected 5149 untreated non-B subtype sequences from the Stanford DB, and calculated the variation frequency at each position with respect to the consensus subtype B sequence (Fig. 4A). (More detailed information on the variation for each individual non-B subtype isolates can be found in Fig. 2 of Gonzales et al., 2001). This suggests a phylogenetic origin for the observed covariance, which can well be obtained simply based on few neutral substitution events in the evolution of the HIV subtypes. These residues do not necessarily possess important functional/structural associations (Noivirt et al., 2005). We will refer to this cluster of residues as the phylogenetic variation cluster (PhVC).
Fig. 4.
Comparison with experimental data. (A) Sequence variation profile compiled from experimental data for the non-B subtype HIV (from Stanford DB). Note the correspondence between peaks (most variable sites) and the phylogenetic variation sites (red in the alignment) identified in the present study. (B) Comparison with drug resistance profile (based on data in Stanford DB http://hivdb.stanford.edu/cgi- bin/PIResiNote.cgi). Dark blue lines refer to residues that exhibit major drug resistance; light blue, to minor drug resistance sites.
It should be noted, however, that sequence variations between subtypes are not necessarily functionally insignificant. This is reflected for example by the fact that different subtypes have different tendencies for acquisition of resistance mutations (Kantor et al., 2005). Indeed, residues related to drug resistance can be found in this cluster. Positions 20 and 36 exhibit enhanced mutation rates in the presence of PIs (Wu et al., 2003, Table 2; Hoffman et al., 2003, Fig. 1A). It is possible that the evolution of HIV subtypes is partially related to the exposure to natural or unnatural PIs. Residue Leu89 in the PhVC is known, for example, as a minor drug-resistant residue (meaning that a mutation at this position contributes to drug resistance only in the presence of a major resistant mutation, whereas a major resistant mutation reduces drug susceptibility by itself; Shafer, 2002). Yet, overall, the members of the PhVC are best characterized as those demonstrating sequence variability between subtypes with no clear functional relation between them.
Table 2.
Results from k-way clustering
| k | Cluster |
|---|---|
| 3 | C1: 30, 75, 88 |
| C2: 1– 9, 12–15, 17, 19, 20, 22, 25, 26, 28, 31, 35–42, 45, 49, 52, | |
| 56, 57, 59, 61, 65, 68–70, 77, 83, 87, 89, 96–99 | |
| 4 | C1: 1, 2, 9, 26, 30, 40, 45, 56, 59, 75, 81, 88, 98 |
| C2: 13–15, 20, 35–38, 41, 42, 49, 57, 69, 70, 77, 83, 89 | |
| C3: 10, 23, 24, 27, 32–34, 43, 46–48, 50, 53–55, 58, 71, 76, 80, 82 | |
| 5 | C1: 30, 75, 88 |
| C2: 1, 2, 9, 26, 40, 45, 59, 87, 98 | |
| C3: 13–15, 20, 35–38, 41, 49, 57, 69, 70, 77, 83, 89 | |
| C4: 10, 23, 24, 27, 32–34, 42, 43, 46–48, 50, 53–55, 58, 71,76, 80, 82 |
For clarity, the largest cluster that includes all the remaining residues in each case, is not shown.
In contrast to the PhVC, the DRCs identified for datasets 1, 3, 4 and 6 mostly contain drug-resistant mutations (Fig. 4B). In particular, some residues belonging to these clusters are associated with mutations involved in multi-drug cross-resistance, such as Leu10, Met46, Ile54, Ala71, Val82, Ile84 and Leu90 (Hertogs et al., 2000; Kozal, 2004). In a previous study (Ohtaka et al., 2003), Leu10, Met46, Ile54, Val82, Ile84 and Leu90 were shown to exert a cooperative effect in lowering the affinity of multiple PIs. Leu10, although not causing resistance alone (it is a minor resistance residue), plays a critical role in eliciting the cooperative response along with Leu90 (Ohtaka et al., 2003), consistent with the high correlation detected here among these residues in the DRC. We also note that some major mutation sites in the DRC are not active in MDR; or say, they are specific to one PI, like Leu24 and Asp30 (Shafer, 2002). Still, their participation in the DRC suggests that the resistance mechanism cooperatively involves several residues.
