Abstract
A major problem in predicting the enantioselectivity of an enzyme toward substrate molecules is that even high selectivity toward one substrate enantiomer over the other corresponds to a very small difference in free energy. However, total free energies in enzyme-substrate systems are very large and fluctuate significantly because of general protein motion. Candida antarctica lipase B (CALB), a serine hydrolase, displays enantioselectivity toward secondary alcohols. Here, we present a modeling study where the aim has been to develop a molecular dynamics-based methodology for the prediction of enantioselectivity in CALB. The substrates modeled (seven in total) were 3-methyl-2-butanol with various aliphatic carboxylic acids and also 2-butanol, as well as 3,3-dimethyl-2-butanol with octanoic acid. The tetrahedral reaction intermediate was used as a model of the transition state. Investigative analyses were performed on ensembles of nonminimized structures and focused on the potential energies of a number of subsets within the modeled systems to determine which specific regions are important for the prediction of enantioselectivity. One category of subset was based on atoms that make up the core structural elements of the transition state. We considered that a more favorable energetic conformation of such a subset should relate to a greater likelihood for catalysis to occur, thus reflecting higher selectivity. The results of this study conveyed that the use of this type of subset was viable for the analysis of structural ensembles and yielded good predictions of enantioselectivity.
Keywords: Molecular dynamics, enantioselectivity, enzyme catalysis, transition state, free energy
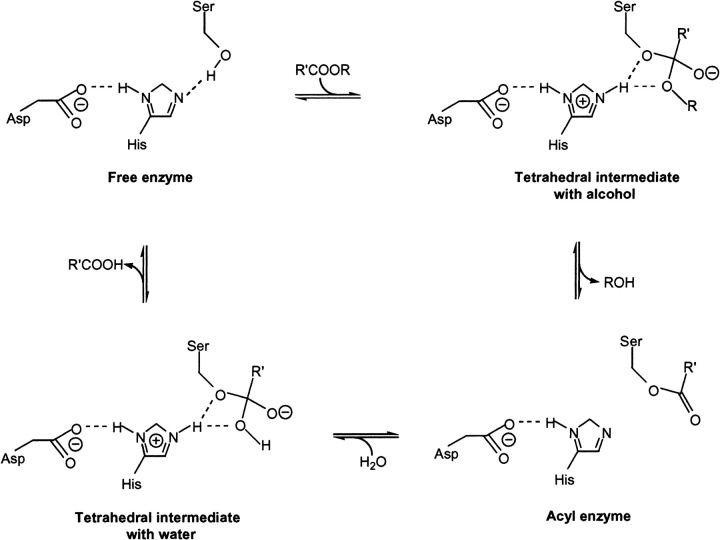
A number of modeling studies have been performed with the aim of rationalizing and/or predicting enantioselectivity of triacylglycerol lipases toward substrate molecules. Examples of such studies include those on triacylglycerol lipases from Candida antarctica (Hæffner et al. 1998), Candida rugosa (Holmquist et al. 1996), Humicola lanuginosa (Berglund et al. 1999), Pseudomonas cepacia (Tuomi and Kazlauskas 1999; Schulz et al. 2000), Rhizopus oryzae, and Rhizomucor miehei (Scheib et al. 1999). These lipases fall into the serine hydrolase class of enzymes and catalyze the reversible hydrolysis of esters to carboxylic acids and alcohols (Fig. 1 ▶). Modeling studies of enzyme enantioselectivity have also been performed on serine proteases, including chymotrypsin (Norin et al. 1993; Ke et al. 1998) and subtilisin (Colombo et al. 1999). All of these studies incorporate molecular dynamics (MD) simulations of modeled transition-state systems and, aside from the work of Hæffner et al. (1998), which is further discussed in the following, enantiopreference is explained or determined from the resulting structural ensembles on the basis of either (1) potential energy differences corresponding to the whole enzyme or (2) distance and angle geometry comparisons at the active site.
Fig. 1.
Reaction mechanism of serine hydrolase catalyzed hydrolysis or esterification.
Enantioselectivity is generally measured by using the enantiomeric ratio, E, which is given by the ratio of kcat/KM values between the enantiomers (Equation 1; Chen et al. 1982). That is,
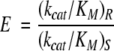 |
1 |
E is related to the difference in free energy of activation between the two enantiomers, ΔΔG# (Equation 2).
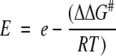 |
2 |
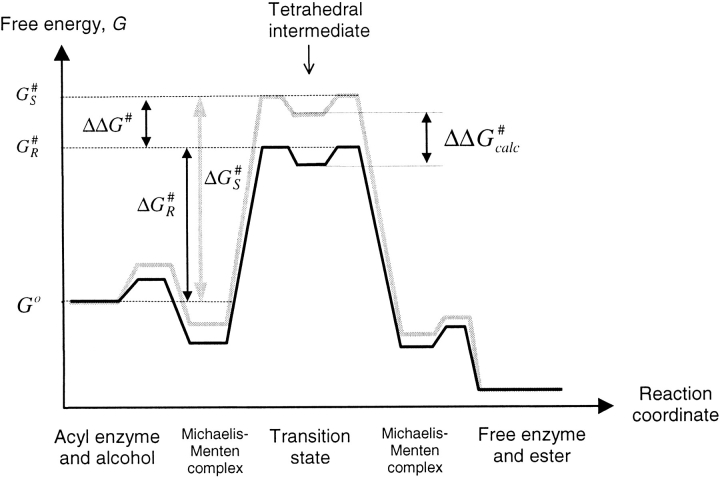
Because the ground-state free energy is the same for both R and S enantiomers, the difference in free energy of activation between the enantiomers equates to the difference in absolute transition state energy between the enantiomers (Fig. 2 ▶; equation 3)
Fig. 2.
