Abstract
Human matrix Gla protein (MGP) is a vitamin K–dependent extracellular matrix protein that binds Ca2+ ions and that is involved in the prevention of vascular calcification. MGP is a 10.6-kD protein (84 amino acids) containing five γ-carboxyglutamic acid (Gla) residues and one disulfide bond. Studies of the mechanism by which MGP prevents calcification of the arterial media are hampered by the low solubility of the protein (<10 μg/mL). Because of solubility problems, processing of a recombinantly expressed MGP-fusion protein chimera to obtain MGP was unsuccessful. Here we describe the total chemical synthesis of MGP by tBoc solid-phase peptide synthesis (SPPS) and native chemical ligation. Peptide Tyr1-Ala53 was synthesized on a derivatized resin yielding a C-terminal thioester group. Peptide Cys54-Lys84 was synthesized on Lys-PAM resin yielding a C-terminal carboxylic acid. Subsequent native chemical ligation of the two peptides resulted in the formation of a native peptide bond between Ala53 and Cys54. Folding of the 1–84-polypeptide chain in 3 M guanidine (pH 8) resulted in a decrease of molecular mass from 10,605 to 10,603 (ESI-MS), representing the loss of two protons because of the formation of the Cys54-Cys60 internal disulfide bond. Like native MGP, synthetic MGP had the same low solubility when brought into aqueous buffer solutions with physiological salt concentrations, confirming its native like structure. However, the solubility of MGP markedly increased in borate buffer at pH 7.4 in the absence of sodium chloride. Ca2+-binding to MGP was confirmed by analytical HPLC, on which the retention time of MGP was reduced in the presence of CaCl2. Circular dichroism studies revealed a sharp increase in α-helicity at 0.2 mM CaCl2 that may explain the Ca2+-dependent shift in high-pressure liquid chromatography (HPLC)-retention time of MGP. In conclusion, facile and efficient chemical synthesis in combination with native chemical ligation yielded MGP preparations that can aid in unraveling the mechanism by which MGP prevents vascular calcification.
Keywords: Matrix Gla protein, vascular calcification, solid phase peptide synthesis, native chemical ligation, protein folding, electrospray ionization mass spectrometry
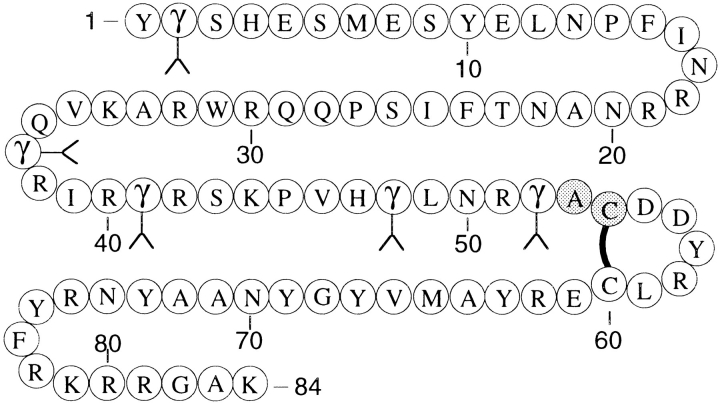
Matrix γ-carboxyglutamic acid (Gla) protein (MGP) is an 84-amino-acid protein that was first described in an extract from bovine bone (Price et al. 1983). Following this report, MGP was detected in rats (Otawara and Price 1986), humans (Kiefer et al. 1988), mice (Ikeda et al. 1991), sharks (Rice et al. 1994), and chickens (Wiedemann et al. 1998) either as a protein or as mRNA. MGP is found in high concentrations in the extracellular matrix of bone and cartilage (Price et al. 1983; Hale et al. 1988) but is also synthesized in a wide variety of other tissues such as lung, heart, kidney, and arterial vessel wall (Fraser and Price 1988). MGP contains five posttranslationally modified γ-carboxyglutamic acids (Price et al. 1983) and, therefore, belongs to the vitamin K–dependent family of proteins (Fig. 1 ▶).
Fig. 1.
Amino acid sequence of matrix Gla protein. The gray residues at position 53–54 represent the alanine-cysteine native chemical ligation site. The solid line represents the disulfide-bond. γ denotes γ-carboxyglutamic acid. Sequence from nucleotide analysis (Kiefer et al. 1988; Chen et al. 1990).
It was reported that deficiencies in MGP caused impaired bone growth and, more importantly, massive calcification of the arterial vessel wall in mice (Luo et al. 1997). To study the molecular mechanisms by which MGP protects against calcification and bone deformation, sufficient amounts of MGP are required. However, the low solubility of MGP limited recombinant production of MGP because an Escherichia coli–expressed dihydrofolate reductase-MGP chimera could not be processed further because of insolubility of the fusion protein under physiological conditions (Braam et al. 2000). Moreover, conventional purification from bone is also hindered by the insolubility of the protein.