The DRCs for datasets 2 and 5 contain a number of sites that depart from those shared by other datasets (Fig. 3). For dataset 2, which contains untreated isolates only, this is clear, and even the observed level of similarity to other datasets is striking. For dataset 5, on the other hand, the result implies that NFV elicits unique responses at specific sites, quite different from that of most other drugs. We note in particular that Asp30 and Asn88 exhibit extraordinarily high MI. As shown before (Rhee et al., 2003), the double mutation D30N and N88D can reduce nelfinavir susceptibility by 50-fold, explaining the selection pressure for their co-variation. When NFV is used in combination with other PIs (dataset 6), the DRC sites shared with other datasets are observed, indicating that the cooperative effect is related to cross-resistance in this case. Most of the residues of the DRC remain unchanged in the IDV set (dataset 3), suggesting that the correlations revealed in our analysis are not only due to individual resistance mutations developed against different drugs, but reflect real cooperativity.
An exhaustive search for correlated mutations among drug-resistant sites in HIV-1 isolates was performed by Wu et al. (2003), which yielded small groups of correlated residues, ranging in size from three to six residues. On the other hand, the present study yields one large cluster providing evidence for the high cooperativity of the residues belonging to these small groups. We also note that the presently detected positions 47 and 48 in the flap region do not appear in the study by Wu et al. as prominent drug resistance sites, but they are known to be major resistant mutations. Wu et al. listed other residues, e.g. Ile62, Leu63 and Ile93, together with known drug resistance residues. We have not detected these residues in our DRC, and neither do they appear in the Stanford PI DB drug resistance notes as drug-induced mutations. Note that our study is based on a larger dataset of isolates, and a major merit of the present work is to identify the DRC sites without prior knowledge of drug-resistant mutation sites, while the study of Wu et al. analyzes the mutations at 45 (out of 99) positions that have been significantly associated with protease inhibitor treatment.
Hoffman et al. 2003 analyzed the correlations between 31 positions in HIV-1 protease, which showed the highest variability in their dataset of HIV-1 isolates (from 648 untreated, and 531 treated persons). These were grouped in three clusters based on the comparison of mutation rates between treated and untreated datasets. This criterion is different from the one (based on MI data) adopted in our study, but it is still tempting to compare the two sets of results. Those residues in Class III therein are similar to those in our DRC, while Class I resembles our phylogenetic cluster PhVC. Notably, residues Lys20, Met36 which are part of our phylogenetic cluster appear in cluster II and cluster III, respectively. These residues exhibit substantial sequence variability between subtypes, and appear to be relevant to drug resistance, but apparently not in a cooperative manner with other residues.
3.3 k-way clustering using more eigenvectors
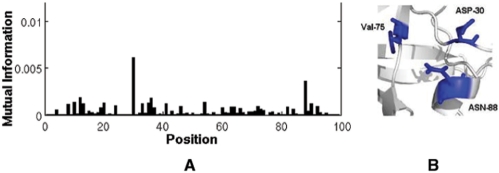
The results from k-way clustering of dataset 1 using k = 3, 4 and 5 are presented in Table 2. The most correlated residues identified above take part in the same clusters, consistent with results from bi-partitioning. Notably, Asp30 and Asn88, which originally belonged to the DRC, exhibited a tendency to form a separate cluster together with Val75. This triplet (Asp30, Asn88, Val75) was also reported to form a cluster in previous work (Wu et al., 2003). It has long been known that co-substitutions at Asp30 and Asn88 are most effective in reducing the susceptibility of nelfinavir; however, little attention has been given to date to their possible association with Val75. As indicated in Figure 5, the high correlation of Val75 with Asp30 and Asn88 (Fig. 5A), consistent with their structural proximity (Fig. 5B), may originate from a cooperative mechanism for drug resistance between these three sites.