Free energy profile for the conversion of the serine hydrolase acyl enzyme and alcohol to the free enzyme and ester. The figure shows the energy profiles for the fast R enantiomer (dark) and the slow S enantiomer (light) of the substrate. The ground-state energy of the reactants, Go, is the same for both enantiomers. The activation free energies for R and S enantiomers are given by ΔG#R and ΔG#S, respectively, and the absolute free energies for the corresponding transition states are given by G#S and G#R. The difference in free energy between the transition state and its corresponding tetrahedral intermediate is negligible (Hu et al. 1998). Therefore, ΔΔG# can be approximated to the difference in absolute free energies between the tetrahedral intermediates of the R and S enantiomers, ΔΔG#calc.
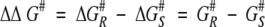 |
3 |
It is clear from equation 2 that even a large value for E corresponds to a very small free energy difference in the order of a few kJ/mole. In comparison to this, the energy of the whole enzyme-substrate system, several thousand kJ/mole, is very large and undergoes fluctuations associated with general protein motion. This highlights the difficulty in achieving quantitatively reliable predictions of enantioselectivity.
The entropic contribution to enantioselectivity can be significant (Overbeeke et al. 1999), and the experimental results of Ottosson and Hult (in press) convey that focusing on the enthalpic contribution alone will not be sufficient for correctly predicting enantioselectivity. Therefore, in the present study, the investigative analyses were performed on ensembles of nonminimized structures from the MD simulations to reflect the inclusion of the entropic component of the system, and so relate the potential energies of the system to free energies.
Experimental work by Ottosson and Hult (in press) on Candida antarctica lipase B (CALB) in organic solvent (hexane) reveals that the enantioselectivity toward the alcohol 3-methyl-2-butanol is affected by the chain length of the acyl substrate in the hydrolysis/esterification reaction. In the present modeling study, we have focused on the prediction of enantioselectivity of CALB toward 3-methyl-2-butanol with various acyl moieties as well as toward 2-butanol and 3,3-dimethyl-2-butanol with the octanoyl moiety (Fig. 3 ▶).
Fig. 3.
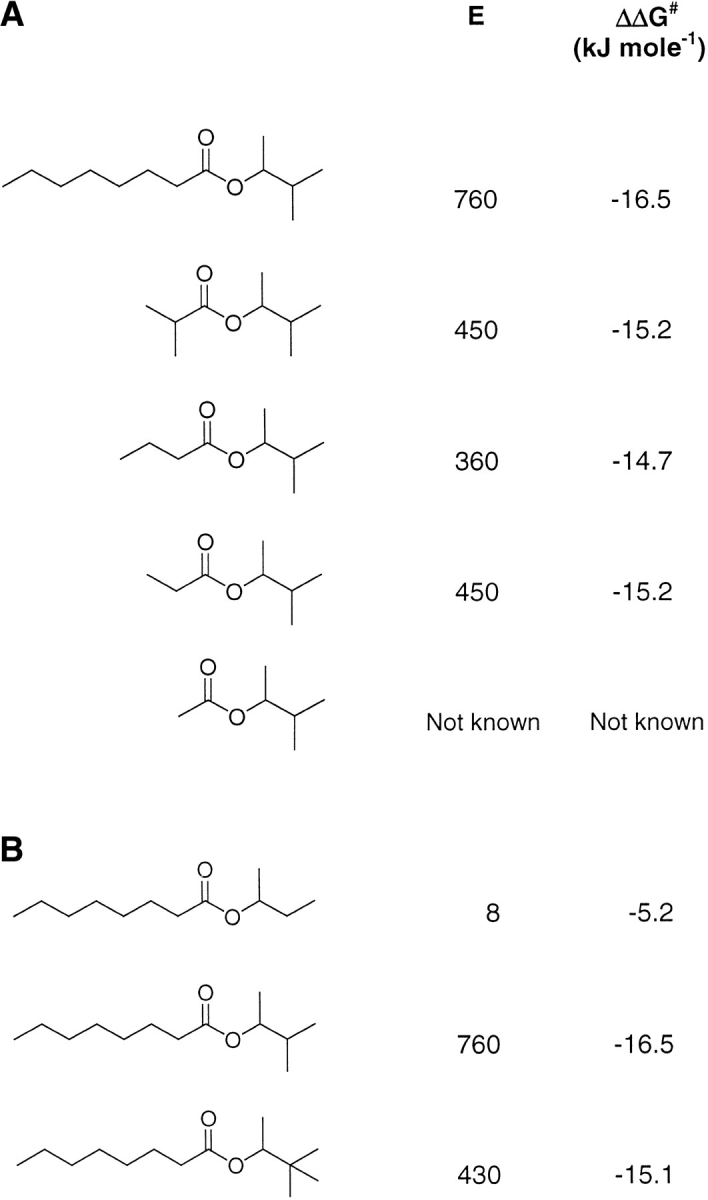
Substrates modeled for present study on Candida antarctica lipase B. (A) Variation in acyl moiety with alcohol moiety remaining constant. (B) Variation in alcohol moiety with acyl moiety remaining constant. The experimentally derived enantiomeric ratios, E, for temperature 300 K with hexane as solvent (Ottosson and Hult, in press; J. Ottosson, unpubl.) and corresponding values of ΔΔG# are also specified.