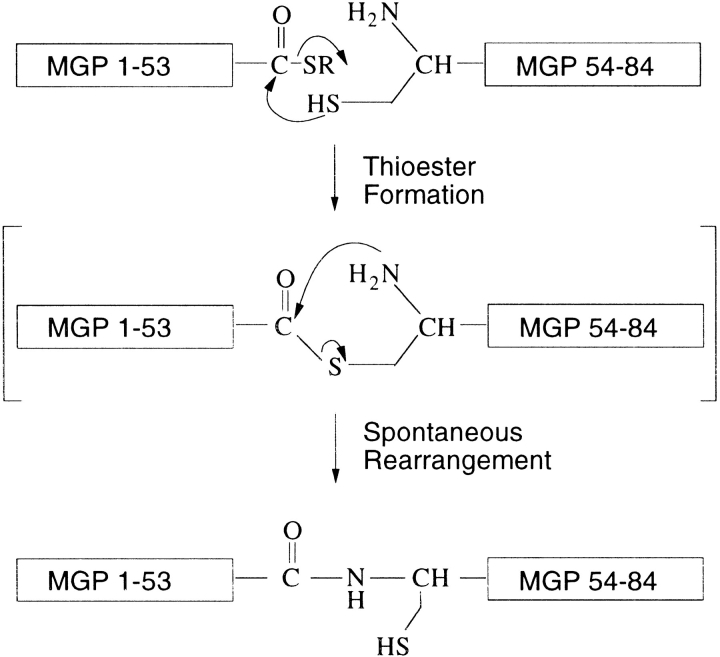
When biologically active proteins cannot be generated by recombinant expression techniques because of insolubility or misfolding, chemical protein synthesis is an excellent alternative that, because of its versatility, can greatly add to structure–function analysis of the protein of interest. Because polypeptide chains of 40–60 amino acids realistically represent the efficiency limit of solid-phase peptide synthesis, native chemical ligation was introduced to join protein segments through native peptide bonds (Dawson et al. 1994). The native chemical ligation technique uses the principle of a chemoselective reaction between two unprotected peptides in aqueous solution in which a carboxyterminal thioester undergoes a thiol-exchange with an aminoterminal cysteine sulfhydryl side chain. A subsequent, rapid, intramolecular rearrangement yields a native peptide bond at the site of ligation (Fig. 2 ▶).
Fig. 2.
Native chemical ligation strategy for the total chemical synthesis of human matrix Gla protein. The 53-residue N-terminal peptide-thioester (COSR) fragment and the 30-residue C-terminal fragment of MGP were synthesized by stepwise SPPS techniques using Boc-chemistry protocols. The two fragments are initially joined by thioester formation (not observed as a discrete intermediate), and a subsequent spontaneous, rapid rearrangement results in the formation of a native peptide bond at the site of ligation.
Here we report the total chemical synthesis of MGP by solid-phase peptide synthesis and native chemical ligation, yielding highly pure MGP with a full amide backbone and native biochemical features.
Results
Synthesis of matrix Gla protein peptides
To establish chemical access to the 84-residue human matrix Gla protein, a two-fragment synthesis was chosen with a suitable ligation site at Ala53-Cys54, conducted by the need for an N-terminal cysteine on the C-terminal peptide at the site of ligation (Fig 1 ▶).
The synthesis of the 84-residue MGP therefore required synthesis of two polypeptides: NH2-Tyr1-Ala53-COSR (MGP 1–53) and NH2-Cys54-Lys84-COOH (MGP 54–84). MGP 1–53 was synthesized on C-terminal thioester generating resin (TAMPAL) to yield the C-terminal mercaptopropionic acid-Leu (MPAL)–activated thioester. MGP 54–84 was synthesized on Lys-PAM resin to yield an N-terminal free cysteine and a C-terminal carboxylic acid group after cleavage of the peptide from the resin.
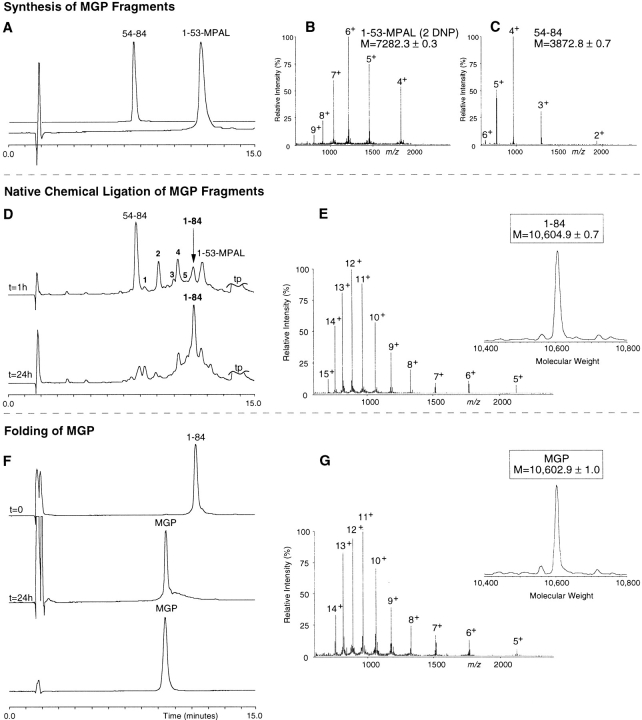
The synthesis (0.3 mmol) of MGP 1–53 on TAMPAL resin yielded 2.08 g peptide resin (theoretical yield: 2.27 g corrected for resin sampling), with an average coupling efficiency of 99.56% (range 99.33%–99.96%) based on ninhydrin assays after each coupling step. After the Nα-Boc group was removed, 330 mg peptide-resin was cleaved with anhydrous HF, yielding 216 mg dry crude peptide that was further purified on preparative high-pressure liquid chromatography (HPLC; Fig. 3A ▶). From preparative HPLC, 29 mg of pure MGP 1–53 was obtained. ESI-MS revealed a molecular mass of 7282.3 ± 0.3 D (Fig. 3B ▶), which agreed well with the theoretical average mass of 7282.6 D of MGP 1–53-MPAL, with dinitrophenyl (Dnp) groups attached to His4 and His47 (Fig. 3B ▶).