Fig. 5.
(A) The MI profile of Val75 with other residues in the treated dataset (dataset1). (B) The structural vicinity of Asp30, Asn88 and Val75. The figure was made with PyMOL (http://www.pymol.org).
3.4 Interpretation with respect to protein dynamics
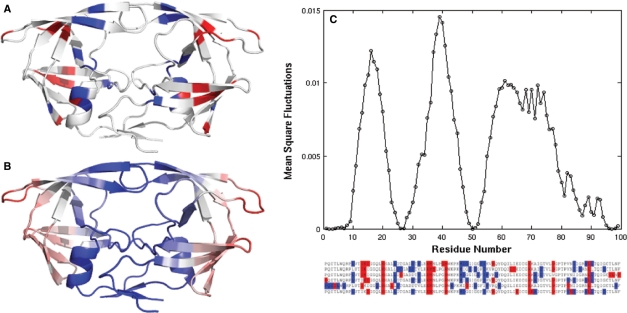
The examination of HIV-1 protease 3D-structure reveals that the residues participating in the DRC tend to occupy the flap region (Met46, Ile47, Ile54), the close neighborhood of the active site (Asp30, Val32, Val82, Ile84), and the dimerization interface (Leu10, Leu90). Most of PhVC residues, on the other hand, are located away from the interface, toward the exterior of the protein (Fig. 6A). Interestingly, both groups of residues assume regular secondary structures (helices or strands), although their relative positions with respect to the interfacial region differs.
Fig. 6.
Comparison of results from CMA and GNM dynamics. (A) The location of the two clusters identified for dataset 1 on the 3D-structure of HIV-1 protease. The DRC is colored blue, and the PVC is colored red. We displayed the residues that have appeared at least three times (out of six examined datasets) in the same cluster in Figure 3. (B) Ribbon diagram color-coded after the mobilities of residues in the first slow mode predicted by the GNM. The residue mobility increases from blue to red. (C) GNM slow-mode profile as a function of residue index. Note that calculations are performed for the dimer, but results are shown for a monomer, the curves for the two monomers being identical. The HIV-1 protease mutant bound with IDV (PDB id: 2B7Z) was used. Ribbon diagrams were made with PyMOL (http://www.pymol.org).
We also examined the distance separation between the closest atoms of residue pairs belonging to the two clusters. Table S2 in the Supplementary Material lists the distances between top-ranking residue pairs in the two clusters (corresponding to the lower left and upper right of the MI map in Fig. 2A). For each pair two values have been considered: intra-molecular (monomers A–A or B–B contacts) or intermolecular (A–B contacts). The distances between the closest atoms for each case are listed. These data clearly demonstrate that the correlated pairs essentially refer to intra-molecular interactions, rather than inter-molecular. Note that the MI method cannot detect the correlations between the fully conserved residues at the interface between the monomers (e.g. P1-F99 and D29-R8).
A further comparison between the results from CMA and the mobilities of residues predicted by the GNM (Bahar et al., 1997) elucidates the close correspondence between the global dynamics of the enzyme and its function. The lowest frequency GNM mode usually defines the global dynamics of the enzyme accessible under native state conditions, and such cooperative motions intrinsically favored by the structure have been shown to relate to enzymatic function (Yang and Bahar, 2005). In particular, the global hinge regions (minima in the mobility profiles driven by global modes) play a critical role in conferring the mechanical properties of enzymes that complement their chemical (catalytic) activities.
In order to examine the dynamics of residues belonging to the DRCs and PhVCs, we performed GNM calculation for an HIV-1 protease mutant bound to IDV (PDB file 2B7Z). This structure contains 10 mutations, most of which belong to the DRC presently identified for the IDV-treated dataset. The color-coded ribbon diagram in panel B of Figure 6, and the slow-mode profile in panel C, display the mobilities in the lowest frequency mode predicted by the GNM for this structure. Comparison of panels A and B shows that the DRC residues tend to occupy positions that are highly constrained in the global mode, whereas PhVC residues are located at relatively flexible positions. These distinctive dynamics of the two groups of residues explains the fact that the PhVCs are accommodated without altering the structure and function; whereas mutations at the DRC sites that are more buried and spatially constrained have functional consequences. Calculations repeated for the substrate-bound complex (PDB id: 2FNS) confirmed that the slow-mode profile is insensitive to structural asymmetry and yielded the almost identical profiles for the two subunits, while the 2nd mode exhibited a stronger dependence on structural asymmetry (see Supplementary Material).
