Abstract
Nonspecific lipid transfer protein from wheat is studied by liquid-state NMR in the presence of xenon. The gas–protein interaction is indicated by the dependence of the protein proton chemical shifts on the xenon pressure and formally confirmed by the first observation of magnetization transfer from laser-polarized xenon to the protein protons. Twenty-six heteronuclear nOes have allowed the characterization of four interaction sites inside the wheat ns-LTP cavity. Their locations are in agreement with the variations of the chemical shifts under xenon pressure and with solvation simulations. The richness of the information obtained by the noble gas with a nuclear polarization multiplied by ∼12,000 makes this approach based on dipolar cross-relaxation with laser-polarized xenon promising for probing protein hydrophobic pockets at ambient pressure.
Keywords: Laser-polarized xenon, SPINOE, wheat nonspecific lipid transfer protein, protein hydrophobic cavity
The catalytic sites of many enzymes are located in hydrophobic pockets. The content of these cavities in the absence of substrate is still subject to debate because the presence of water molecules may play a fundamental role in the thermodynamics of the enzymatic activity (Ernst et al. 1995; Matthews et al. 1995; Quillin et al. 2000). Currently, only two physical methods allow direct characterization of these hydrophobic cavities at the atomic level: i) X-ray diffraction of a protein crystal under noble gas pressure (xenon, krypton,. . .; Montet et al. 1997; Prangé et al. 1998; Quillin et al. 2000). The disadvantages of this method are that medium-to-high pressures must be used, and the dynamic properties of the interaction cannot easily be understood; ii) hydration studies using liquid-state 1H NMR, which involve solvent–protein protons cross-relaxation. These measurements can be delicate because exchange between water and hydroxyl protons can mask the through-space interactions between the solvent and the protein protons (Otting et al. 1991). A variation of this method is the use of small organic molecules that can probe cavities. Then the dependence of the internuclear distances on the intermolecular cross-relaxation rates between the protons of the protein and of the organic compound can be exploited (Otting et al. 1997). However, as these nOe signals are usually proportional to the concentration of the small organic compounds (fast-exchange conditions), improving the sensitivity of this approach requires very high pressures (e.g., 200 bars of methane). This may induce structural modifications of the protein (Prangé et al. 1998) or its unfolding (Inoue et al. 1998), hence limiting the application of this method to only highly stable proteins.
For a few years it appears that xenon could be a good candidate to replace these small organic molecules in liquid-state NMR. According to Wolfenden and Radzicka (1994), because the partial pressure of xenon in the vapor phase is larger than that of water, xenon occupancy in apolar cavities of a protein is expected to be higher. The most attractive feature of this noble gas is that its nuclear spin can be polarized by optical pumping, which enables huge signal enhancements (polarization multiplied by 104 relative to the thermal polarization at high field and room temperature). In fact, since the first detection of cross-relaxation between laser-polarized xenon and solvent protons of benzene, it has been suggested that protein hydrophobic cavities could be characterized based on the so-called SPINOE experiment (Navon et al. 1996). Depending on the xenon magnetization, the intermolecular nOe signals would then become observable without needing to increase the xenon pressure. However, because the magnetization of laser-polarized xenon is far from thermal equilibrium, the signal decreases in a relatively short time (tenths to hundreds of seconds), making the cross-relaxation between 129Xe and nuclei of a solute molecule difficult to detect. Thus, until now, direct observation of 129Xe–1H nOes was reported only on medium-sized cage-molecules (Song et al. 1997; Lühmer et al. 1999; Desvaux et al. 2000).