Fig. 3.
Total synthesis of human matrix Gla protein by native chemical ligation. Synthesis of MGP fragments: (A) HPLC chromatogram (C18: 22.5%–41% acetonitrile, 1.23%/min) of the starting synthetic peptide segments MGP 54–84 and MGP 1–53. (B) ESI-MS spectrum of MGP 1–53 shows the m/z ratios (fourth through ninth ionized states) with an calculated mass of 7282.3 ± 0.3 D (theoretical average mass of 7282.6). (C) ESI-MS spectrum of MGP 54–84 shows the m/z ratios (second through sixth ionized states) with an calculated mass of 3872.8 ± 0.7 D (theoretical average mass: 3873.4 D). Native chemical ligation of MGP fragments: (D) HPLC chromatograms of the ligation reaction, started by the addition of 2% (v/v) thiophenol and 2% benzylmercaptan to the peptide mixture MGP 1–53 and MGP 54–84. At t = 1 h, ligated product (1–84) is shown as well as the C-terminal segment starting material. The unreacted N-terminal material can be accounted for as multiple intermediates eluting between 7 and 13 min (see text). At t = 24 h, ligation was almost complete. (E) ESI-MS spectrum shows the m/z pattern of the ligated material (fifth through fifteenth ionized states) with a calculated mass of 10,604.9 ± 0.7 D (theoretical average mass of the reduced 84-residue MGP: 10,604.8 D). Folding and disulfide formation of MGP: (F) HPLC chromatograms of the purified, reduced MGP polypeptide (t = 0), the crude, folded material (t = 24 h), and the purified final product (MGP). (G) ESI-MS spectrum shows the m/z pattern (fifth through fourteenth ionized state) with a calculated molecular mass of 10,602.9 ± 1.0 D (theoretical average mass of MGP containing one disulfide: 10,602.8 D).
The synthesis of MGP 54–84 (0.35 mmol) on Lys-PAM resin yielded 2.06 g peptide-resin (theoretical yield: 2.11 g) with an average coupling efficiency of 99.52% (range 99.23%–99.83%). Yielding 185 mg dry crude peptide, 300 mg peptide resin was cleaved with anhydrous HF. From a preparative HPLC run of 80 mg of crude MGP 54–84, 21 mg of pure MGP 54–84 was obtained (Fig. 3A ▶). ESI-MS showed an apparent molecular mass of 3872.8 ± 0.7 D (Fig. 3C ▶), which fitted well with the calculated average isotopic mass of 3873.4 D.
Synthesis of matrix Gla protein by native chemical ligation
Native chemical ligation of MGP peptides was performed with 27.8 mg (3.8 μmol) MGP 1–53 and 17.1 mg (4.4 μmol) MGP 54–84. The peptides were dissolved in 2 mL ligation buffer with 2% thiophenol and 2% benzylmercaptan, resulting in a final peptide concentration of 1.9 mM MGP 1–53 and 2.2 mM MGP 54–84 at pH ≈ 7. After 1 h at 37°C, multiple peaks could be observed on analytical HPLC (Fig. 3D ▶). In addition to the starting materials (54–84 and 1-53-MPAL[2 Dnp]), the following fragments were identified: a +26-D derivative of 54–84 (peak 1); 1-53-MPAL without Dnp (peak 2); 1-53-MPAL with 1 His-Dnp (peak 3); MGP 1-53-benzylmercaptan thioester exchange (peak 4); 1-53-MPAL with 1 His-Dnp (peak 5); and the ligated 84-residue polypeptide (1–84; Fig. 3D ▶). The difference in retention time of peaks 3 and 5 can be explained by Dnp groups that are attached to different His residues in fragment 1–53. ESI-MS revealed a mass for the 1–84 MGP polypeptide of 10,604.9 D, which fitted well with the theoretical average isotopic mass of 10,604.8 D (Fig. 3E ▶). After 24 h, ligation was almost complete, and the mixture was applied to preparative HPLC (C18: 22.5%–41% acetonitrile, 0.31%/min) to isolate the 1–84-polypeptide chain, yielding 17 mg (42% yield) dry peptide after lyophilization.
Folding of matrix Gla protein
The 84-residue polypeptide chain of MGP (16 mg) was folded in 3 M guanidine (pH 8.0) at a final concentration of 0.2 mg/mL polypeptide. Overnight stirring exposed to air at 4°C resulted in a quantitative shift of the peptide peak on analytical HPLC from 11.36 to 9.55 min (Fig. 3F ▶). This shift was likely because of a conformational change caused by the formation of the disulfide loop between Cys54 and Cys60. ESI-MS showed a decrease in mass of the folded polypeptide from 10,604.9 D to 10,602.9 D, the loss of 2.0 D representing the loss of two hydrogen atoms from the free sulfhydryl groups on formation of the disulfide bond (Fig. 3G ▶). After purification on preparative HPLC (C18: 22.5%–41% acetonitrile, 0.31%/min), 6 mg of MGP containing five Gla residues and an internal disulfide were isolated (35% yield).