In serine proteases the structures of the transition states closely resemble those of the tetrahedral intermediates (Kraut 1977). This should also be the case for lipases that possess a similar "catalytic triad" of residues. The present MD study was based on the modeled CALB-substrate transition states of the acyl alcohol, which were described by the corresponding tetrahedral intermediates (see "tetrahedral intermediate with alcohol" in Fig. 1 ▶). Additionally, the free energy difference between the transition state and the corresponding tetrahedral intermediate is very small (Hu et al. 1998). Thus, the difference between the free energies of the tetrahedral intermediates approximates to ΔΔG# (Fig. 2 ▶). Similarly, we have taken the difference between the average MD ensemble potential energy, EMD, for each enantiomeric configuration to be the modeling-derived value for ΔΔG# (Equation 4) (The dynamics simulations performed under constant temperature and volume actually give Helmholz free energy F. The relation between G and F is given by G=F+pV.)
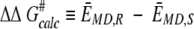 |
4 |
As in the study of Hæffner et al. (1998), investigative analyses of MD simulations in the present study focused on the potential energies described by subsets within each system. The subsets presented in this study were classed into three categories based on the method by which their constituent atoms were selected. These categories are (1) function-based subsets, which focused on the small number of atoms in the system that comprise the core elements of the transition state; (2) structure-based subsets, which focused on the substrate as well as enzyme residues in close proximity to it; and (3) energy-based subsets, which focused on the substrate as well as regions with differences in interaction energy between the enzyme and substrate when switching the enantiomeric configuration (R/S) of the substrate. The last two subsets were the focal points of the study of Hæffner et al. (1998), who found that structure-based subsets were not good predictors of enantioselectivity, whereas the use of energy-based subsets yielded better predictions. For the present study, function-based subsets provided a new way of representing the system, and yielded good predictions of enantioselectivity.
The present study has developed on the work of Hæffner et al. (1998) and, aside from the experimental data set on which the modeling was based, there are three major differences between these studies. These are as follows: (1) the present study focused on the analysis of structures nonminimized to reflect the inclusion of the entropic component within the MD simulations, whereas Hæffner et al. (1998) focused on analyzing energy-minimized structures; (2) the present work was based on the analysis of 250 structures taken at regular intervals from each simulation whereas Hæffner et al. (1998) selected 10 low energy structures from each simulation, which were subsequently energy minimized; and (3) although both studies are based on modeling of the enzyme-substrate system in the transition state, the present study focused explicitly on the key components of the transition state in the analysis of structures.
Results
The modeled transition state
The alcohol moieties were modeled in both R and S configurations. Their modeled positions coincided with the productive binding mode for each enantiomer as described by Orrenius et al. (1998). For the sake of completion, the nonproductive binding modes of the alcohols, as described by Orrenius et al. (1998), were also built as a test set for all three alcohols in both R and S configurations, with octanoyl being used as the acyl moiety. For these nonproductive binding modes, one of the following two outcomes were observed in each case: (1) the hydrogen bond between His 224 and the substrate alcohol oxygen was lost; (2) the alcohol moiety switched conformation to that of the productive binding mode. For the remaining substrates (i.e., 3-methyl-2-butanol with other acyl moieties), only the productive binding mode was modeled for each alcohol enantiomer.
For the case where a propanoyl group was used as the acyl moiety, the terminal aliphatic carbon of the acyl chain was poorly packed and subject to a high degree of mobility within the acyl cavity when positioned in the same conformation as longer acyl chains. Therefore, this terminal carbon of the propanoyl group was remodeled so that it pointed out of the acyl binding cavity where it packed closely to the alcohol moiety and between residues from both of the side walls of the substrate-binding cavity. The results presented for the propanoyl substrate refer to the latter case.
Potential energies relating to the whole enzyme
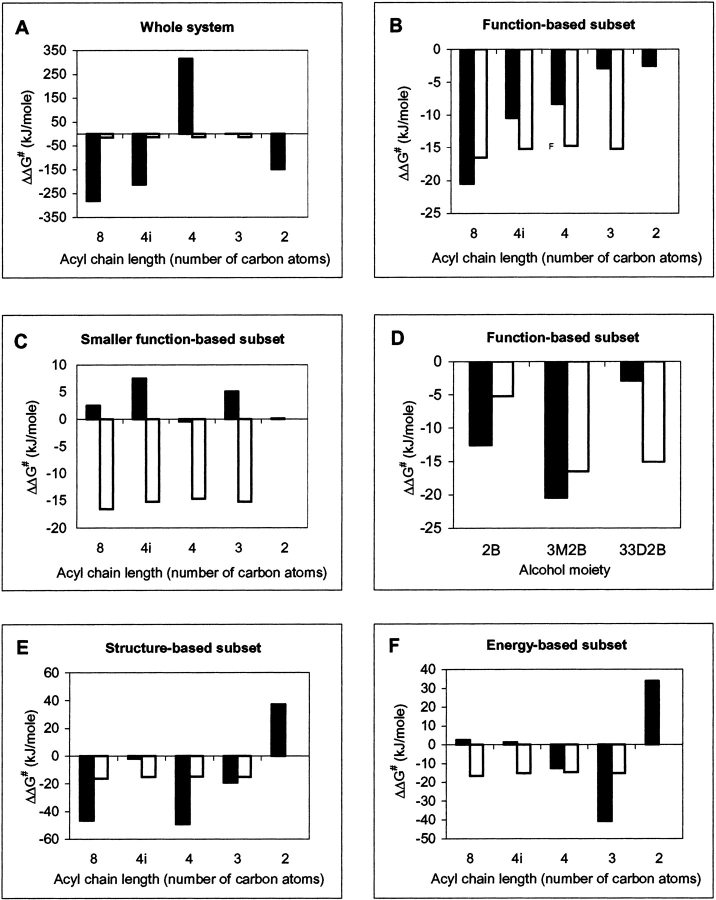
The calculated values for ΔΔG#, derived by taking the potential energies of the whole of the modeled systems, are shown in Figure 4A ▶. Examples of the mean energies and standard deviations for R and S enantiomers are given in Table 1. The results conveyed R-preference (negative values of ΔΔG#) for some of the substrates and S-preference (positive values of ΔΔG#) for others. The exclusion of water molecules in the calculation of ΔΔG# yielded similar results (data not shown).
