Abstract
Summary: In structural biology and -genomics, nuclear magnetic resonance (NMR) spectroscopy and crystallography are the methods of choice, but sample requirements can be hard to fulfil. Valuable structural information can also be obtained by using a combination of limited proteolysis and mass spectrometry, providing not only knowledge of how to improve sample conditions for crystallization trials or NMR spectrosopy by gaining insight into subdomain identities but also probing tertiary and quaternary structure, folding and stability, ligand binding, protein interactions and the location of post-translational modifications. For high-throughput studies and larger proteins, however, this experimentally fast and easy approach produces considerable amounts of data, which until now has made the evaluation exceedingly laborious if at all manually possible. MTMDAT, equipped with a browser-like graphical user interface, accelerates this evaluation manifold by automated peak picking, assignment, data processing and visualization.
Availability: MTMDAT can be downloaded from the following page: http://www.cms.liu.se/chemistry/molbiotech/maria_sunnerhagens_group/mtmdat by clicking on the corresponding links (windows- or unix-based) together with the manual and example files. The program is free for academic/non-commercial purposes only.
Contact: janhe@ifm.liu.se
1 INTRODUCTION
Limited proteolysis detected by SDS-page has long been used to gain information on domain identities and quaternary rearrangements in larger proteins (Carey, 2000). The introduction of highly resolved mass spectrometry (MS), especially matrix-assisted laser desorption/ionization (MALDI)-time-of-flight (TOF)-MS, enabled more detailed protein characterization, including per-residue-mapping of protein–DNA (Cohen et al., 1995), protein–protein (Lacy et al., 2005) and protein-surface interactions (Lundqvist et al., 2005) by following the degradation of unstable or accumulation of stable protein fragments during a time-course experiment. Despite these promising results, the richness in detail is limited and larger proteins were hardly accessible using this method due to the amount of data generated. Recently, we published an algorithm where a residue-wise relative cleavage propensity is calculated from MS spectra (Hennig et al., 2005). The cleavage propensity visualizes the tertiary or quaternary structure probing of proteins and/or their complexes in more detail by localized mapping of proteolytic protection in protein intra- and interactions. The required data can be rapidly recorded by MALDI-TOF-MS in a matter of hours. However, the large body of data acquired in such experiments presents a problem for manual evaluation since it increases exponentially with the size of the proteins being studied. To this end we developed MTMDAT to assist the data evaluation process. The time needed from recording the data to presenting the information content is thereby reduced from weeks to hours. In a most recent study (Hennig et al., 2008), we used this method and tool to determine subdomain interaction sites. The identified residues were used as input for data-driven docking of these subdomains and yielded valuable results. Thus, this method in conjunction with MTMDAT provides a high-throughput method to screen for protein complex structures without the time-consuming process of sample conditioning and optimizations. In addition, domain identification can be done very quickly, which can facilitate successful crystallization trials in structural genomics, and the identification of inter-domain interactions can picture their arrangement. The usefulness of MTMDAT can even be exploited in proteomic studies, since it provides the user with the possibility to screen for post-translational modifications and to determine their exact location on the protein.
2 WORKFLOW
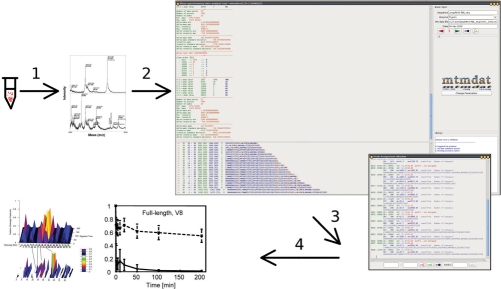
The general workflow is depicted in Figure 1. After performing the actual limited proteolysis experiment, each sampled time point is analysed by MS. Spectra need then to be converted to standard text-(ASCII) files for MTMDAT.
Fig. 1.
Workflow scheme of limited proteolysis experiments analysed by MS and MTMDAT. (1) From the reaction tube to the mass spectrometer, evaluated by MTMDAT (2), doing the peak picking and the peak assignment (3) and produces output for plotting in various ways (4).
MTMDAT requires the sequence of the protein to be analysed, which can be uploaded or pasted into the appropriate field in one-letter-code, where any other characters than those assigned to the 20 essential amino acids are automatically filtered out. The protease used can be chosen from a pulldown menu. All following steps are simply performed by pressing the browser-like forward button five times. First, your protein will be cleaved virtually, and all possible fragments are listed from the shortest to the uncleaved full-length protein, together with the position in the entire sequence, the average mass, the monoisotopic mass and the sequence of the fragment with highlighted cleavage sites. It is also possible to include protein modifications, such as thiol groups modified with mercaptoethanol or acrylamide, acetylation and many more. Modifications not present in the current version can easily be implemented upon request.