When MGP was brought into Tris buffer (50 mM Tris, 100 mM NaCl at pH 7.4) at a concentration of 0.2 mg/mL at room temperature, MGP initially dissolved, but it precipitated within minutes after addition of buffer (data not shown). Addition of guanidine to the Tris buffer prevented precipitation at guanidine concentrations >2.5 M. The insolubility of synthetic MGP resembles that of native, purified MGP, which precipitates at concentrations >10 μg/mL (Price et al. 1987).
However, when MGP was brought into Boric acid buffer (25 mM boric acid at pH 7.4) at 0.2 mg/mL, no precipitation was observed, and peak area analysis by analytical HPLC showed that >90% of MGP was still in solution after overnight incubation (data not shown).
Ca2+ binding to matrix Gla protein
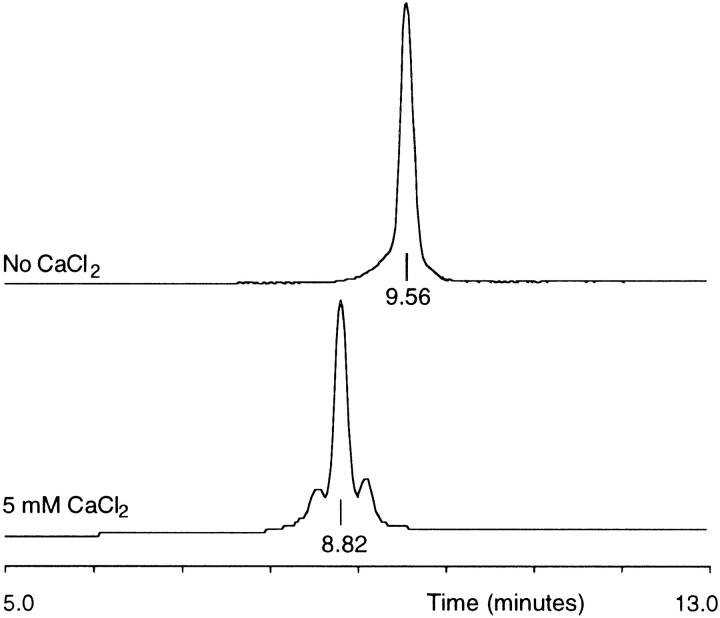
Evidence of Ca2+ binding to MGP was obtained by analytical HPLC in the absence or presence of 5 mM CaCl2. The retention time of MGP decreased from 9.6 min in the absence of CaCl2 to 8.8 min in the presence of 5 mM CaCl2 (Fig. 4 ▶). This shift in retention time was either caused by a charge effect or by a conformational change of MGP on binding of Ca2+ ions. The two additional peaks that elute before and after the main peak may represent MGP molecules that have bound a different number of Ca2+ ions (Fig. 4 ▶).
Fig. 4.
Ca2+ binding to synthetic MGP. HPLC chromatograms (C18: 22.5%–41% acetonitrile, 1.23%/min) of synthetic MGP in the absence or in the presence of 5 mM CaCl2.
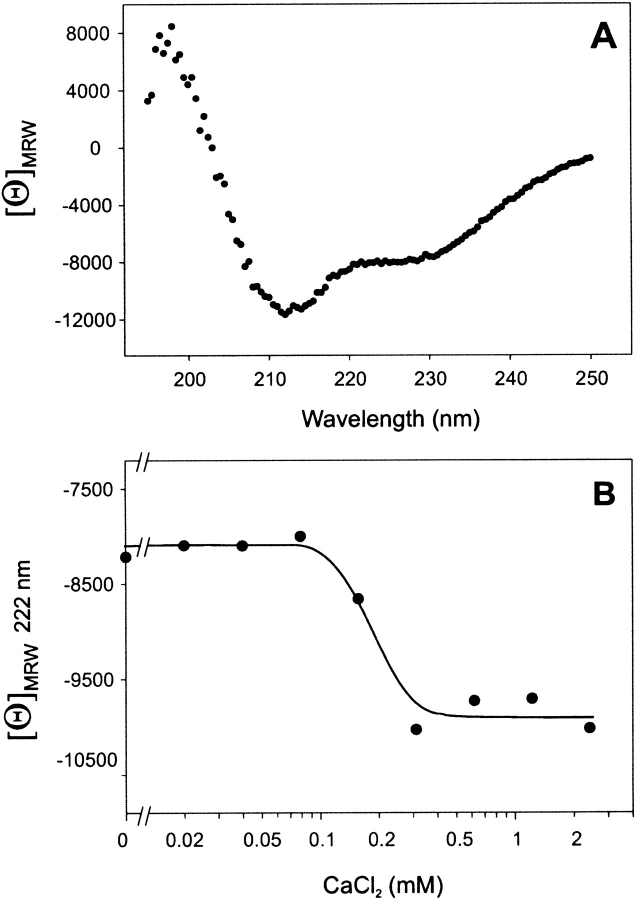
To investigate a possible change in secondary structure contents of MGP on binding of Ca2+ ions, folded MGP was subjected to circular dichroism (CD) studies. In the absence of CaCl2, CD spectroscopy showed a typical protein structure spectrum composed of a mixture of α-helix, β-structure, and random coil (Fig. 5 ▶). From the mean residual weight rotation at 222 nm, it was estimated that MGP contained 21% α-helix in the absence of CaCl2. However, addition of varying amounts of CaCl2 (20 μM–2.4 mM) to MGP resulted in a sharp increase in mean residual weight rotation at 222 nm at ∼0.2 mM CaCl2 (Fig. 5B ▶). This effective concentration of CaCl2 lies in the range of Gla-dependent Ca2+ binding to bone Gla protein (Hauschka and Carr 1982) and to Gla-containing coagulation factors (Nelsestuen et al. 2000, and references therein), which occurs between 0.1 and 1 mM CaCl2. It was calculated that MGP contained 21% α-helix in the absence of CaCl2, which increased to 26% at 0.3 mM CaCl2, representing an increase in α-helicity of ∼25%.