Fig. 4.
Comparison of calculated ΔΔG# values for various subsets (▪, black bars) with experimental values from Candida antarctica lipase B-catalyzed esterification in hexane (□, white bars). Parts A, B, C, E, and F refer to 3-methyl-2-butanol with variation in the acyl chain moiety. '4i' refers to iso-butanoyl; all other acyl moieties are straight chained. Part D refers to variation in the alcohol moiety (2B, 2-butanol; 3M2B, 3-methyl-2-butanol; 33D2B, 3,3-dimethyl-2-butanol); in each case octanoyl is used as the acyl moiety. The graphs denote potential energies for the following cases. (A) Whole of modeled system including waters. (B) and (D) Function-based subset describing the core structural elements of the transition state, as illustrated in Figure 5 ▶, for variation in acyl and alcohol moieties of the substrate, respectively. (C) Smaller function-based subset describing the substrate alcohol oxygen to His 224 hydrogen bond only, and consisting of the alcohol oxygen of the substrate, His 224:Nε and the corresponding hydrogen atom. (E) Structure-based subset consisting of full substrate and residues lining the active-site cavity, as given in Figure 6 ▶. (F) Energy-based subset consisting of the full substrate and those residues possessing a difference in interaction energy of ≥0.42 kJ mole−1 (0.1 kcal mole−1) between R and S enantiomers of the substrate.
Table 1.
Examples of potential energy mean and standard deviation values for selected subsets
| Potential energy ĒMD, Mean ± standard deviation (kJ mol−1) | ||
| Subset | R-enantiomer | S-enantiomer |
| Whole system | −23370 ± 180 | −23090 ± 192 |
| Function-based | −102 ± 10 | −81 ± 12 |
| Structure-based | +515 ± 65 | +562 ± 66 |
| Energy-based | +600 ± 54 | +596 ± 52 |
The substrate here comprises of 3-methyl-2-butanol and octanoyl moieties. A set of 250 non-minimized structures, taken over the last 50 ps of the corresponding MD simulation, were used for each calculation. "Whole system" refers to the full enzyme-substrate structure with waters included. The "function-based" subset refers to that described in Figure 5 ▶. the "structure-based" and "energy-based" subsets refer to those described in Figure 6 ▶.
These results were not in line with the experimental values, which show R-preference toward the alcohol.
The experimentally derived ΔΔG# was not available for 3-methyl-2-butanol with the ethanoyl moiety. However, it is reasonable to consider that the enantiopreference toward the alcohol in this case is the same as for 3-methyl-2-butanol with the longer chained acyl moieties.
Potential energies relating to subsets within the enzyme
Function-based subsets
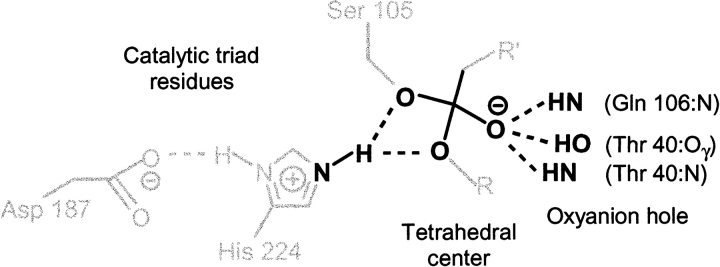
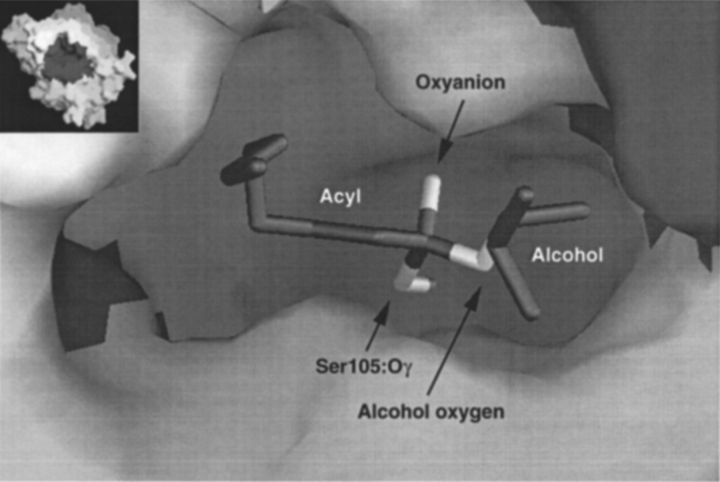
We have defined a function-based subset to be one that represents the core structural elements of the modeled transition state. For the system under study, we have taken one instance of a function-based subset to be that illustrated in Figure 5 ▶, which consists of the sum of the following two groups of atoms. (1) The "tetrahedral center," that is, the substrate's tetrahedral carbon (the first carbon of the acyl moiety) and the four atoms to which it is covalently bound and which produce the tetrahedral configuration, namely, the second carbon of the acyl moiety, the substrate oxyanion, the substrate alcohol oxygen, and the Ser 105 side-chain oxygen. (2) The hydrogen bond donor atoms, including the corresponding hydrogen atoms, associated with the tetrahedral center, namely, His 224:Nε, Gln 106:N, Thr 40:N, and Thr 40:Oγ. The calculated values for ΔΔG# by using this function-based subset are given in Figure 4B ▶. Examples of mean energies and standard deviations for R and S enantiomers are given in Table 1. The results indicate that this subset is a good predictor of enantioselectivity.
Fig. 5.
Function-based subset (dark) that describes the core structural elements of the modeled transition state in Candida antarctica lipase B.
