Abstract
GTP cyclohydrolase I feedback regulatory protein (GFRP) mediates feedback inhibition of GTP cyclohydrolase I activity by 6R-l-erythro-5,6,7,8-tetrahydrobiopterin (BH4), which is an essential cofactor for key enzymes producing catecholamines, serotonin, and nitric oxide as well as phenylalanine hydroxylase. GFRP also mediates feed-forward stimulation of GTP cyclohydrolase I activity by phenylalanine at subsaturating GTP levels. These ligands, BH4 and phenylalanine, induce complex formation between one molecule of GTP cyclohydrolase I and two molecules of GFRP. Here, we report the analysis of ligand binding using the gel filtration method of Hummel and Dreyer. BH4 binds to the GTP cyclohydrolase I/GFRP complex with a Kd of 4 μM, and phenylalanine binds to the protein complex with a Kd of 94 μM. The binding of BH4 is enhanced by dGTP. The binding stoichiometrics of BH4 and phenylalanine were estimated to be 10 molecules of each per protein complex, in other words, one molecule per subunit of protein, because GTP cyclohydrolase I is a decamer and GFRP is a pentamer. These findings were corroborated by data from equilibrium dialysis experiments. Regarding ligand binding to free proteins, BH4 binds weakly to GTP cyclohydrolase I but not to GFRP, and phenylalanine binds weakly to GFRP but not to GTP cyclohydrolase I. These results suggest that the overall structure of the protein complex contributes to binding of BH4 and phenylalanine but also that each binding site of BH4 and phenylalanine may be primarily composed of residues of GTP cyclohydrolase I and GFRP, respectively.
Keywords: Tetrahydrobiopterin, GTP cyclohydrolase I, feedback regulation, allosteric regulation, GTP cyclohydrolase I feedback regulatory protein, phenylalanine
GTP cyclohydrolase I is widely distributed from bacteria to humans. GTP cyclohydrolase I (EC 3.5.4.16) hydrolyzes GTP to produce dihydroneopterin triphosphate (Fig. 1 ▶), which is further converted to a variety of pteridines including BH4 in animals and tetrahydrofolate in plants and microorganisms (Nichol et al. 1985; Kaufman 1993). BH4 serves as an essential cofactor for phenylalanine hydroxylase, tyrosine hydroxylase, tryptophan hydroxylase, and nitric oxide synthase. In the biosynthesis of BH4, GTP cyclohydrolase I is the most important control site. The catalytic activity of GTP cyclohydrolase I is directly regulated by GFRP (Harada et al. 1993; Milstien et al. 1996). GFRP mediates the feedback inhibition of the enzyme activity by BH4. The inhibition of the enzyme activity by BH4 and GFRP is reversed by phenylalanine. These control mechanisms assure that BH4 is not needlessly formed when the level of phenylalanine is low and that supply of BH4 for phenylalanine hydroxylase increases when the level of phenylalanine is high. The occurrence of these mechanisms in vivo is supported by many studies conducted with normal individuals and patients with phenylketonuria (Nichol et al. 1985, and references therein).
Fig. 1.
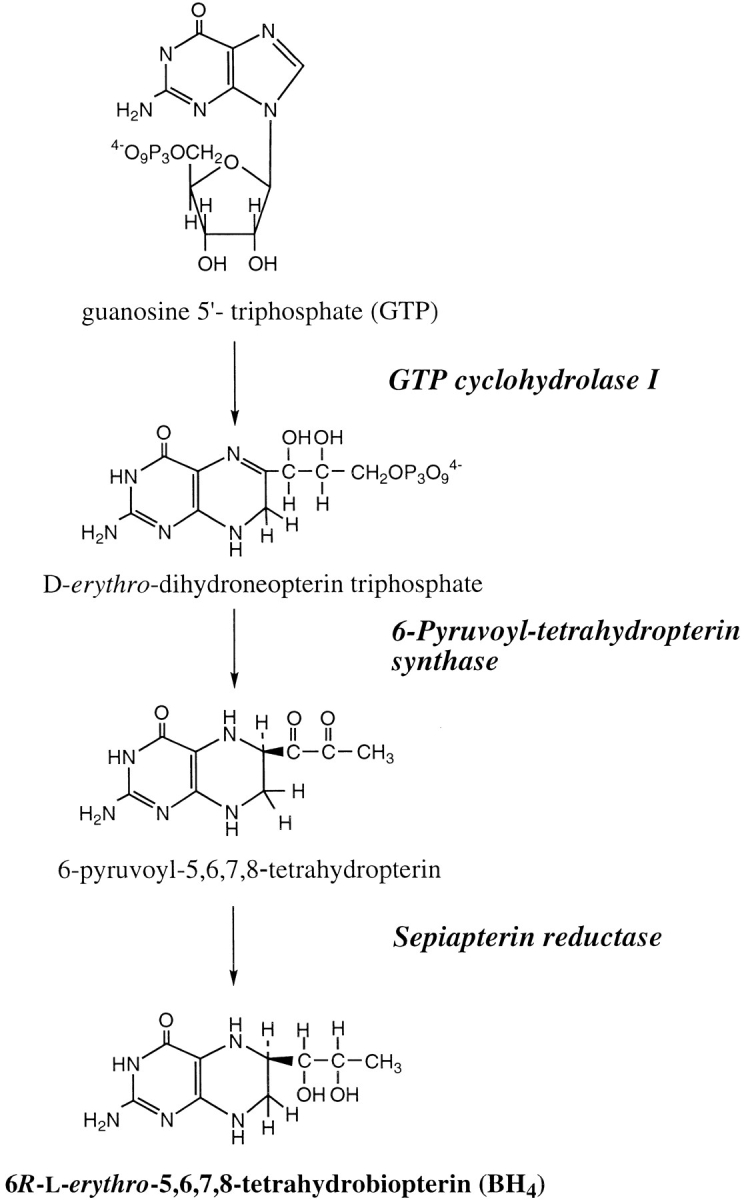
The biosynthetic pathway of BH4.