Fig. 5.
Circular dichroism spectrum of synthetic matrix Gla protein. (A) The spectrum for matrix Gla protein at 25°C in boric acid shows 21% α-helical structure as can be estimated from the mean residual weight (MRW) ellipticity at 222 nm. (B) The increase (25%) of mean residual weight ellipticity of MGP at 222 nm as a function of CaCl2 concentration.
To compare synthetic MGP with native MGP, SDS-PAGE was performed under denaturing conditions. After Western blotting, MGP was detected with monoclonal antibodies against a synthetic peptide 3–15 from MGP (Braam et al. 2000). Figure 6 ▶ shows MGP isolated from human bone (kindly donated by Dr. Reidar Wallin from Wake Forest University) in comparison with synthetic MGP (Mr: 10.6 kD) and the fragments from which MGP was synthesized. As can be observed, the synthetic MGP in lane 1 comigrates with a purified sample of human MGP in lane 2 (barely visible) and a crude preparation of human MGP in lane 4. The MGP fragment 1–53 with a mass of 7.3 kD in lane 3 has increased mobility compared to full-length MGP, and because of the single free cysteine in MGP 1–53, a dimer of MGP 1–53 is visible at ∼15 kD. MGP fragment 54–84 could not be detected with the monoclonal antibody. This SDS-PAGE observation provides additional proof for the native-like condition of chemically synthesized MGP.
Fig. 6.
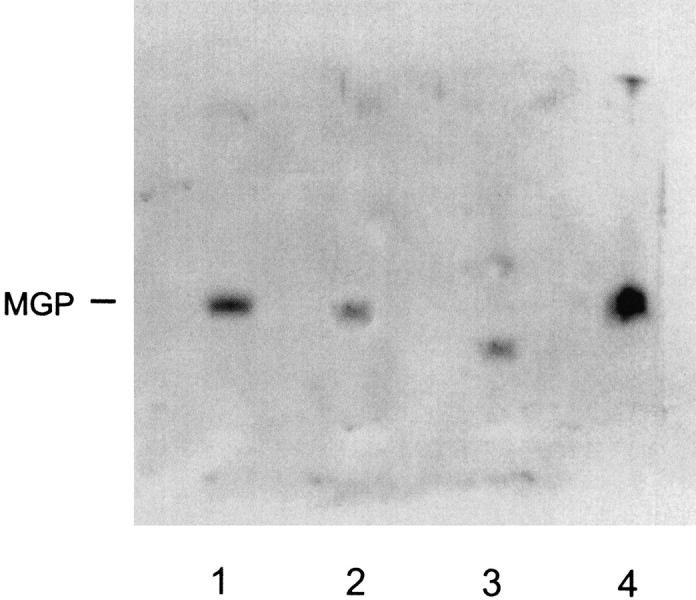
SDS-PAGE of MGP. Synthetic MGP (lane 1), purified MGP from human bone (lane 2), MGP peptide 1–53-COSR (lane 3), and crude MGP from human bone (lane 4) were applied to 12.5% SDS-PAGE, Western blotting, and detection with a monoclonal antibody against peptide 3–15 from human MGP.
Discussion
Matrix Gla protein, an essential Ca2+ chelator that prevents vascular calcification, was successfully synthesized using solid-phase synthesis and native chemical ligation. The 84-residue polypeptide was folded to obtain the native-like (low-solubility) conformation containing one disulfide bond. Synthetic MGP comigrated with MGP derived from human bone on SDS-PAGE under denaturing conditions. The mass of the folded matrix Gla protein as measured using electrospray ionization mass spectrometry was identical to the theoretical average isotopic mass. The ability of MGP to form its internal disulfide shows that the initial thioester bond between Ala53 and Cys54 has rearranged into a peptide bond as depicted in Figure 2 ▶, resulting in the regeneration of the functional Cys54 sulfhydryl side chain.
When proteins fold, ESI-MS shows a characteristic shift of maximal intensity from the higher m/z states in the reduced protein to the lower m/z states in the folded protein. This is because the native, folded conformation of proteins typically can carry fewer charges under ESI-MS ionization conditions than the unfolded, extended polypeptides (Chowdhury et al. 1990). The minor shift from the twelfth charged state for the reduced MGP to the eleventh charged state for the folded, oxidized MGP (Fig. 3E,G ▶) is most likely a result of the relatively high amount of positively chargeable residues arginine (14) and asparagine (7) in MGP. This means that these residues stay exposed to the bulk solvent in the folded MGP, and this might play a role in the observed low solubility of MGP.