Finally, we compare the global mobility profile (panel C) with the sequence position of the two clusters (Fig. 3) reproduced in Figure 6 to ease the visual comparison. The residues in the DRC are seen to usually lie close to global hinge regions (minima), while those in the PhVC are distributed in high mobility regions. Calculations were repeated for the 2nd and 3rd GNM modes as well. Comparison of the minima and maxima in these modes with the PhVC (red) and DRC (blue) sites along the sequence shows that PhVC modes exhibit relatively high mobilities in modes 2 and/or 3 as well, whereas the confinement of DRC residues to hinge sites is characteristic of the first (global) mode. The DRC residues located at the flap region (residues 46–54) show a high mobility in modes 2 and 3. See the Supplementary Material for the counterpart of Figure 6C for these two modes. Co-localization of MDR sites with global hinge regions thus emerges as an effective means of impacting the cooperative dynamics, and hence the function of the enzyme (Bahar et al., 1998) and on the catalysis.
4 CONCLUSION
In the present study, we analyzed the covariance patterns in HIV-1 protease sequences using a simple metric, MI, followed by spectral clustering. The approach proved to discriminate between two groups of correlated mutation sites, shortly referred to as DRC and PhVC. Mutations in the DRC tend to confer MDR while those in the PhVC seem to differentiate between different HIV-1 protease subtypes. We have further explored the biophysical basis of the observed differences between the two clusters of correlated sites. The two clusters were found to significantly differ with regard to their role in the intrinsic structural dynamics of the enzyme. The DRC sites select key mechanical regions, near the global hinges that control the most cooperative motions of the enzyme; PhVC residues, on the other hand, preferentially occupy flexible regions that can easily accommodate residue substitutions.
Covariance analysis of related protein sequences is known to be problematic in many aspects (Fodor and Aldrich, 2004; Halperin et al., 2006). Many options exist to improve the basic method presented here. For example, MI treats all substitutions of amino acids equally, ignoring physicochemical preferences. In the future, it may be worth considering different essential covariance measures for further analysis. Methods for assigning significant scores using the original MI scores and shuffling of the original data (Hoffman et al., 2003; Shackelford and Karplus, 2007) can also help in obtaining more meaningful results.
One major goal here was, however, to draw attention to the utility of clustering the covariance data. We utilized a relatively less detailed, but objective and theoretically robust approach. Significantly, this approach allowed us to separate the sequence covariance arising from functional pressures (e.g. MDR) from those evolutionarily selected within the examined phylogeny. Both groups of correlations exhibit strong signals when covariance properties are quantified in terms of MI. Yet, the distinctive character of the two groups, confirmed by experiments (Fig. 4), and rationalized by comparison with structural dynamics (Fig. 6), supports the utility of adopting a spectral bi-clustering method for efficiently discriminating between potential correlations of fundamentally different nature/origin. It will be of interest to further explore the utility of spectral bi-clustering for differentiating between correlated mutations that reflect ‘real’ inter-residue interactions and those reflecting other evolutionary signals, often considered as noise for most analyses purposes (Noivirt et al., 2005).
Notably, some of the sites for potential MDR, indistinguishable in the untreated sequences (Fig. 2B), can be detected upon rank ordering the residues via spectral clustering of MI data; furthermore, treated sequences subjected to different regimens share common DRC residues (Fig. 3). These two observations invite attention to the intrinsic tendency of the enzyme to potentially select those effective sites to develop mutations that confer MDR, irrespective of treatment.