In the case of proteins, interactions involving xenon were detected, but always indirectly through the influence of the protein concentration on the xenon magnetization decay (Bowers et al. 1999; Stith et al. 1999; Rubin et al. 2000). In this article, we have extended our original experiment, in which we detected heteronuclear cross-relaxation between laser-polarized xenon and protons of a cage-molecule in water (Desvaux et al. 2000), to the first direct characterization of cavities of a protein, the ns-LTP from wheat. This 90-amino acid protein, which belongs to the ubiquitous ns-LTP family that is present in the plant kingdom (Kader 1996), is able to transfer amphiphilic molecules of various sizes between membranes in vitro. The protein functions in vivo, which are not yet precisely understood, seem to be related to the cuticle formation (Sterk et al. 1991) and/or to the plant defense against microbial agents (Terras et al. 1992). The common structural fold of the lipid transfer protein is stabilized by four disulfide bridges and contains four α-helices packed against a C-terminal arm (Gincel et al. 1994; Shin et al. 1995; Gomar et al. 1996; Heinemann et al. 1996; Lee et al. 1998; Poznanski et al. 1999). This scaffold exhibits a large hydrophobic cavity with a volume of ∼400 Å3 for wheat ns-LTP (Gincel et al. 1994). When the aliphatic part of a lipid is located in the hydrophobic cavity, the volume of this cavity increases up to 750 Å3 (Sodano et al. 1997; Tassin-Moindrot et al. 1999), and adopts a tunnel-like shape (Shin et al. 1995; Gomar et al. 1996; Lerche et al. 1997; Sodano et al. 1997; Lerche and Poulsen 1998; Charvolin et al. 1999; Tassin-Moindrot et al. 1999). At its entrance, defined by the aromatic ring of the amino acid Tyr 79, the protein–lipid complex can be stabilized by interactions between the polar head group of the lipid, the protein surface, and the solvent. Hydrophobic interactions between amino acids of the protein pocket and the amphiphilic molecule are difficult to characterize by 1H NMR because of the spectral overlap that impedes the quantitative use of the intermolecular nOes. Generally, only the polar head group can be located relative to the protein (Tassin-Moindrot et al. 1999). Interactions between small probe molecules and the protons of the hydrophobic cavity can be detected either using high pressures (methane or ethane) or with saturated solutions (benzene, cyclohexane, or cyclopropane), but it is hardly possible to define their specific location inside the cavity (Liepinsh et al. 1999). Here, we present a new method in which xenon atoms bind selectively at ambient pressure to four sites of the wheat ns-LTP cavity. These sites are characterized by solvation simulations, the observation of chemical shift variations under xenon pressure, and the exploitation of laser-polarized xenon–proton dipolar cross-relaxation.
Results
Solvation simulations of wheat ns-LTP by xenon
The capability of the hydrophobic cavity to incorporate xenon atoms without large modifications of its overall structure is explored by simulating solvation of the protein by xenon. This procedure consists in a grid search with van der Waals exclusions; the protein coordinates are kept fixed. Because of the large van der Waals diameter of xenon (4.5 Å) and the lack of precision of the location of the amino acid side chains, this simulation is performed on the 15 wheat ns-LTP NMR structures deposited at the PDB. Between one and five xenon atoms (average number 2.9, root mean square deviation = 1.2) are found inside the cavity (see Fig. 1 ▶ for two representative examples). Xenon is preferentially located on two sites (labeled A and A′) that are deeply embedded in the protein pocket. No lipidic group had been detected in these areas on the wheat ns-LTP–lipids complexes (Sodano et al. 1997; Charvolin et al. 1999) except prostaglandin B2, which is found in site A′ and 3 Å distant from site A (Tassin-Moindrot et al. 1999). On average, two xenon atoms occupy sites around these positions (the minimum number of sites is 1, the maximum is 4). On the structure of Fig 1b ▶, two other sites labeled B and C are observed. Statistically they are occupied in 46% of the 15 NMR structures. Site B is enclosed in the tunnel-like cavity that appears when large lipids are bound in the swelled structures of ns-LTP. Site C is the closest to the entrance of the protein pocket. It is in the proximity of the Tyr 79 aromatic group, which closes the cavity in the absence of lipid and undergoes a rotation of ∼100° when ns-LTP binds amphiphilic molecules (Shin et al. 1995; Gomar et al. 1996; Lerche et al. 1997; Sodano et al. 1997; Lerche and Poulsen 1998; Charvolin et al. 1999; Tassin-Moindrot et al. 1999).
Fig. 1.