GFRP forms a physically stable protein complex with GTP cyclohydrolase I (Harada et al. 1993). BH4 induces formation of the inhibitory GTP cyclohydrolase I/GFRP complex, and phenylalanine induces formation of the stimulatory GTP cyclohydrolase I/GFRP complex (Harada et al. 1993). BH4 is the most potent of biopterins with different oxidative states. Formation of the inhibitory complex requires GTP as well as BH4 (Harada et al. 1993; Yoneyama and Hatakeyama 1998). GTP can be replaced with dGTP or guanosine 5`-O-(3`-thiotriphosphate) (Yoneyama and Hatakeyama 1998). In contrast, phenylalanine alone is capable of inducing formation of the stimulatory complex (Harada et al. 1993; Yoneyama and Hatakeyama 1998). Phenylalanine reduces the positive cooperativity exhibited by GTP cyclohydrolase I, increasing enzyme activity at subsaturating GTP concentrations (Harada et al. 1993; Yoneyama et al. 1997). Stimulating the enzyme activity and inducing formation of the stimulatory complex are highly specific to l-phenylalanine; neither d-phenylalanine nor tyrosine have any effect (Harada et al. 1993; Yoneyama and Hatakeyama 1998).
Rat GTP cyclohydrolase I is a decameric protein with a subunit molecular weight of 25 kD (Hatakeyama et al. 1989, 1991; Yoneyama and Hatakeyama 1998). The quaternary structure of the rat enzyme is predicted to be a dimer of pentamers, because the corresponding Escherichia coli enzyme, which shows a high degree of amino acid sequence similarity to the rat enzyme, has such a structure, determined crystallographically (Nar et al. 1995). Rat GFRP is a pentameric protein with a subunit molecular weight of 9.5 kD (Yoneyama et al. 1997). Gel filtration experiments as well as enzyme activity measurements established that two molecules of GFRP interact with one molecule of GTP cyclohydrolase I both in the presence of GTP and BH4 and in the presence of phenylalanine (Yoneyama and Hatakeyama 1998). Gel filtration analysis indicated that the complex has a radius of gyration similar to that of the enzyme itself. Because the shape of the enzyme is a torus (Nar et al. 1995), we proposed a model of a quaternary structure of the protein complex in which a GFRP pentamer binds to each of the outer faces of two pentamers of GTP cyclohydrolase I associated face to face (Yoneyama et al. 1997; Yoneyama and Hatakeyama 1998).
Thus, the protein stoichiometry of both types of complexes and the ligand specificity for complex formation have been determined. However, the binding of ligands to the protein complexes remained to be investigated. For this purpose, we used the gel filtration procedure of Hummel and Dreyer (1962). The experimental procedure enabled us to simultaneously measure the extent of GTP cyclohydrolase I/GFRP complex formation and the binding of ligands. We demonstrate that the GTP cyclohydrolase I/GFRP complex consisting of 10 subunits each of GTP cyclohydrolase I and GFRP binds 10 molecules of ligand. Experiments on ligand binding to free GTP cyclohydrolase I and GFRP provided information on the locations of the binding sites of the ligands.