Using HPLC and CD spectroscopy, it was shown that synthetic MGP could bind Ca2+, inducing a shift in the HPLC retention time of MGP and an increase in α-helicity as measured as a change in mean residual weight ellipticity at 222 nm. From the increase in mean residual rotation at 222 nm, it was calculated that the content of α-helix increased by 25% in the presence of CaCl2. Therefore, the shift in retention time of synthetic MGP on HPLC in the presence of CaCl2 can be explained by an increase in α-helicity of MGP on binding of Ca2+ ions.
Similar behavior was shown for bone Gla protein (osteocalcin), in which α-helicity was observed to increase from 8%–38% as a result of Ca2+ binding (Hauschka and Carr 1982). Although no homology between the proteins is obvious, some similar functional properties during bone metabolism can be anticipated, and it is therefore interesting that in both proteins, ∼20 residues are in α-helical conformation in the presence of CaCl2.
The synthetic, native-like MGP generated in our study will be used for structural studies and as an antigen to raise antibodies required for the development of immunoassays. Determination of MGP in plasmas from patients with cardiovascular disease and healthy individuals will give the necessary insight into a possible role of MGP in the development of atherosclerosis.
Materials and methods
Materials
Most Boc-amino acids and 2–(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyl-uronium hexafluorophosphate (HBTU) were from NovaBiochem. Boc-Lys(Cl-Z)-OH was obtained from Bachem Bioscience; Boc-Arg(p-toluenesulfonyl)-OH and Boc-Asn(xanthyl)-OH were from Midwest Bio-Tech. S-trityl-mercaptopropionic acid was obtained from Peptides International. Boc-Lys-OCH2Pam-resin and Boc-Leu-OCH2Pam-resin were obtained from Applied Biosystems. N,N,-diisopropylethylamine (DIEA), p-cresol, thiophenol, and benzylmercaptan were obtained from Aldrich. N,N-Dimethylformamide (DMF) and HPLC-grade acetonitrile were purchased from Fischer. Trifluoroacetic acid (TFA) was obtained from Halocarbon, and HF was purchased from Matheson Gas.
Nα-Boc-γ, γ-di-cyclohexyl-L-γ–carboxyglutamic acid–OH synthesis
Ṅ-Boc-γ,γ-di-cyclohexyl-L-γ-carboxyglutamic acid-OH (Boc-Gla[dcHx]-OH) was synthesized as described (Nishiuchi et al. 1993).
Peptide synthesis
The sequence of MGP to be synthesized (Kiefer et al. 1988; Chen et al. 1990) is shown in Figure 1 ▶. Peptides were prepared by manual solid-phase peptide synthesis (SPPS) on a 0.3-mmol scale (MGP 1–53) and 0.35-mmol scale (MGP 54–84) using the in situ neutralization/HBTU activation procedure for Boc chemistry essentially as previously described (Schnölzer et al. 1992). Each synthetic cycle consisted of Ṅ-Boc removal by a 1–2-min treatment with neat TFA, a 1-min DMF-flow wash, and a 10-min coupling (Arg, Asn: 20 min) with 1.0 mmol preactivated Boc-amino acid in the presence of excess DIEA, followed by a second DMF-flow wash. For 3 min, 1.1 mmol Boc amino acids (except Boc-Gla[dcHx]-OH) were preactivated with 1.0 mmol HBTU (0.5 M in DMF)in the presence of 3 mmol DIEA. For 3 min, 0.44 mmol Ṅ-Boc Gla(dcHx)-OH was preactivated with 0.4 mmol HBTU in the presence of 1.5 mmol DIEA. After each coupling step, yields were determined by measuring residual free amine with the quantitative ninhydrin assay (Sarin et al. 1981). After coupling of Gln residues, a DCM flow wash was used before and after deprotection using TFA to prevent possible high-temperature (TFA/DMF)-catalyzed pyrrolidonecarboxylic acid formation (Schnölzer et al. 1992). Side-chain-protected amino acids were Boc-Arg (p-toluenesulfonyl)-OH, Boc-Asn(xanthyl)-OH, Boc-Asp(O-cyclohexyl)-OH, Boc-Cys(4-methylbenzyl)-OH, Boc-Gla(di-O-cyclohexyl)-OH, Boc-Glu(O-cyclohexyl)-OH, Boc-His(dinitrophenyl)-OH, Boc-Lys(2-Cl-Z)-OH, Boc-Ser(benzyl)-OH, Boc-Thr(benzyl)-OH, and Boc-Tyr(2-Br-Z)-OH. Other amino acids were used without side-chain protection. After chain assembly was completed, the peptides were deprotected and cleaved from the resin by treatment with anhydrous HF for 1 h at 0°C using 4% anisole (MGP 1–53) or p-cresol(MGP 54–84) as a scavenger. The imidazole side chain–dinitrophenyl (Dnp)–protecting groups remained on His residues on peptide MGP 1–53 because the Dnp-removal procedure is incompatible with C-terminal thioester groups. However, Dnp is gradually removed by thiols during the ligation reaction yielding unprotected His. After cleavage, both peptides were precipitated and washed with ice-cold diethylether. Subsequently, the precipitate was dissolved in aqueous acetonitrile and lyophilized.