A challenging, yet important task, which is a natural continuation to this work, is to detect correlations between protease residues and residues of other mature/pre-mature proteins of HIV-1. A recent work demonstrates how such correlations can be detected between a protease mutation (V82A) and a mutation at the nucleocapsid-p1 cleavage site (Prabu-Jeyabalan et al., 2004). It remains to be seen if current methodology can be extended to investigating the relation between the protease and other cleavage sites as well as the correlations with other regions in HIV-1 pre-proteins, toward shedding more light on the late stages of the virus maturation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Rieko Ishima for useful discussions. I.B. gratefully acknowledges support from NIH grants 5U54 MH074411 02 and R01 LM007994 03.
Conflict of Interest: none declared.
REFERENCES
- Atchley WR, et al. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol. Biol. Evol. 2000;17:164–178. doi: 10.1093/oxfordjournals.molbev.a026229. [DOI] [PubMed] [Google Scholar]
- Bahar I, et al. Direct evaluation of thermal fluctuations in protein using a single parameter harmonic potential. Fold. Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- Bahar I, et al. Vibrational dynamics of proteins: significance of slow and fast modes in relation to function and stability. Phys. Rev. lett. 1998;80:2733–2736. [Google Scholar]
- Cecconi F, et al. Molecular dynamics studies on HIV-1 protease drug resistance and folding pathways. Proteins. 2001;43:365–372. [PubMed] [Google Scholar]
- Chen Y, et al. Evolutionarily conserved allosteric network in the Cys loop family of ligand-gated ion channels revealed by statistical covariance analyses. J. Biol. Chem. 2006;281:18184–18192. doi: 10.1074/jbc.M600349200. [DOI] [PubMed] [Google Scholar]
- Chung F. Spectral Graph Theory. Washington: Conference board of the Mathematical Sciences; 1997. [Google Scholar]
- Clarke ND. Covariation of residues in the homeodomain sequence family. Protein Sci. 1995;4:2269–2278. doi: 10.1002/pro.5560041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TA, Thomas JA. Elements of Information Theory. New York: John Wiley and Sons; 1991. [Google Scholar]
- Eyal E, et al. A pair-to-pair amino acids substitution matrix and its applications for protein structure prediction. Proteins. 2007a;67:142–153. doi: 10.1002/prot.21223. [DOI] [PubMed] [Google Scholar]
- Eyal E, et al. Rapid assessment of correlated amino acids from pair-to-pair (P2P) substitution matrices. Bioinformatics. 2007b;23:1837–1839. doi: 10.1093/bioinformatics/btm256. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, et al. An evolutionarily conserved network of amino acids mediates gating in voltage-dependent potassium channels. J. Mol. Biol. 2001;340:307–318. doi: 10.1016/j.jmb.2004.04.064. [DOI] [PubMed] [Google Scholar]
- Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- Gobel U, et al. Correlated mutations and residue contacts in proteins. Proteins. 1994;18:309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- Gonzales MJ, et al. Human immunodeficiency virus type 1 reverse-transcriptase and protease subtypes: classification, amino acid mutation patterns, and prevalence in a northern California clinic-based population. J. Infect. Dis. 2001;184:998–1006. doi: 10.1086/323601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haliloglu T, et al. Gaussian dynamics of folded protein. Phys. Rev. Lett. 1997;79:3090–3093. [Google Scholar]
- Halperin I, et al. Correlated mutations: advances and limitations. A study on fusion proteins and on the Cohesin-Dockerin families. Proteins. 2006;63:832–845. doi: 10.1002/prot.20933. [DOI] [PubMed] [Google Scholar]
- Hatley ME, et al. Allosteric determinants in guanine nucleotide-binding proteins. Proc. Natl Acad. Sci. USA. 2003;99:33–54. doi: 10.1073/pnas.1835919100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertogs K, et al. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross resistance: a survey of over 6000 samples. AIDS. 2000;14:1203–1210. doi: 10.1097/00002030-200006160-00018. [DOI] [PubMed] [Google Scholar]
- Hoffman NG, et al. Covariation of amino acid positions in HIV-1 protease. Virology. 2003;314:536–548. doi: 10.1016/s0042-6822(03)00484-7. [DOI] [PubMed] [Google Scholar]