Results
Ligand binding to the inhibitory complex
In the gel filtration technique originally developed by Hummel and Dreyer (1962), a gel column is equilibrated with a solution containing ligands at desired concentrations. A small sample of protein solution in which the total ligand concentration equals that already in the column is then injected into the column. The chromatographic profile shows a leading peak corresponding to the ligated protein, followed by a trough emerging at the elution volume of the ligand. The area of the trough represents the depletion of ligand that resulted from the bound ligand that eluted with the protein.
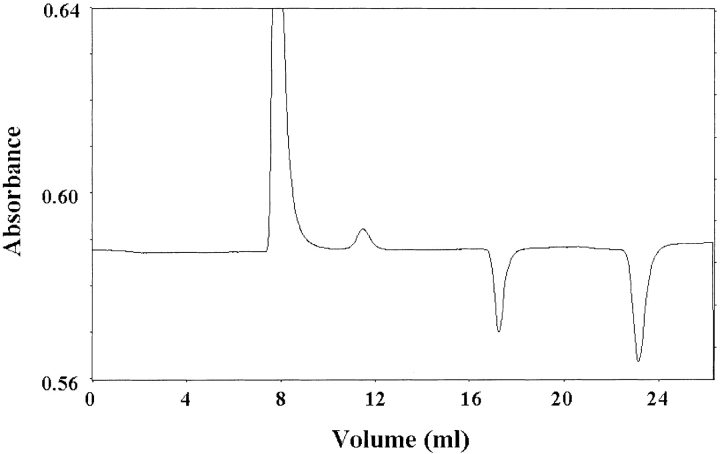
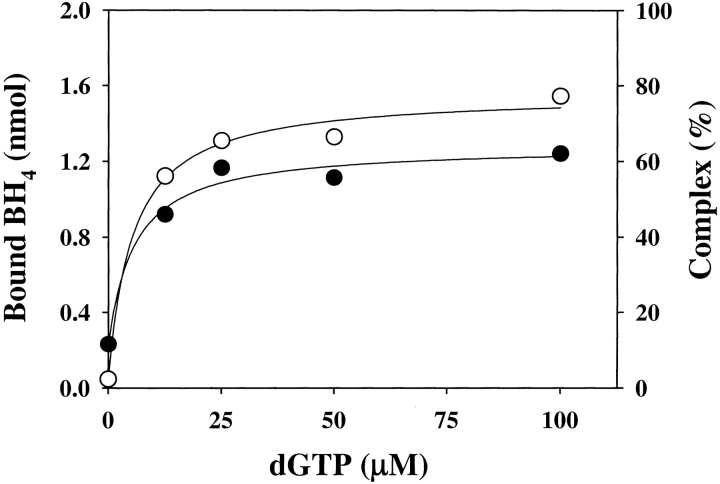
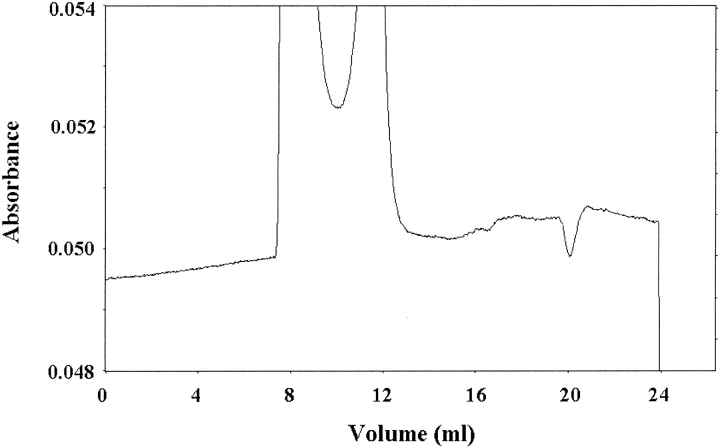
Under the conditions described in Materials and Methods, BH4 eluted at a volume different from that of dGTP (Fig. 2 ▶); dGTP was used for the experiments because it has the same potency of inducing inhibitory complex formation as GTP but is not hydrolyzed (Yoneyama and Hatakeyama 1998). Moreover, the method allowed us to simultaneously measure the extent of the association of GFRP to GTP cyclohydrolase I, because free GTP cyclohydrolase I and the protein complex elute at almost the same positions and, accordingly, the estimation of the extent of protein complex formation was made from the decrease in free GFRP (Yoneyama and Hatakeyama 1998). As shown in Figure 3 ▶, we measured the binding of BH4 to the protein complex, which was dependent on the presence of dGTP. Similarly, in the presence of 100 μM dGTP, BH4 bound to the protein complex in a hyperbolic manner (Fig. 4 ▶). The curve was fitted to the equation,
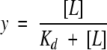 |
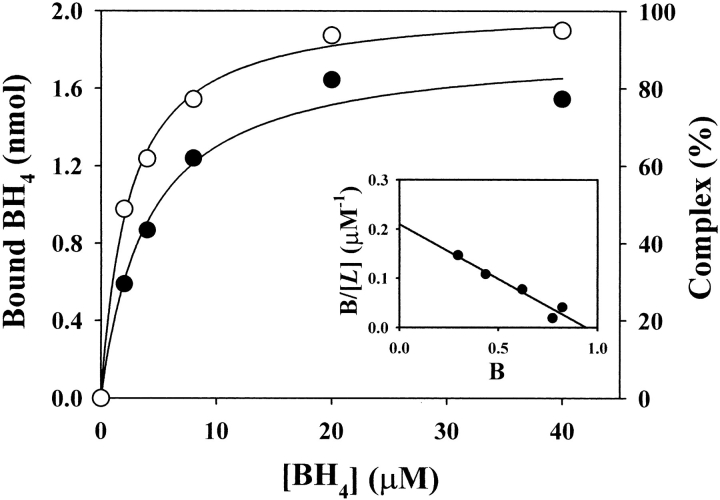
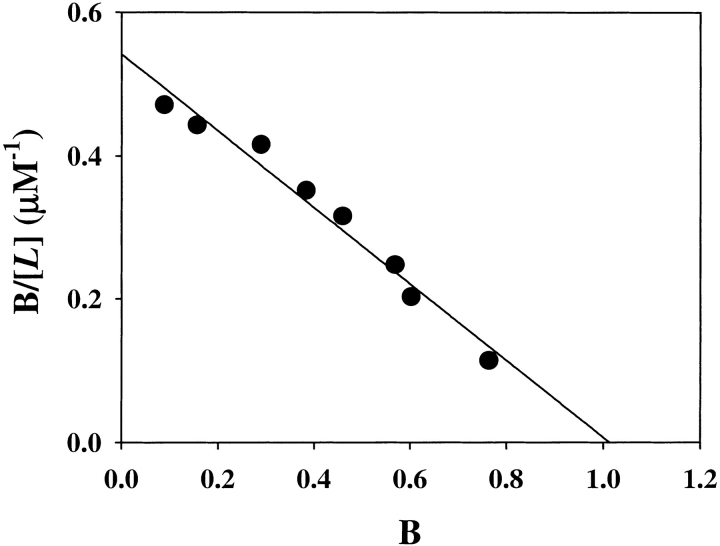
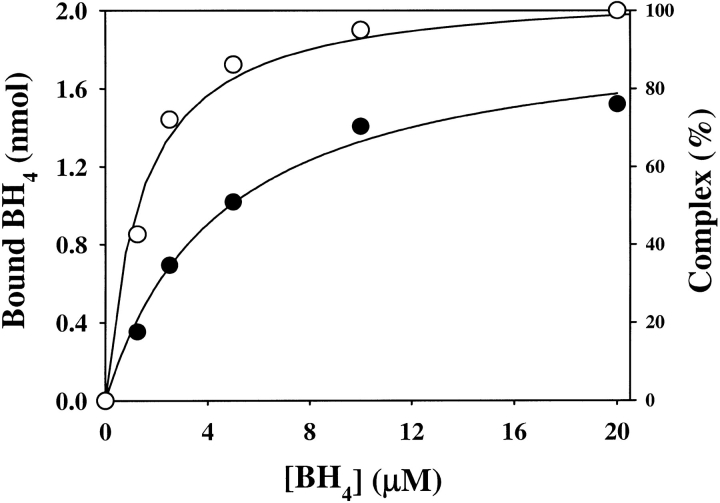
where y is the saturation fraction and [L] is the concentration of free ligand. The number of moles of BH4 bound at saturation was thus determined to be 9.1 ± 0.5 moles bound per mole of GTP cyclohydrolase I/GFRP complex. This suggests that one BH4 molecule is bound per pair of GTP cyclohydrolase I and GFRP subunits. The Kd of BH4 binding to the GTP cyclohydrolase I/GFRP complex in the presence of 100 μM dGTP was 4.0 ± 0.8 μM. These observations were confirmed by equilibrium dialysis experiments, although the values were slightly different (Fig. 5 ▶).