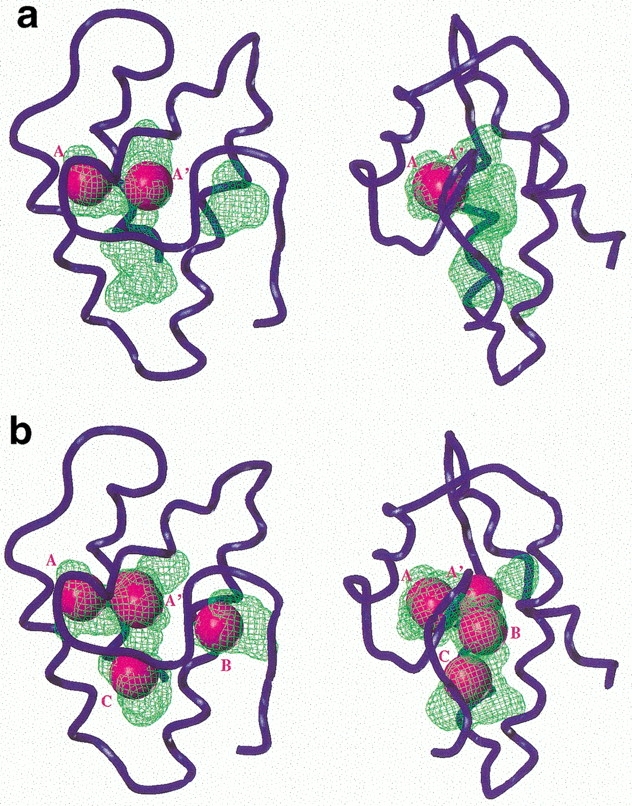
Two representative examples of the solvation simulation of the wheat ns-LTP by xenon. Orthographic views of PDB code 1GH1 model 4 (a) and 2 (b) calculated and represented using the SYBYL software (the same point of view is used for all figures). The protein backbone is shown as a blue tube. The cavity, calculated with the home-written program CAVITE (Gomar et al. 1998), is represented as green volumes. The location of the xenon atoms found by SYBYL are represented by spacefill atoms colored in magenta and named A, A′ B, and C.
Analysis of the chemical shifts variation
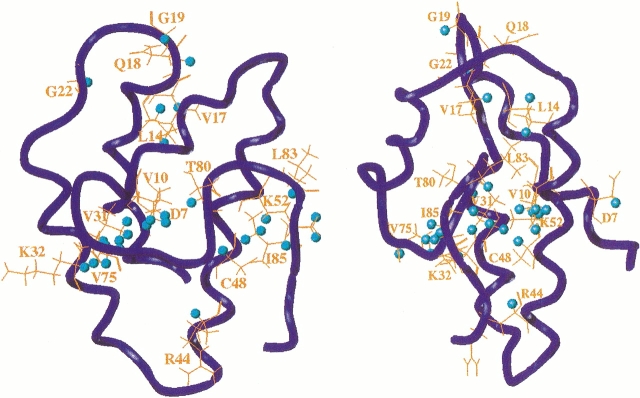
An interaction between xenon and wheat ns-LTP is clearly revealed by monitoring the 390 chemical shifts of the nonexchangeable protons as a function of xenon pressure. Indeed even with a pressure of 1 bar, which represents 4.8 mM of dissolved xenon in pure water at 293°K (Clever 1979), 2 proton chemical shifts (Hα of Val 31 and Hα of Thr 80) vary by >0.02 ppm. When the pressure reaches 5 bars, 22 variations >0.02 ppm (maximum shift = 0.17 ppm for Hα of Val 31) are detected. They correspond to 16 amino acids (see Fig. 2 ▶ and Supplemental Material), eight of which are located inside the ns-LTP cavity. They are mainly hydrophobic residues (Val 10, Leu 14, Val 17, Val 31, Val 75, Leu 83, and Ile 85), the exception being Thr 80. The other amino acids are more polar and are either scattered on the surface of the protein (Asp 7, Gln 18, Gly 19, Gly 22, Lys 32, Cys 48, and Lys 52) or located at the entrance of the cavity (Arg 44). As a comparison, when a nitrogen pressure of 5 bars is used, the variations of the proton chemical shifts are reduced. Only Hα of Val 31 and Hα of Thr 80 present variations >0.02 ppm. This indicates that the structural modifications that are induced by the applied pressure cannot explain all the observed shifts in the case of xenon and that at least some of the observed changes result from an interaction between the protein and the xenon.
Fig. 2.
Representation of the amino acids for which variation of the proton chemical shifts >0.02 ppm is observed under a xenon pressure of 5 bars (in orange, with the nuclei in cyan). The protein backbone is represented by the blue tube (drawn with SYBYL).