The user can now upload MS spectra to be analysed. All information available from the file will be displayed. A noise filter makes it possible for MTMDAT to pick peaks and distinguish them from noise. This noise-filter can be applied automatically and manually. The automatic noise filter is based on common statistical analysis of all data pairs the mass spectrometer generates. If, for any reason (e.g. spectra without baseline correction), the user chooses to manually apply a noise filter (e.g. raise or lower the noise level) it can be done in a separate parameter window. Then the peaks are picked and listed in the main window with their corresponding intensity and mass-to-charge ratio. MTMDAT compares the generated peak list with the virtual fragment list made in the beginning and assigns the experimentally derived peaks to their corresponding fragment mass. A peak assignment window appears, where all assigned peaks are listed. The user can interactively check the assignment and correct it in case of need.
3 THE OUTPUT AND 3D VISUALIZATION
For each fragment in the assignment list, a file will be generated. The file itself consists of xy-data, namely the time point of the sample and the calculated intensity of the fragments’ peak relative to all other peaks in the assignment list. For all subsequently processed spectra of following time points, the xy-data will be saved automatically into the same files given that the user uses the same file name root. Thus, for each fragment MTMDAT produces files, which can be used in any plotting program to plot the change in relative abundance of this peptide fragment over time.
After evaluating all time-resolved MALDI spectra in a single limited proteolysis experiment, MTMDAT can produce a plot file for 3D visualization of relative cleavage propensities. Here, MTMDAT takes all the previously produced time-course plot files and calculates the relative cleavage propensity based on the algorithm in Hennig et al. (2005) (an example of such a plot is shown in Fig. 1). MTMDAT can produce these plot files in several different arrangements for different plotting programs. Furthermore, the user can choose if only cleavable residues are shown on the horizontal axis or the entire sequence.
4 GRAPHICAL USER INTERFACE
A graphical user interface (GUI) is used to simplify and streamline the setup of an evaluation. The GUI enables one to modify all relevant program parameters, such as the noise filter parameters or protein modifications (resulting in different masses in the virtual fragment list, e.g. with β-mercaptoethanol modified cysteines, etc). The browser-like usability enables the user to go back a step if one would like to change parameters. MTMDAT comes as a software written in Java, relying on jre1.6.0 or later.
5 APPLICABILITY
Evaluating the easily accessible but overwhelmingly large body of MALDI data from proteolysis experiments has until now been hampered by the lack of appropriate software. Recently, we successfully applied MTMDAT not only to protein subdomain interactions and protein–ligand interactions, but also determined the origin of stability increase due to Zn2+-binding (Hennig et al., 2008). This provides a high-throughput approach to access limited proteolysis data for larger proteins. We are convinced that MTMDAT will prove helpful to a wide range of scientists in the structural genomics field.
ACKNOWLEDGEMENTS
The authors thank Martin Karlsson for helping in the validation process of MTMDAT.
Funding: This work was supported by grants to M. S. from the Swedish Research Council, Swedish Foundation for Strategic Research, and Linköping University. Instrumentation was funded by the Wallenberg foundation.
Conflicts of Interest: none declared.
REFERENCES
- Carey J. A systematic and general proteolytic method for defining structural and functional domains of proteins. Methods Enzymol. 2000;328:499–514. doi: 10.1016/s0076-6879(00)28415-2. [DOI] [PubMed] [Google Scholar]
- Cohen SL, et al. Probing the solution structure of the DNA-binding protein Max by a combination of proteolysis and mass spectrometry. Protein Sci. 1995;4:1088–1099. doi: 10.1002/pro.5560040607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig J, et al. Structural organization and Zn2+-dependent subdomain interactions involving autoantigenic epitopes in the RING-B-box-Coiled-coil (RBCC) region of Ro52. J. Biol. Chem. 2005;280:33250–33261. doi: 10.1074/jbc.M503066200. [DOI] [PubMed] [Google Scholar]
- Hennig J, et al. The fellowship of the RING: the RING-B-box linker region 5 (RBL) interacts with the RING in TRIM21/Ro52, contains a native autoantigenic epitope in Sjgren syndrome, and is an integral and conserved region in TRIM proteins. J. Mol. Biol. 2008;377:431–449. doi: 10.1016/j.jmb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Lacy ER, et al. Molecular basis for the specificity of p27 toward cyclin-dependent kinases that regulate cell division. J. Mol. Biol. 2005;349:764–773. doi: 10.1016/j.jmb.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Lundqvist M, et al. Proteolytic cleavage reveals interaction patterns between silica nanoparticles and two variants of human carbonic anhydrase. Langmuir. 2005;21:11903–11906. doi: 10.1021/la050477u. [DOI] [PubMed] [Google Scholar]