Fig. 2.
Chromatographic profile of the binding of GFRP, dGTP, and BH4 to GTP cyclohydrolase I. A Superdex 75 column was equilibrated with buffer containing 50 mM Hepes-KOH (pH 7.2), 0.2 M KCl, 1 mM EDTA, 1 mM dithiothreitol, 20 μM BH4, and 50 μM dGTP. GTP cyclohydrolase I (2 nmole) plus GFRP (2 nmole) was dissolved in the same buffer and applied to the column. The eluate was monitored at 280 nm until the volume of 21 mL eluted and thereafter at 300 nm. Elution positions of the proteins and ligands are GTP cyclohydrolase I (7.8 mL), GFRP (11.3 mL), dGTP (17.3 mL), and BH4 (23.1 mL).
Fig. 3.
Effect of dGTP concentration on GFRP binding to GTP cyclohydrolase I (open circles) and BH4 binding to the complex (filled circles) in the presence of 8 μM BH4. GTP cyclohydrolase I (2 nmole) plus GFRP (2 nmole) was applied to the column. The results shown are from a representative experiment that was repeated twice.
Fig. 4.
Binding of BH4 by the complex at pH 7.2. Effect of BH4 concentration on GFRP binding to GTP cyclohydrolase I (open circles) and BH4 binding to the complex (closed circles) in the presence of 100 μM dGTP. GTP cyclohydrolase I (2 nmole) plus GFRP (2 nmole) was applied to the column. The results shown are from a representative experiment that was repeated twice. Scatchard plot is also shown.
Fig. 5.
Binding of BH4 by the complex. Equilibrium dialysis experiments were performed as described in Materials and Methods. Scatchard analysis of the data reveals that the binding of BH4 to the complex is characterized by a Kd of 1.9 μM and a stoichiometry of 1.0.
We tested whether BH4 interacted with free GTP cyclohydrolase I or GFRP using the same gel filtration method (Table 1). GTP cyclohydrolase I bound BH4 very weakly, whereas GFRP did not bind BH4 at all. The binding of BH4 to GTP cyclohydrolase I was enhanced by dGTP (Table 1). When a higher concentration of BH4 (40 μM) was examined in the presence of dGTP (100 μM), more BH4 bound to the enzyme (0.36 moles/mole of GTP cyclohydrolase I subunit). The interaction of BH4 with GTP cyclohydrolase I, however, was much weaker than that with the GTP cyclohydrolase I/GFRP complex (cf. Fig. 4 ▶ and Table 1). By contrast, free GFRP did not bind BH4 even in the presence of dGTP (Table 1).
Table 1.
BH4 binding to free GTP cyclohydrolase I or GFRP at pH 7.2 and 6.0 a
| Bound BH4 | |||
| Ligand concentration | Protein | pH 7.2 | pH 6.0 |
| mol/mol protein subunit | |||
| 20 μM BH4 | None | 0.000 ± 0.000 | −0.015 ± 0.021 |
| 20 μM BH4 | GTP cyclohydrolase I | 0.023 ± 0.005b | 0.097 ± 0.011b |
| 20 μM BH4 | GFRP | 0.004 ± 0.005c | −0.015 ± 0.011c |
| 20 μM BH4 + 100 μM dGTP | None | 0.000 ± 0.000 | ND |
| 20 μM BH4 + 100 μM dGTP | GTP cyclohydrolase I | 0.105 ± 0.010b | ND |
| 20 μM BH4 + 100 μM dGTP | GFRP | 0.015 ± 0.011c | ND |
a A Superdex 75 column was equilibrated with 50 mM Hepes-KOH (pH 7.2 or 6.0), 0.2 M KCl, 1 mM EDTA, 1 mM dithiothreitol plus 20 μM BH4 or 20 μM BH4 and 100 μM dGTP. GTP cyclohydrolase I (2 nmol) or GFRP (2 nmol) were dissolved in the same buffer and applied to the column. Binding of BH4 was determined as described in Materials and Methods. The data are represented as mean ± S.D. (n = 3). ND, not determined.
bP < 0.03 vs. control by Student's t test.
cP > 0.1 vs. control by Student's t test.