Smaller function-based subsets focusing exclusively on one or two of the catalytic hydrogen bonds were also investigated. For example, one subset consisted of only the alcohol oxygen of the substrate, its hydrogen bond donor, that is, His 224:Nε, and the corresponding hydrogen atom. The calculated values for ΔΔG# by using this subset are shown in Figure 4C ▶. Such smaller instances of function-based subsets were not found to be satisfactory predictors of enantioselectivity.
The function-based subset illustrated in Figure 5 ▶ was also tested for its ability to predict enantioselectivity for two additional alcohol substrates in CALB, namely, 2-butanol and 3,3-dimethyl-2-butanol (both illustrated in Fig. 3B ▶) by using the octanoyl group for the acyl moiety in each case. As with 3-methyl-2-butanol, the productive and nonproductive binding modes were modeled for both 2-butanol and 3,3-dimethyl-2-butanol, and it was determined that the favourable conformation for each alcohol enantiomer corresponded to the productive binding mode. Each of the nonproductive binding modes fell into one of the following two outcomes: either the position of the alcohol moiety was unstable and switched conformation to that of the productive binding mode, or the hydrogen bond between His 224:Nε and the alcohol oxygen was lost. The selected function-based subset was able to correctly predict enantiopreference for all of the tested alcohols (Figure 4D ▶).
Structure-based subsets
A structure-based subset is one where the system is represented by the selection of the substrate in conjunction with those regions of the system in close spatial proximity to it. Here we have defined two instances of a structure-based subset. The first of these consisted of the full substrate and residues that line the active site. The residues defining this subset are given in Figure 6 ▶. The values for ΔΔG# yielded by this subset are given in Figure 4E ▶. Examples of mean energies and standard deviations for R and S enantiomers are given in Table 1. The second structure-based subset investigated was defined as consisting of the full substrate along with those residues in close proximity to the alcohol moiety (results not shown). Based on these results, structure-based subsets were not considered to be satisfactory predictors of enantioselectivity.
Fig. 6.
Molecular surface representation of the Candida antarctica lipase B active-site cavity. The inset figure illustrates the size of the active-site (dark patch) as compared with the whole enzyme. The residues selected as lining the active site are as follows: 38–42, 47, 73, 104–109, 132–134, 138, 140, 141, 144, 150, 151, 153, 154, 157, 188–190, 224, 225, 278, 281, 282, 285. The main figure shows the modeled transition state for 3-methyl-2-butyl octanoate. The residues forming the energy-based subset with a difference in interaction energy of ≥0.42 kJ mole−1 (0.1 kcal mole−1) with the whole of this substrate were as follows: 38–40, 73, 104–106, 134, 140, 144, 153, 154, 157, 188–190, 224, 278, 285. The dark patches of the molecular surface in the main figure correspond to residues excluded from the latter subset.
Energy-based subsets
This category of subset is selected on the basis of a difference in interaction energy between enzyme and substrate for each enantiomeric configuration of the substrate. In this study interaction energies were calculated between the substrate and individual amino acids lining the active site for each enantiomeric configuration of the substrate (the selected active site residues are given in Fig. 6 ▶). Of these interactions, the amino acids with differences greater than a given cutoff constituted a particular subset. Subsets based on different cutoff values (0.21, 0.42, and 1.26 kJ mole−1; that is, 0.05, 0.1, and 0.3 kcal mole−1, respectively) were investigated. Here we defined two subtypes of an energy-based subset. The first was generated on the basis of interaction energies with the full substrate. The residues making up such a subset with an interaction energy cutoff of 0.42 kJ mole−1 (0.1 kcal mole−1) are given in Figure 6 ▶. The values of ΔΔG# for this subset are given in Figure 4F ▶. Table 1 gives examples of mean energies and standard deviations for R and S enantiomers for this subset. The second subtype of an energy-based subset was based on interaction energies with the alcohol moiety only (results not shown). On the basis of these results, energy-based subsets were not considered to satisfactorily predict enantioselectivity. Additionally, different levels of cutoff energy for each subtype of energy-based subset yielded significantly different patterns of ΔΔG# (results not shown).
Discussion
The aim of this study has been to develop a methodology for predicting enantioselectivity in CALB, a serine hydrolase, by using an MD approach. The study focused on the CALB-substrate system in the transition state. The structures of the transition states for the catalyzed acyl transfer reaction in serine proteases are closely approximated by those of the tetrahedral intermediates (Kraut 1977). This must be equally true for lipases that possess a similar catalytic mechanism.
Because the enantiomeric ratio is dependent on temperature (equation 2), we simulated at a temperature that corresponded to the experimental conditions (300 K). The analysis of structures following the MD simulations was performed directly on structures from the dynamics simulation, because they can be considered to include the entropic contribution within the modeled systems. The entropic contribution to enantioselectivity can be significant (Overbeeke et al. 1999) and, according to the results of Ottosson and Hult (in press), focusing on the enthalpic contribution alone (i.e., using energy-minimized structures) will not be sufficient for the correct prediction of enantioselectivity. An incidental benefit of using nonminimized rather than minimized structures is that computation times are significantly reduced.
Although the experimentally derived enantiomeric ratio was not available for 3-methyl-2-butanol with the ethanoyl moiety, we have included this substrate in the present modeling study. It seems very unlikely that the enantiopreference toward the alcohol in this case would be the reverse of that corresponding to 3-methyl-2-butanol with the longer chained acyl moieties.