The increase of the chemical shift variations with xenon pressure indicates that interaction sites are not fully occupied at 1 bar of xenon pressure. Attempts to estimate the occupancy by comparing the shifts at 1 bar and 5 bars were unsuccessful because the observed variations at 5 bars are compatible at our level of precision, with five times the observed shifts at 1 bar. The deduced occupancy at 1 bar is then lower than ∼10%. Using higher xenon pressures to evaluate the binding constants is unreliable because this could induce small conformational modifications of the protein, which in the case of ns-LTP is known to be a problem because it is sufficiently flexible to adapt its shape to the size of the ligand.
Cross-relaxation between protons of ns-LTP and laser-polarized xenon
Almost independent of temperature or pressure, only one peak for xenon in an aqueous solution of ns-LTP is observed (189 ppm at 1 bar and 293K, reference at 0 ppm for the gas phase). Xenon is consequently in fast exchange on the NMR time scale between its bound and free states. The best experimental conditions are thus defined by the properties of the SPINOE experiment (Lühmer et al. 1999; Desvaux et al., 2000, Berthault et al. 2001). Formally, in conditions of fast exchange and neglecting cross-correlation phenomena, a fine theoretical description of this experiment consists of expressing the generalized Solomon equations (Solomon 1955) in terms of the global xenon magnetization MS and of the proton magnetization MIk (see Desvaux et al. 2000 for an analytical description in the case of a spin system composed of n protons). The solution of this system when considering a SPINOE experiment (MIk[τm = 0] = 0, difference ΔMIk[τm] of two proton spectra, one acquired with MS and one with −MS) reveals that the best detection of the proton signal enhancement, ΔMIk/MIk, requires the maximization of MS/MIk. In the case of wheat ns-LTP, improving MS by increasing the xenon concentration is not reliable. For xenon pressures greater than ∼1.5 bar, the large amount of foam, which appears above the solution, reduces the magnetic field homogeneity and impedes reliable peak assignment. Consequently, the SPINOE experiment is performed with ∼1 bar of laser-polarized xenon and the MS/MIk ratio is increased by lowering the protein concentration to 0.8 mM.
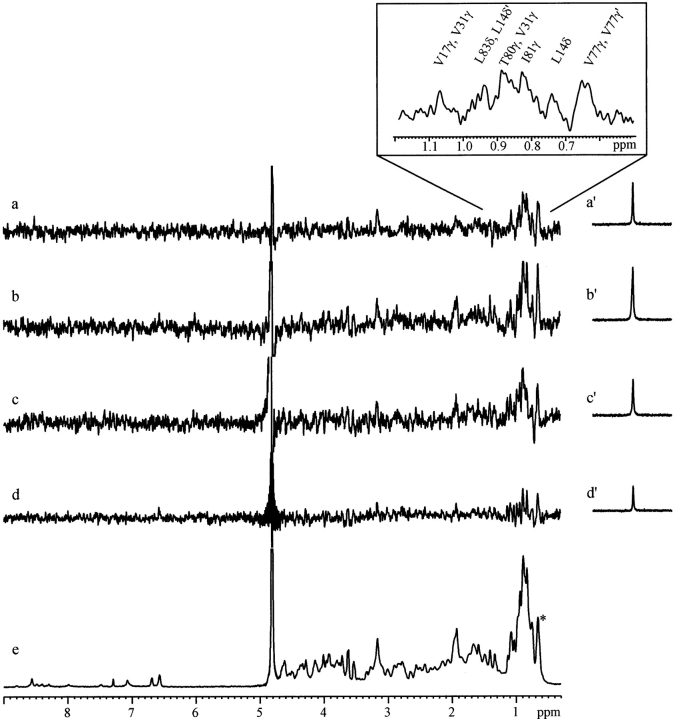
Even if the analysis of the wheat ns-LTP chemical shift variations under xenon pressure already indicates an interaction, the capability of detecting dipolar cross-relaxation between laser-polarized xenon and particular protons of the cavity is a definite proof of a regioselective binding (Fig. 3 ▶). Characterization of the interaction sites requires the transformation of dipolar cross-relaxation peaks into xenon–proton distance restraints. From all the SPINOE spectra, corresponding to seven mixing times that ranged between 100 ms and 800 ms, we have mainly used the spectra acquired at τm = 200 ms for peak assignment. For this value, the proton magnetization has already built up from zero, and the influence of the proton spin diffusion because of efficient homonuclear cross-relaxation is still minor, thus allowing qualitative characterization. Longer mixing times are used to detect remote heteronuclear xenon–proton cross-relaxation, possibly enhanced by proton–proton spin diffusion, and to characterize unambiguously the involved amino acids (e.g., by detecting the corresponding Hα proton). In addition, detection of sites associated with small transfers, for instance because of a low occupancy, can be achieved.