During the course of the experiments, we found that a small change in pH affects the binding behavior of BH4 significantly. At pH 7.2, formation of the GTP cyclohydrolase I/GFRP complex was not induced at all by BH4 alone at a concentration of 8 μM (Fig. 3 ▶) and was only partially induced by BH4 when 20 μM BH4 was used. In contrast, at pH 6.0, much lower concentrations of BH4 alone induced formation of the protein complex (Fig. 6 ▶). The EC50 value of BH4 for formation of the protein complex at pH 6.0 was 1.4 ± 0.2 μM. The value is even lower than the value of 2.4 ± 0.2 μM obtained at pH 7.2 for formation of the protein complex that is enhanced by the presence of dGTP. The binding of BH4 at pH 6.0 had a Kd of 4.5 ± 0.7 μM. The maximum binding was estimated to be 9.5 ± 0.5 moles/mole of GTP cyclohydrolase I/GFRP complex. Again, almost one molecule of BH4 is bound to the protein complex for each pair of GFRP and GTP cyclohydrolase I subunits.
Fig. 6.
Binding of BH4 by the complex at pH 6.0. Effect of BH4 concentration on GFRP binding to GTP cyclohydrolase I (open circles) and BH4 binding to the complex (closed circles) in the absence of dGTP. GTP cyclohydrolase I (2 nmole) plus GFRP (2 nmole) was applied to the column. The results shown are from a representative experiment that was repeated twice.
When free GTP cyclohydrolase I (2 nmole) was examined for BH4 binding at pH 6.0 with a BH4 concentration of 20 μM, the enzyme bound about 0.1 moles of BH4/mole of GTP cyclohydrolase I subunit. In contrast, no BH4 binding was observed for free GFRP with a BH4 concentration of 20 μM (Table 1).
Ligand binding to the stimulatory complex
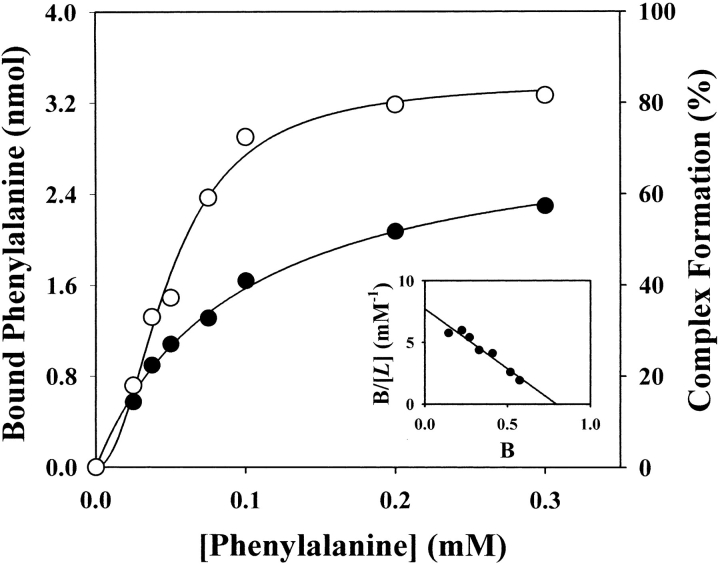
Phenylalanine binding to the GTP cyclohydrolase I/GFRP complex was also measured using this method (Fig. 7 ▶). We examined the effects of varying concentrations of phenylalanine on the amount of protein complex formed and on the amount of phenylalanine bound to the protein complex. The resulting curve for the protein complex formation was sigmoidal (Fig. 8 ▶), as we described previously (Yoneyama and Hatakeyama 1998). In contrast, the binding isotherm of phenylalanine was hyperbolic (Fig. 8 ▶), indicating that the binding of phenylalanine is not cooperative. Nine molecules of phenylalanine bind per molecule of the GTP cyclohydrolase I/GFRP complex with a Kd of 94 ± 8 μM (Fig. 8 ▶). These observations were confirmed by equilibrium dialysis experiments, although the values were slightly different (Fig. 9 ▶).
Fig. 7.
Chromatographic profile of the binding of GFRP and phenylalanine to GTP cyclohydrolase I. A Superdex 75 column was equilibrated with 50 mM Hepes-KOH (pH 7.2), 0.2 M KCl, 1 mM EDTA, 1 mM dithiothreitol, and 0.1 mM phenylalanine. GTP cyclohydrolase I (4 nmole) plus GFRP (4 nmole) was dissolved in the same buffer and applied to the column. The eluate was monitored at 280 nm until the volume of 15 mL eluted and thereafter at 257 nm. Elution positions of the proteins and ligands are GTP cyclohydrolase I (7.8 mL), GFRP (11.3 mL), and phenylalanine (20.2 mL).
Fig. 8.
Binding of phenylalanine by the complex at pH 7.2. Effect of phenylalanine concentration on GFRP binding to GTP cyclohydrolase I (open circles) and phenylalanine binding to the complex (filled circles). GTP cyclohydrolase I (4 nmole) plus GFRP (4 nmole) was applied to the column. The results shown are from a representative experiment that was repeated twice. Scatchard plot is also shown.
Fig. 9.
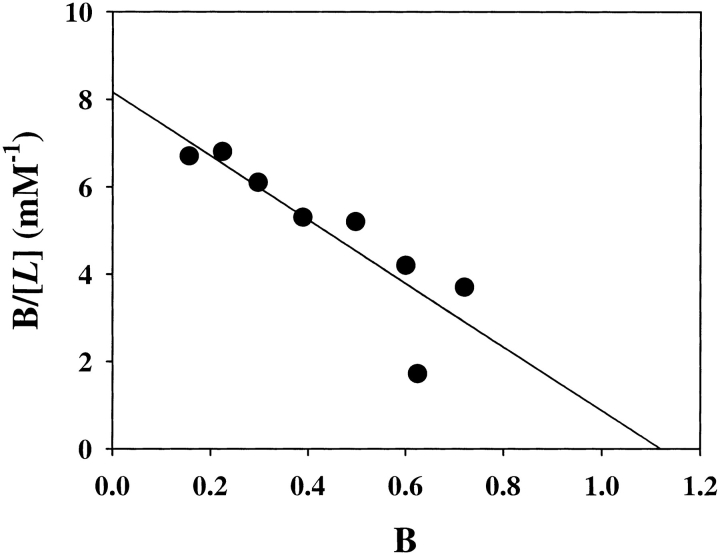
Binding of phenylalanine by the complex. Equilibrium dialysis experiments were performed as described in Materials and Methods. Scatchard analysis of the data reveals that the binding of phenylalanine to the complex is characterized by a Kd of 137 μM and a stoichiometry of 1.1.