- Hornak V, et al. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proc. Natl Acad. Sci. USA. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass I, Horovitz A. Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins. 2002;48:611–617. doi: 10.1002/prot.10180. [DOI] [PubMed] [Google Scholar]
- Kantor R, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res. Hum. Retro. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- Kozal M. Cross-resistance patterns among HIV protease inhibitors. AIDS Patient Care STDs. 2004;18:199–208. doi: 10.1089/108729104323038874. [DOI] [PubMed] [Google Scholar]
- Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- Martin LC, et al. Using information theory to search for co-evolving residues in proteins. Bioinformatics. 2005;21:4116–4124. doi: 10.1093/bioinformatics/bti671. [DOI] [PubMed] [Google Scholar]
- Noivirt O, et al. Detection and reduction of evolutionary noise in correlated mutation analysis. Protein Eng. Des. Sel. 2005;18:247–253. doi: 10.1093/protein/gzi029. [DOI] [PubMed] [Google Scholar]
- Ohtaka H, et al. Multidrug resistance to HIV-1 protease inhibition requires cooperative coupling between distal mutations. Biochemistry. 2003;42:13659–13666. doi: 10.1021/bi0350405. [DOI] [PubMed] [Google Scholar]
- Olmea O, et al. Effective use of sequence correlation and conservation in fold recognition. J. Mol. Biol. 1999;293:1221–1239. doi: 10.1006/jmbi.1999.3208. [DOI] [PubMed] [Google Scholar]
- Perryman AL, et al. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: Possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004;13:1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabu-Jeyabalan M, et al. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 2004;78:12446–12454. doi: 10.1128/JVI.78.22.12446-12454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader AJ, et al. The Gaussian Network Model: Theory and Applications. In: Cui Q, Bahar I, editors. Normal Mode Analysis. Theory and Applications to Biological and Chemical Systems. Taylor & Francis Group, London: Chapman & Hall/CRC; 2006. pp. 41–64. [Google Scholar]
- Rhee SY, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford G, Karplus K. Contact prediction using mutual information and neural nets. Proteins. 2007;69:159–164. doi: 10.1002/prot.21791. [DOI] [PubMed] [Google Scholar]
- Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev. 2002;15:247–277. doi: 10.1128/CMR.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Malik J. Normalized cut and image segmentation. IEEE Tran. Pattern Anal. Mach. Intell. 2000;22:888–905. [Google Scholar]
- Shindyalov IN, et al. Can three-dimensional contacts in protein structures be predicted by analysis of correlated mutations? Protein Eng. 1994;7:349–358. doi: 10.1093/protein/7.3.349. [DOI] [PubMed] [Google Scholar]
- Shulman AI, et al. Structural determinants of allosteric ligand activation in RXR Heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- Süel GM, et al. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, et al. The prediction of protein contacts from multiple sequence alignments. Protein Eng. 1996;9:941–948. doi: 10.1093/protein/9.11.941. [DOI] [PubMed] [Google Scholar]
- Wold S, et al. Principal component analysis. Chemometr. Intell. Lab. Syst. 1987;2:37–52. [Google Scholar]
- Wu TD, et al. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 2003;77:4836–4847. doi: 10.1128/JVI.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LW, Bahar I. Coupling between catalytic site and collective dynamics: a requirement for mechanochemical activity of enzymes. Structure. 2005;13:893–904. doi: 10.1016/j.str.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LW, et al. oGNM: online computation of structural dynamics using the Gaussian Network Model. Nucleic Acids Res. 2006;34:W24–W31. doi: 10.1093/nar/gkl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V, et al. Relation between sequence and structure of HIV-1 protease inhibitor complexes: a model system for the analysis of protein flexibility. J. Mol. Biol. 2002;315:21–52. doi: 10.1006/jmbi.2001.5173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.