TAMPAL resin
0.6 mmol S-trityl mercaptopropionic acid was activated with 0.54 mmol HBTU in the presence of 2 mmol DIEA and coupled for 10 min to Boc-Leu-OCH2Pam-resin (99.61% coupling). The resulting TAMPAL resin was used as a starting resin for polypeptide chain assembly following removal of the trityl protecting group and formation of the thioester bond with any desired amino acid using standard peptide coupling protocols. Treatment of the final peptide with anhydrous HF yielded the C-terminal activated mercaptopropionic acid-leucine (MPAL) thioester peptide that can directly participate in native chemical ligation (Hackeng et al. 1999). Trityl-deprotection was achieved by 2 × 1-min treatment with a solution of 1.25% triisopropylsilane and 1.25% H20 in TFA.
HPLC
Analytical reversed-phase HPLC was performed on a Hewlett Packard HPLC 1050 system using Vydac C-18 columns (5 μm, 0.46 × 5 cm). Semipreparative reversed-phase HPLC was performed on a Rainin HPLC system using a Vydac C-18 column (10 μm, 2.5 × 25 cm). Linear gradients of acetonitrile in water/0.1% TFA were used to elute bound peptides. The flow rates used were 1 mL/min (analytical) and 15 mL/min (semipreparative).
Mass spectrometry
ESI-MS was performed on an API-III triple quadrupole mass spectrometer (PE-Sciex). Peptide masses were calculated from the experimental m/z ratios from all the observed protonation states of a peptide using MacSpec software (Sciex). Theoretical masses of peptides and proteins were calculated using MacProMass software (Beckman Research Institute).
Native chemical ligation
The ligation of unprotected synthetic peptide segments was performed as described earlier (Hackeng et al. 1999). In short, peptides were dissolved in 0.1 M phosphate buffer containing 6 M guanidine, 1% (v/v) benzylmercaptan, and 1% (v/v) thiophenol (pH 7). The ligation reaction was performed at 37°C under frequent vortexing, and the reaction was monitored by HPLC and ESI-MS until completion.
CD spectroscopy
CD spectra were recorded on an AVIV stopped-flow CD spectrometer Model 202SF at 25°C. MGP was dissolved in 25 mM boric acid at pH 7.4, and the concentration was determined by quantitative amino acid analysis. CD spectra are presented as the molar ellipticity versus wavelength in 0.5-nm increments. The mean residual weight ellipticity was calculated using Equation (1).
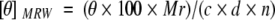 |
1 |
where [θ]MRW is the mean residue ellipticity (deg cm2 dmol−1), θ the observed ellipticity at 222 nm, Mr the molecular weight of MGP, c the concentration in mg/mL, d the pathlength in centimeters, and n the number of residues (Schmid 1989).
The ellipticity of MGP at 222 nm was recorded as a function of the CaCl2 concentration. To a volume of 250 μL of MGP (3.4 μM), CaCl2 was added to a final concentration of 20, 40, 79, 157, 311, 618, 1225, and 2411 μM. MGP concentrations were corrected for dilution of the sample by CaCl2 (20 μL maximal addition). For the calculation of α-helical content of MGP, the averages of the mean residual weight ellipticities at 223, 222.5, 222, 221.5, and 221 nm were used.
The percentage of α-helix in MGP was determined as the ratio of the mean residual weight ellipticity at 222 nm and the maximal theoretical mean residual weight ellipticity at 222 nm ([θ]max; Chen et al. 1974) using Equation (2).
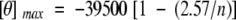 |
2 |
SDS-PAGE and Western blot analysis
Protein samples in SDS sample buffer (40 mM Tris-HCl at pH 6.7, 4% [w/v] SDS, 10% [v/v] glycerol, 2% [v/v] 2-mercaptoethanol, 0.01% [w/v] bromophenol blue) were applied to polyacrylamide gels 20% (w/v) in SDS under denaturing conditions. After electrophoresis, the gels were transferred to Immobilon-P PVDF membranes (Millipore). After washing the membranes in PBS for 15 min, nonspecific binding sites were blocked with blocking buffer (3% w/v nonfat dry milk in PBS) for 1 h at room temperature and incubated at 4°C overnight with an α 3–15 MGP antibody in blocking buffer (Braam et al. 2000). After washing three times with 0.3% (v/v) Tween 20 in phosphate-buffered saline (PBS-Tween), the membrane was incubated for 1 h at room temperature with HRP-conjugated rabbit anti-mouse IgG in PBS-Tween. The membrane was washed with PBS-Tween, followed by a final wash step of 15 min with PBS and development with ECL Western blotting detection reagents (Amersham Pharmacia). Finally, protein bands were visualized on Hyperfilm ECL (Amersham Pharmacia).
Acknowledgments
We thank Philip E. Dawson for providing access to his chemical protein synthesis facility, Kevin Judice for his excellent advice during synthesis of Nα-tBoc-γ,γ-dicyclohexyl-L-γ-carboxyglutamic acid, and Reidar Wallin for providing us with purified human matrix Gla protein.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Boc, tert-butoxycarbonyl
DIEA, N,N,-diisopropylethylamine
DMF, N,N,-Dimethyl-formamide
Dnp, 2,4-Dinitrophenyl
ESI-MS: electrospray ionization mass spectrometry
m/z, mass to charge ratio
HBTU, 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
SPPS, solid phase peptide synthesis
TAMPAL, trityl-associated mercaptopropionic acid leucine
TFA, trifluoroacetic acid
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.44701.