In contrast to BH4, phenylalanine bound to free GFRP but not to free GTP cyclohydrolase I (Table 2). The affinity of phenylalanine binding to GFRP was much weaker than that to the GTP cyclohydrolase I/GFRP complex. The binding of phenylalanine to the complex was higher at pH 6.0 than at pH 7.2 (data not shown). The EC50 value for formation of the protein complex with phenylalanine at pH 6.0 was 10 ± 2 μM (data not shown). This value is much lower than the value of 51 ± 3 μM obtained at pH 7.2 (Fig. 8 ▶). Phenylalanine did not bind to free GTP cyclohydrolase I even at pH 6.0 (Table 2).
Table 2.
Phenylalanine binding to free GTP cyclohydrolase I or GFRP at pH 7.2 and 6.0a
| Bound phenylalanine | |||
| Ligand concentration | Protein | pH 7.2 | pH 6.0 |
| mol/mol protein subunit | |||
| 400 μM phenylalanine | None | 0.095 ± 0.023 | −0.030 ± 0.004 |
| 400 μM phenylalanine | GTP cyclohydrolase I | 0.063 ± 0.016c | −0.038 ± 0.110c |
| 400 μM phenylalanine | GFRP | 0.221 ± 0.021b | 0.182 ± 0.019b |
a A Superdex 75 column was equilibrated with 50 mM Hepes-KOH (pH 7.2 or 6.0), 0.2 M KCl, 1 mM EDTA, 1 mM dithiothreitol plus 400 μM phenylalanine. GTP cyclohydrolase I (4 nmol) or GFRP (4 nmol) were dissolved in the same buffer and applied to the column. Binding of phenylalanine was determined as described in Materials and Methods. The data are represented as mean ± S.D. (n = 3).
bP < 0.005 vs. control by Student's t test.
cP > 0.05 vs. control by Student's t test.
Discussion
We demonstrate by using the Hummel and Dreyer method the binding of BH4 and phenylalanine to the inhibitory and stimulatory GTP cyclohydrolase I/GFRP complexes, respectively. The Kd values at pH 7.2 were determined to be 4 and 94 μM for BH4 and phenylalanine, respectively, and the stoichiometry of each ligand binding is almost one per pair of GTP cyclohydrolase I and GFRP subunits. Because both of the complexes consist of 10 subunits of each of GTP cyclohydrolase I and GFRP, the complexes are constructed of 10 molecules of each protein subunit and ligand.
We observed previously that formation of the inhibitory complex requires the presence of dGTP or GTP as well as BH4. Here, we show that dGTP acts by enhancing the binding of BH4 (Fig. 3 ▶). The binding of BH4, however, is not totally dependent on the presence of dGTP. BH4 alone was able to partially (40%) induce formation of the inhibitory complex at a higher concentration (20 μM). At pH 6.0, BH4 alone fully induced formation of the inhibitory complex (Fig. 6 ▶). In contrast, phenylalanine did not require any additional factor for its binding to the protein complex and for its ability to induce formation of the stimulatory complex at both pH 7.2 and 6.0. The affinity of phenylalanine to the GTP cyclohydrolase I/GFRP complex was higher at pH 6.0 than 7.2 (data not shown), and formation of the stimulatory complex was enhanced at pH 6.0 compared with pH 7.2.
Thus, the affinities of the individual binding of BH4 and phenylalanine to GTP cyclohydrolase I and GFRP were both enhanced at pH 6.0 compared with pH 7.2. Because both ligands do not change charge in that pH region, there may be some ionizable group(s) perhaps in the binding sites of each ligand that affect their interaction with the ligands. Alternatively, some ionizable group(s) located at the interface between the two proteins may be involved in the association between the two proteins.
We infer that the binding site for BH4 is primarily composed of residues of GTP cyclohydrolase I based on two lines of evidence. First, BH4 bound to free GTP cyclohydrolase I but not to free GFRP (Table 1). Second, the binding of BH4 to free GTP cyclohydrolase I was enhanced by dGTP (Table 1). This inference is consistent with the kinetic data that BH4 inhibited GTP cyclohydrolase I activity at a higher concentration (100 μM) with 27% inhibition in the absence of GFRP (data not shown).
BH4 binding to free GTP cyclohydrolase I was much weaker than to the GTP cyclohydrolase I/GFRP complex. This suggests two possible ways of involvement of GFRP in the binding of BH4 to the GTP cyclohydrolase I/GFRP complex. First, no part of GFRP directly interacts with BH4, but GFRP stabilizes an altered conformation of GTP cyclohydrolase I resulting from BH4 binding. Second, in addition to the stabilization by GFRP, part of the BH4-binding site is formed by GFRP and the other by GTP cyclohydrolase I. GFRP interacts only with relatively small part of BH4 with an affinity too low to be detected using the gel filtration procedure.
In contrast, phenylalanine bound to free GFRP but not to GTP cyclohydrolase I, and phenylalanine binding to free GFRP was much weaker than to the GTP cyclohydrolase I/GFRP complex. These suggest that the phenylalanine-binding site is completely or primarily composed of GFRP residues in the GTP cyclohydrolase I/GFRP complex and the binding of phenylalanine to GFRP is stabilized by GTP cyclohydrolase I.