The transition-state models and the importance of free energy
The crystal structure of free CALB (PDB code 1TCA; Uppenberg et al. 1994) was used as the starting point for modeling the CALB-substrate transition states. The conformations of the substrates were based on the structures of CALB with a phosphonate inhibitor that mimics the transition state (PDB code 1LBS; Uppenberg et al. 1995) and with an ester that contains an acyl chain long enough on which to base the acyl moiety of the substrate to be modeled (PDB code 1LBT; Uppenberg et al. 1995). The free enzyme crystal structure of CALB (1TCA) is the CALB crystal structure with highest resolution (1.55 Å). Furthermore, there is no significant difference between the structures of free and complexed CALB (the Cα RMSD is 0.3 Å and the all-atom RMSD is 0.4 Å between 1TCA and 1LBS).
It has been shown that in CALB the alcohol moiety can be modeled into two binding modes, one that is productive (possessing all catalytically essential hydrogen bonds) and one that is nonproductive (one or more of the catalytically essential hydrogen bonds absent) for each alcohol enantiomer (Orrenius et al. 1998). In the present study, manual docking of each alcohol enantiomer revealed that the best fit for each enantiomer of the alcohol moiety coincided with the productive binding mode described by Orrenius et al. (1998). The nonproductive binding mode was also investigated for each alcohol enantiomer (with the octanoyl group used as the acyl moiety) but was found either to undergo conformational repositioning of the substrate to that of the productive mode or to lose the catalytic alcohol oxygen to His 224:Nε hydrogen bond.
In the case where a propanoyl group was used for the acyl moiety (and 3-methyl-2-butanol for the alcohol moiety), the third carbon of the propanoyl chain did not fit snugly into the acyl binding cavity when modeled in the same configuration as the longer acyl chains but was subject to a relatively high degree of conformational freedom. The propanoyl moiety was therefore remodeled with its third carbon pointing out of the cavity such that it packed closely against the alcohol moiety and residues forming the side walls of the CALB active site cavity.
Subsets within the modeled system
The potential energies of the whole of the modeled enzyme and their fluctuations (Table 1) are very large compared to the ΔΔG# values for enantioselectivity, which are only a few kJ mole−1. Thus, it is hardly surprising that the calculated values of ΔΔG# based on the whole of the modeled system (Figure 4A ▶) or the modeled system without waters (results not shown) were not satisfactory and yielded incorrect predictions for enantiopreference in some cases. Much of the energy fluctuation corresponds to general protein motion away from the catalytic region. Even though this general motion may average out over time, the MD simulations performed in this study are too short to allow for this result. The use of subsets to represent the system in the analysis of MD structures addresses this issue and allows us to focus on the structural regions that are most important in determining enantioselectivity.
Function-based subsets
Selectivity of the enzyme toward one enantiomeric configuration of the substrate over the other corresponds to a lower free energy of activation and therefore to a more favorable conformation of the transition state. The transition state of an enzyme–substrate system for a particular reaction can be thought of in terms of its essential or core structural elements. These structural elements reflect the mechanistic function of the enzyme. We use this region of the transition state system to define what we term a "function-based subset". The positions of all of the remaining atoms in the system outside the selected subset contribute in varying degrees, either directly or indirectly, to the positions of these core atoms. Because this subset focuses explicitly on the bond-making and bond-breaking regions, we consider that a more favorable energetic conformation of this subset should correspond to a greater likelihood for catalysis to occur, thus reflecting enantiopreference.
The validity of function-based subsets is supported by the idea that enzyme–substrate interactions can be thought of in terms of "dynamic" and "passive" binding (Kirby 1996). Dynamic binding refers to the interactions at the reaction center that are associated with the mechanistic bond-making and bond-breaking processes. These interactions change substantially as bonds are made and broken during the enzymatic reaction. Passive binding refers to interactions involved in ordinary molecular recognition and which include hydrophobic interactions and noncatalytic hydrogen bonds. Thus, passive interactions focus primarily on the general enzyme–substrate fit, whereas dynamic interactions focus primarily on the catalytic mechanism. If we consider selectivity in terms of dynamic and passive binding, then for the fast-reacting enantiomer, a better fit between enzyme and substrate through passive interactions corresponds to conformationally (and therefore energetically) more favorable dynamic interactions, thereby relating to higher catalytic turnover. However, for the slow-reacting enantiomer, a better fit through passive interactions can be seen to correspond to conformationally (and energetically) less favorable dynamic interactions, thus relating to a lower catalytic turnover.
We have defined one instance of a function-based subset to be that illustrated in Figure 5 ▶, consisting of the substrate's tetrahedral center and the atoms involved in the catalytic hydrogen bonds. The His 224:Nδ to Asp 187:Oγ2 hydrogen bond in CALB is not directly involved with the tetrahedral center and was excluded from this subset. The selected subset is a good predictor of enantioselectivity (Figure 4B ▶).
Smaller function-based subsets focusing exclusively on one or two of the catalytic hydrogen bonds were not satisfactory predictors of enantioselectivity. For example, Figure 4C ▶ conveys the results for the subset consisting of three atoms that form a single catalytic hydrogen bond; the comprising atoms are the alcohol oxygen of the substrate (hydrogen bond acceptor), His 224:Nε (hydrogen bond donor), and the associated hydrogen atom. This particular hydrogen bond corresponds to that which Schulz et al. (2000) use as the focal point for MD-derived enantioselectivity predictions in Pseudomonas cepacia lipase (their enantioselectivity prediction is based on the HNε-Oalcohol hydrogen bond distance in an average minimized structure for the slow enantiomer). We consider that the poor predictions we observed in the present study for the smaller function-based subsets were due to inadequate representation of the key elements of the transition state.
The function-based subset described in Figure 5 ▶ also correctly predicts enantiopreference when varying the alcohol moiety (Figure 4D ▶). The substrates tested here are 2-butanol, 3-methyl-2-butanol and 3,3-dimethyl-2-butanol, with the octanoyl group as the acyl moiety in each case (see Figure 3B ▶).