References
- Braam, L.A.J.L.M., Dissel, P., Gijsbers, B.L.M.G., Spronk, H.M.H., Hamulyák, K., Soute, B.A.M., Debie, W., and Vermeer, C. 2000. Assay for human matrix Gla protein in serum. Arterioscler Thromb. Vasc. Biol. 20 1257–1261. [DOI] [PubMed] [Google Scholar]
- Chen, Y.H., Yang, J.T., and Chau, K.H. 1974. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13 3350–3359. [DOI] [PubMed] [Google Scholar]
- Chen, L., O'Brian, J.P., Smith, H.S., and Liu, E. 1990. Overexpression of matrix Gla protein mRNA in malignant human breast cells, isolation by differential cDNA hybridization. Oncogene 5 1391–1395. [PubMed] [Google Scholar]
- Chowdhury, S.K., Katta, V., and Chait, B.T. 1990. Probing conformational changes in proteins by mass spectrometry. J. Am. Chem. Soc. 112 9012–9013. [Google Scholar]
- Dawson, P.E., Muir, T.W., Clark-Lewis, I., Kent, S.B.H. 1994. Synthesis of proteins by native chemical ligation. Science 266 776–779. [DOI] [PubMed] [Google Scholar]
- Fraser, J.D. and Price, P.A. 1988. Lung, heart, and kidney express high levels of mRNA for the vitamin K–dependent matrix Gla protein. J. Biol. Chem. 263 11033–11036. [PubMed] [Google Scholar]
- Hackeng, T.M., Griffin, J.H., and Dawson, P.E. 1999. Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. 96 10068–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, J.E., Fraser, J.D., and Price, P.A. 1988. The identification of matrix Gla protein in cartilage. J. Biol. Chem. 263 5820–5824. [PubMed] [Google Scholar]
- Hauschka, P.V. and Carr, S.A. 1982. Calcium-dependent α-helical structure in osteocalcin. Biochemistry 21 2538–2547. [DOI] [PubMed] [Google Scholar]
- Ikeda, T., Yamaguchi, A., Icho, T., Tsuchida, N., and Yoshiki, S. 1991. cDNA and deduced amino acid sequence of mouse matrix gla protein: One of five glutamic acid residues potentially modified to gla is not conserved in the mouse sequence. J. Bone. Miner. Res. 6 1013–1017. [DOI] [PubMed] [Google Scholar]
- Kiefer, M.C., Bauer, D.M., Young, D., Hermsen, K.M., Masiarz, F.R, and Barr, P.J. 1988. The cDNA and derived amino acid sequences for human and bovine matrix Gla protein. Nucleic Acids Res. 16 5213–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, G., Ducy, P., McKee, M.D., Pinero, G.J., Loyer, E., Behringer, R.R., and Karsenty, G. 1997. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature 386 78–81. [DOI] [PubMed] [Google Scholar]
- Nelsestuen, G.L., Shah, A.M., and Harvey, S.B. 2000. Vitamin K-dependent proteins. Vitam. Horm. 58 355–389. [DOI] [PubMed] [Google Scholar]
- Nishiuchi, Y., Masayuki, N., Nakata, M., Kimura, T., and Sakakibara, S. 1993. Synthesis of γ-carboxyglutamic acid-containing peptides by the Boc strategy. Int. J. Peptide Protein Res. 42 533–538. [DOI] [PubMed] [Google Scholar]
- Otawara, Y. and Price, P.A. 1986. Developmental appearance of matrix GLA protein during calcification in the rat. J. Biol. Chem. 261 10828–10832. [PubMed] [Google Scholar]
- Price, P.A., Urist, M.R., and Otawara, Y. 1983. Matrix Gla protein, a new γ-carboxyglutamic acid-containing protein which is associated with the organic matrix of bone. Biochem. Biophys. Res. Commun. 117 765–771. [DOI] [PubMed] [Google Scholar]
- Price, P.A., Fraser, J.D., and Metz-Virca, G. 1987. Molecular cloning of matrix Gla protein: Implications for substrate recognition by the vitamin K-dependent γ-carboxylase. Proc. Natl. Acad. Sci. 84 8335–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, J.S., Williamson, M.K., and Price, P.A. 1994. Isolation and sequence of the vitamin K−dependent matrix Gla protein from the calcified cartilage of the soupfin shark. J. Bone Miner. Res. 9 567–576. [DOI] [PubMed] [Google Scholar]
- Sarin, V.K., Kent, S.B.H., Tam, J.P, and Merrifield, R.B. 1981. Anal. Biochem. 117 147–157. [DOI] [PubMed] [Google Scholar]
- Schmid, F.X. 1989. Protein structure, a practical approach (ed. T.E. Creighton), pp. 251–285. IRL Press, Oxford.
- Schnölzer, M., Alewood, P., Jones, A., Alewood, D., and Kent, S.B.H. 1992. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Int. J. Pept. Protein Res. 40 180–193. [DOI] [PubMed] [Google Scholar]
- Wiedemann, M., Trueb, B., and Belluoccio, D. 1998. Molecular cloning of avian matrix Gla protein. Biochim. Biophys. Acta 1395 47–49. [DOI] [PubMed] [Google Scholar]