Materials and methods
Materials
BH4 was a generous gift from the Suntory Institute for Medicinal Research and Development (Gunma, Japan). Phenylalanine and dGTP were obtained from Sigma. Recombinant rat GFRP and GTP cyclohydrolase I were prepared as described previously (Harada et al. 1993; Yoneyama et al. 1997). The molar concentrations of GFRP and GTP cyclohydrolase I are expressed as those of their subunits (Yoneyama and Hatakeyama 1998).
Gel filtration analyses of complex formation and ligand binding
Measurements of complex formation between GFRP and GTP cyclohydrolase I and ligand binding to the protein complex were simultaneously performed using gel filtration (Hummel and Dreyer 1962; Ackers 1973). The following procedures for the preparation of solution were followed to ensure that concentrations of every constituent were the same in the protein sample solution injected and the buffer used for column equilibration, except for the protein injected itself. The protein samples were initially filtered through a 1 × 10 cm column of Sephadex G-25 superfine to equilibrate them with the buffer that contained all of the constituents used for the Superdex 75 gel filtration analysis except for the ligands (50 mM Hepes-KOH at pH 7.2, 0.2 M KCl, 1 mM EDTA, 1 mM dithiothreitol). Then the protein concentrations were adjusted to a concentration that was twofold higher than that used for the analysis. The resulting protein solution was injected into a 1 × 30-cm column of Superdex 75 (Amersham Pharmacia Biotech) after being mixed with an equal volume of equilibration buffer that contained a twofold higher concentration of ligand than that used for the analysis. The injected volume was 200 μL. The Superdex 75 column was itself equilibrated with a solution made by mixing an equal volume of the equilibration buffer and the same equilibration buffer, containing a twofold higher concentration of ligand, used for the preparation of protein sample.
Elution was performed at a flow rate of 0.8 mL/min at room temperature. The eluate was monitored on a Shimadzu SPD-10AVP absorbance detector with a 1-cm light path cell at different wavelengths at different elution periods for the detection of protein and each ligand as described below and in the legends for Figs. 2 and 7 ▶ ▶. The protein complex formation was estimated from a decrease in the peak height of free GFRP, as described previously (Yoneyama and Hatakeyama 1998). The amount of ligand bound by proteins was estimated by the area of a trough observed at the elution volume of each ligand. Phenylalanine and BH4 eluted at a volume of 20.2 and 23.1 mL, respectively. Phenylalanine and BH4 were monitored at 257 and 300 nm, respectively. The trough areas (O.D. × seconds) were acquired by a Shimadzu Class VP chromatography data system version 4.2 and then the values were converted to moles using the flow rate and the molar extinction coefficients of each ligand. We used a reported molar extinction coefficient of phenylalanine (195 M−1 cm−1 at 257 nm) (Dawson et al. 1986) and the molar extinction coefficient of BH4 that we determined to be 9.74 × 103 M−1 cm−1 at 300 nm. The data were analyzed by nonlinear regression curve fitting using SigmaPlot scientific graph (Jandel Scientific).
The recovery of BH4 and phenylalanine on the Superdex 75 column chromatography was dose-dependent and 100 ± 2%. The protein dose dependency on the ligands bound by proteins was linear.
Equilibrium binding analysis
Assessment of ligand binding by equilibrium dialysis was performed in an acrylic 8-place equilibrium type cell (Bel-Art Products, Paquannock, NJ). Each cell contained eight pairs of chambers (100 μL each) separated by a semipermeable dialysis membrane. Dialysis membranes were prepared from Spectra/Por dialysis tubing (molecular weight cut-off for permeability: 12,000–14,000) (Spectrum Medical Industries, Inc. Laguna Hills, CA), which had been boiled in 2% NaHCO3–1 mM EDTA. For experiments of BH4 binding, the same buffer as that used for gel filtration experiments was used except for 5 mM dithiothreitol in the presence of 100 μM dGTP; concentrations of both GTP cyclohydrolase I and GFRP used were 2 μM, and BH4 concentrations used were in the range of 0.25–8 μM. For experiments of phenylalanine binding, the same buffer as that used for gel filtration experiments was used; both GTP cyclohydrolase I and GFRP concentrations used were 20 μM, and phenylalanine concentrations used were in the range of 25–400 μM. The experiments were initiated by adding 100 μL of protein solution to one chamber and 100 μL of a solution without protein to another. Chambers were sealed with bolts. An air bubble was enclosed in each chamber to facilitate mixing. The cell was agitated on a rocking platform at a speed of 100 rpm. The cell was incubated for 2 or 4 h at room temperature to reach equilibrium for phenylalanine and BH4; respectively. Ligand and protein concentrations were then determined from aliquots of each chamber in the cell. Protein was not detected from aliquots from the chamber to which protein was not added.
Protein concentration was determined by performing Superdex 75 gel filtration as described above. Aliquots were directly injected into the column equilibrated in the buffer described above without ligand, and the eluate was monitored at 280 nm.
BH4 concentration was determined by the method of Fukushima and Nixon (1980) with modification. Briefly, 25 μL of the solution was mixed with 250 μL of 0.2% (w/v) iodine and 0.4% (w/v) potassium iodide in 0.1 N HCl. The mixture was incubated for 1 h at room temperature and then mixed with 25 μL of 2% ascorbic acid. After centrifugation, 50 μL of the supernatant solution was applied to a Partisil 10 ODS-1 column (4.6 × 250 mm) (Whatman Inc. Clifton, NJ) fitted with a Nova-Pak C18 guard column (Waters Corporation, Milford, MA). Biopterin was eluted isocratically with a solvent of 50 mM sodium acetate buffer (pH 5) containing 0.1 mM EDTA and 2% methanol at a flow rate of 0.8 mL/min and detected fluorometrically by a Waters 474 Fluorescence Detector (excitation, 350 nm; emission, 440 nm).