Other subsets
Hæffner et al. (1998) have previously studied two categories of subset, the first of which is based on the selection of regions that are spatially close to the substrate (i.e., structure-based subsets), and the other of which is based on difference in interaction energies between the substrate and individual residues in the enzyme with respect to each substrate enantiomer (i.e., energy-based subsets). They found that energy-based subsets (using minimized structures) were more useful than structure-based subsets for predicting enantioselectivity.
In the present study, where the analysis of structures was performed on nonminimized structures, we found that neither structure-based subsets nor energy-based subsets were good predictors of enantioselectivity (for example, see Figure 4 ▶, E and F, respectively). However, it should be noted that the energy-based subset results of Hæffner et al. (1998) and those of this study are not directly comparable as a result of the aforementioned differences in the methodologies used.
Nevertheless, a serious setback when attempting to use energy-based subsets as a method for predicting enantioselectivity is knowing where to take the cutoff energy for any given substrate, especially because the resulting calculated values for ΔΔG# are very sensitive to this cutoff (results not shown). Furthermore, calculating interaction energies between the substrate and individual amino acids within the enzyme involves a computational time investment that is not required when using structure- or function-based subsets.
Materials and methods
The modeling was performed by using the SYBYL molecular modeling package version 6.6 (Tripos Inc.) on an SGI Octane UNIX workstation. The enzyme–substrate system was described by using the Kollman All Atom force field (Weiner et al. 1984, 1986) for the purposes of energy-minimization calculations, MD simulations, and potential energy calculations when analyzing structures. The Kollman All Atom types for the substrate were assigned as follows: carbon, CT; hydrogen, HC; oxyanion oxygen, OH; nonoxyanion oxygen, OS. Nonstandard partial charges were calculated by using the Pullman method (Berthod and Pullman 1965; Berthod et al. 1967) with a formal charge of –1 for the substrate oxyanion.
A nonbonded cutoff distance of 8 Å and a distance-dependent dielectric function with a scaling factor of one were used in all calculations. An NTV ensemble (i.e., constant number of atoms, temperature, and volume), a temperature of 300 K (aside from a short warm-up phase; see following) and a time step of 1 fs was used in all MD simulations. Energy minimizations were performed by using the Powell method (Powell 1977). Nonstandard Kollman All Atom forcefield parameters were assigned in analogy to the existing parameters for angle bending as follows: (1) OH-CT-OS, 109.5°, force constant 250 kJ mole−1 degree−2; (2) OS-CT-OS, 109.5°, force constant 250 kJ mole−1 degree−2.
All crystal structure coordinates were from the Protein Data Bank (Berman et al. 2000).
Preparation of the free enzyme
The crystal structure of the free CALB enzyme (PDB code 1TCA; Uppenberg et al. 1994) was used as the starting point for the modeled transition-state system. The substrate was built into the free enzyme rather than into the transition state analog crystal structure (PDB code 1LBS; Uppenberg et al. 1995) because the crystal structure of the former has been solved to a higher resolution (i.e., 1.55 Å in 1TCA compared with 2.6 Å in 1LBS), and there is no significant conformational difference between the two structures (Cα RMSD 0.3 Å, all atom RMSD 0.4 Å). The two N-acetyl-D-glucosamine (NAG) moieties in the free CALB structure were removed.
Hydrogen atoms were then added to the enzyme and water molecules. The positions of the water hydrogens and then the enzyme hydrogens were optimized by using a consecutive series of short (1 ps) MD runs and energy minimizations. This series of optimization steps was repeated until the energy of the system was stable. Thereafter, an iterative series of energy minimizations were performed on the water hydrogens, enzyme hydrogens, and full water molecules. Finally, the whole system was energy minimized. Such a rigorous optimization of hydrogen atom positions was performed because the positions of many of the enzyme hydrogens and all of the water hydrogens are ambiguous in protein structures that contain only heavy atoms.
Preparation of the transition-state system
The tetrahedral intermediate form of the substrate was manually modeled on the basis of crystal structures of CALB with (1) a phosphonate inhibitor, that is, a transition state analog (PDB code 1LBS; Uppenberg et al. 1995) and (2) an ester with a sufficiently long acyl chain on which to base the conformations of the acyl moieties in the models (PDB code 1LBT; Uppenberg et al. 1995). The alcohol moiety was modeled in both R and S configurations. For each enantiomeric configuration of the alcohol moiety, two binding modes (i.e., "productive" and "nonproductive") described by Orrenius et al. (1998) were modeled for the case where the octanoyl group was used as the acyl moiety.
The catalytic histidine, His 224, was defined as protonated. Four water molecules (HOH130, HOH149, HOH238, and HOH285) were removed from the active site cavity to make room for the substrate. The optimization of the substrate and catalytic Ser 105 side-chain oxygen positions consisted of a consecutive series of short (1 ps) MD runs and energy minimizations. Three other water molecules (HOH136, HOH219, and HOH265) in the active-site cavity close to the acyl chain were ejected from the active site well beyond the surface of the enzyme during subsequent optimization steps for the octanoyl group and were thus excluded from this and all the other systems modeled in this study. The remaining water molecules were considered to be strongly bound to the enzyme. Such waters can play an important role in stabilizing the structure of the enzyme in organic solvent. Finally, energy minimization of the whole system was performed to produce the starting structure for the subsequent MD simulations.
Molecular dynamics simulations
Each energy-minimized CALB-substrate system was run through an MD warm-up phase to a temperature of 300 K in a series of six steps at 50 K intervals, where the simulated duration of each interval was 2 ps. Thereafter, a 100-ps MD simulation was performed at 300 K for each system. A sample structure was extracted at every 0.2 ps from each simulation. Investigative analyses were performed on the last 50 ps of each simulation, yielding an ensemble of 250 nonminimized structures for each system.
Acknowledgments
We thank the Wallenberg Foundation and the Swedish Research Council for Engineering Sciences for their financial support. We are also very grateful to Jenny Ottosson for prepublished data, and to Eduardo Garcia-Urdiales for his support and many helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.33901.
References
- Berglund, P., Vallikivi, I., Fransson, L., Dannacher, H., Holmquist, M., Martinelle, M., Björkling, F., Parve, O., and Hult, K. 1999. Switched enantiopreference of Humicola lipase for 2-phenoxyalkanoic acid ester homologs can be rationalized by different substrate binding modes. Tetrahedron: Asymmetry 10 4191–4202. [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthod, H. and Pullman, A. 1965. Sur le calcul des caractéristiques du squelette σ des molécules conjuguées. J. Chim. Phys. 62 942–946. [Google Scholar]
- Berthod, H., Giessner-Prettre, C.L., and Pullman, A. 1967. Sur les rÔles respectifs des électrons σ et π dans les propriétés des dérivés halogénés des molécules conjuguées. Application Á l'étude de l'uracile et du fluorouracile. Theor. Chim. Acta. 8 212–222. [Google Scholar]
- Chen, C.S., Fujimoto, Y., Girdaukas, G., and Sih, C.J. 1982. Quantitative analysis of biochemical kinetic resolutions of enantiomers. J. Am. Chem. Soc. 104 7294–7299. [Google Scholar]
- Colombo, G., Toba, S., and Merz Jr., K.M. 1999. Rationalization of the enantioselectivity of subtilisin in DMF. J. Am. Chem. Soc. 121 3486–3493. [Google Scholar]
- Hæffner, F., Norin, T., and Hult, K. 1998. Molecular modeling of the enantioselectivity in lipase-catalyzed transesterification reactions. Biophys. J. 74 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist, M., Hæffner, F., Norin, T., and Hult, K. 1996. A structural basis for enantioselective inhibition of Candida rugosa lipase by long-chain aliphatic alcohols. Protein Sci. 5 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.H., Brinck, T., and Hult, K. 1998. Ab initio and density functional theory studies of the catalytic mechanism for ester hydrolysis in serine hydrolases. Int. J. Quantum Chem. 69 89–103. [Google Scholar]
- Ke, T., Tidor, B., and Klibanov, A.M. 1998. Molecular-modeling calculations of enzymatic enantioselectivity taking hydration into account. Biotechnol. Bioeng. 57 741–745. [DOI] [PubMed] [Google Scholar]
- Kirby, A.J. 1996. Enzyme mechanisms, models and mimics. Angew. Chem. Int. Ed. Engl. 35 707–724. [Google Scholar]
- Kraut, J. 1977. Serine-proteases: Structure and mechanism of catalysis. Annu. Rev. Biochem. 46 331–358. [DOI] [PubMed] [Google Scholar]
- Norin, M., Hult, K., Mattson, A., and Norin, T. 1993. Molecular modelling of chymotrypsin-substrate interactions: Calculation of enantioselectivity. Biocatalysis 7 131–147. [Google Scholar]
- Orrenius, C., H|izffner, F., Rotticci, D., Öhrner, N., Norin, T., and Hult, K. 1998. Chiral recognition of alcohol enantiomers in acyl transfer reactions catalysed by Candida antarctica lipase B. Biocatalysis and Biotransformation 16 1–15. [Google Scholar]
- Ottosson, J. and Hult, K. Influence of acyl chain length on the enantioselectivity of Candida antarctica lipase B and its thermodynamics components in kinetic resolution of sec-alcohols. J. Mol. Cat. B.
- Overbeeke, P.L.A., Ottosson, J., Hult,K., Jongejan, J.A., and Duine, J.A. 1999. The temperature dependence of enzymatic kinetic resolutions reveals the relative contribution of enthalpy and entropy to enzymatic enantioselectivity. Biocatalysis and Biotransformation 17 61–79. [Google Scholar]
- Powell, M.J.D. 1977. Restart procedures for the conjugate gradient method. Mathematical Programming 12 241–254. [Google Scholar]
- Scheib, H., Pleiss, J., Kovac, A., Paltauf, F., and Schmid, R.D. 1999. Stereoselectivity of Mucorales lipases toward triradylglycerols – A simple solution to a complex problem. Protein Sci. 8 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, T., Pleiss, J., and Schmid, R.D. 2000. Stereoselectivity of Pseudomonas cepacia lipase toward secondary alcohols: A quantitative model. Protein Sci. 9 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomi, W.V. and Kazlauskas, R.J. 1999. Molecular basis for enantioselectivity of lipase from Pseudomonas cepacia toward primary alcohols. Modeling, kinetics, and chemical modification of Tyr29 to increase or decrease enantioselectivity. J. Org. Chem. 64 2638–2647. [DOI] [PubMed] [Google Scholar]
- Uppenberg, J., Hansen, M.T., Patkar, S., and Jones, T.A. 1994. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 2 293–308. [DOI] [PubMed] [Google Scholar]
- Uppenberg, J., Öhrner, N., Norin, M., Hult, K., Kleywegt, G.J., Patkar, S., Waagen, V., Anthonsen, T., and Jones, T.A. 1995. Crystallographic and molecular-modelling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols. Biochemistry 34 16838–16851. [DOI] [PubMed] [Google Scholar]
- Weiner, S.J., Kollman, P.A., Case, D.A., Singh, U.C., Ghio, C., Alagona, G., Profeta, S., and Weiner, P. 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106 765–784. [Google Scholar]
- Weiner, S.J., Kollman, P.A., Nguyen, D.T., and Case, D.A. 1986. An all atom force for simulations of proteins and nucleic acids. J. Comp. Chem. 7 230–252. [DOI] [PubMed] [Google Scholar]