Phenylalanine concentration was also determined by a HPLC method (Allen et al. 1999) modified as follows. Aliquots were mixed with an equal volume of 12% perchloric acid. After centrifugation, the supernatant solution was applied to a Partisil 10 ODS-1 column (4.6 × 250 mm) (Whatman Inc. Clifton, NJ) fitted with a Nova-Pak C18 guard column (Waters Corporation, Milford, MA). The mobile phase consisted of 15 mM H3PO4/20% methanol pumped through at a flow rate of 1 mL/min. The column eluate was monitored fluorometrically by a Waters 474 Fluorescence Detector (excitation, 215 nm; emission, 283 nm).
All data acquisitions were done using the Shimadzu Class VP chromatography data system version 4.2. From the peak areas obtained, the molar amounts of protein and ligand were determined in reference to the values obtained from the corresponding standard solutions of protein and ligand that were dissolved in the same buffer used in the experiments and treated as the same way described above. A linear relationship was obtained for each of protein and ligands between the peak areas and the amounts used.
The concentration of free ligand ([L]) was the ligand concentration that was determined after incubation from the solution contained in the chamber without protein. The concentration of bound ligand was calculated from the difference in ligand concentration between the two chambers. The average number of ligand molecules bound per subunit molecule of protein (B) was calculated from the concentration of bound ligand and the concentration of protein determined from the solution recovered from the chambers. The data were plotted as B/[L] versus B as recommended by Scatchard (Scatchard 1949). Data are the average of triplicates.
Acknowledgments
This work was supported by National Institutes of Health Research Grant DK51257. We thank Drs. Stephen L. Phillips and John M. Brewer for critical review of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
BH4, 6R-l-erythro-5,6,7,8-tetrahydrobiopterin
GFRP, GTP cyclohydrolase I feedback regulatory protein
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.38501.
References
- Ackers, G.K. 1973. Studies of protein ligand binding by gel permeation techniques. Methods Enzymol. 27 441–455. [DOI] [PubMed] [Google Scholar]
- Allen, K.R., Deggs, T.J., Rushworth, P.A., Smith, M., and Henderson, M.J. 1999. Measurement of phenylalanine and tyrosine in plasma by high-performance liquid chromatography using the inherent fluorescence of aromatic amino acid. Ann. Clin. Biochem. 36 207–211. [DOI] [PubMed] [Google Scholar]
- Dawson, R.M.C., Elliot, D.C., Elliot, W.H., and Jones, K.M. 1986. Data for biochemical research, 3rd ed. Oxford University Press, New York.
- Fukushima, K. and Nixon, J.C. 1980. Analysis of reduced forms of biopterin in biological tissues and fluids. Anal. Biochem. 102 176–188. [DOI] [PubMed] [Google Scholar]
- Harada, T., Kagamiyama, H., and Hatakeyama, K. 1993. Feedback regulation mechanisms for the control of GTP cyclohydrolase I activity. Science 260 1507–1510. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, K., Harada, T., Suzuki, S., Watanabe, K., and Kagamiyama, H. 1989. Purification and characterization of rat liver GTP cyclohydrolase I. J. Biol. Chem. 264 21660–21664. [PubMed] [Google Scholar]
- Hatakeyama, K., Inoue, Y., Harada, T., and Kagamiyama, K. 1991. Cloning and sequencing of cDNA encoding rat GTP cyclohydrolase I. J. Biol. Chem. 266 765–769. [PubMed] [Google Scholar]
- Hummel, J.P. and Dreyer, W.J. 1962. Measurement of protein-binding phenomena by gel filtration. Biochem. Biophys. Acta 63 530–532. [DOI] [PubMed] [Google Scholar]
- Kaufman, S. 1993. New tetrahydrobiopterin-dependent systems. Annu. Rev. Nutr. 13 261–286. [DOI] [PubMed] [Google Scholar]
- Milstien, S., Jaffe, H., Kowlessur, D., and Bonner, T.I. 1996. Purification and cloning of the GTP cyclohydrolase I feedback regulatory protein, GFRP. J. Biol. Chem. 271 19743–19751. [DOI] [PubMed] [Google Scholar]
- Nar, H., Huber, R., Auerbach, G., Fisher, M., Hosl, C., Ritz, H., Bracher, A., Meining, W., Eberhardt, S., and Bacher, A. 1995. Active site topology and reaction mechanism of GTP cyclohydrolase I. Proc. Natl. Acad. Sci. 92 12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol, C.A., Smith, G.K., and Duch, D.S. 1985. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu. Rev. Biochem. 54 729–764. [DOI] [PubMed] [Google Scholar]
- Scatchard, G. 1949. The attractions of proteins for small molecules and ions. Ann. N.Y. Acad. Sci. 51 660–672. [Google Scholar]
- Yoneyama, T. and Hatakeyama, K. 1998. Decameric GTP cyclohydrolase I forms complexes with two pentameric GTP cyclohydrolase I feedback regulatory proteins in the presence of phenylalanine or of a combination of tetrahydrobiopterin and GTP. J. Biol. Chem. 273 20102–20108. [DOI] [PubMed] [Google Scholar]
- Yoneyama, T., Brewer, J.M., and Hatakeyama, K. 1997. GTP cyclohydrolase I feedback regulatory protein is a pentamer of identical subunits. J. Biol. Chem. 272 9690–9696. [DOI] [PubMed] [Google Scholar]