Fig. 3.
(Left) 129Xe → 1H SPINOE spectra of wheat ns-LTP acquired at 600 MHz in D2O for different mixing times (a, 200ms; b, 400ms; c, 600ms; and d 800 ms). On the right (a′–d′), the corresponding 129Xe spectra observed after a small angle flip pulse. The variation of the xenon intensity and the associated proton signal enhancement result from the variable number of spectra added. (e) proton NMR spectrum of the protein. The * indicates γCH3 of Val 75. In insert, subspectrum of (a) with the assignment of unambiguous peaks of this region.
Twenty-six interactions between protons of ns-LTP and laser-polarized xenon are assigned, 13 of which having been already detected with mixing times <200 ms. These 26 interactions, which mainly involve methyl groups, correspond to 16 hydrophobic amino acids all located inside the protein cavity (see Supplemental Material). At the end of the assignment procedure, only three peaks (1.93 ppm, 3.16 ppm, and 3.25 ppm) could not be assigned because of spectral overlap.
Interaction sites of xenon with ns-LTP
On the SPINOE spectra of Figure 3 ▶, the 1H signal enhancements of Val 75 methyl groups because of polarization transfer from xenon range between 1% and 3%. For example, for a mixing time of 200 ms, the enhancement is equal to 2%. In contrast to the study of cross-relaxation between α-cyclodextrin protons and laser-polarized xenon in water (Desvaux et al. 2000), the proton signal enhancement, scaled by the xenon magnetization, is an increasing function of time only for short mixing times (τm < 300 ms) and presents large systematic errors for longer mixing times. This, together with efficient proton–proton spindiffusion phenomena and spectral overcrowding, prevents accurate peak integration. Considering this and the fact that the dynamics of xenon inside the cavity is unknown, we use large distance intervals as constraints during the molecular modeling, while the protein structure is kept rigid.
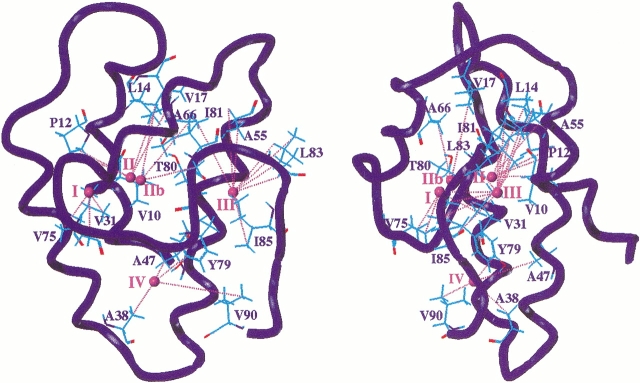
At the end of the molecular modeling procedure, the 16 amino acids having protons in through-space interaction with xenon are clusterized into five areas of the protein cavity (Fig. 4 ▶). The first site (I), which is defined by 4 nOes observed at short mixing time, is in the vicinity of Val 31 and Val 75. The second (II) and third (IIb) sites, which are very close each other, are defined by 5and 3-nOe data, respectively (4 nOe and 2 nOe for short mixing times). They correspond to interactions with Val 10, Pro 12, Leu 14, Val 17, Ala 66, and Thr 80. Because the distance between xenon II and IIb is very short (3.6 Å), a good description seems to be that a unique xenon is in very fast exchange between sites II and IIb (considering only one xenon leads to distance-restraints violation). The fourth site (III) is defined by 10 nOes (3 nOes at short mixing time) and involves Ala 55, Ile 81, Leu 83, and Ile 85. The last site (IV) is close to the entrance of the cavity because four interactions with Ala 38, Ala 47, Tyr 79, and Val 90 are detected. For this last site, heteronuclear nOes can be detected only for long mixing times, but the xenon–proton distances are consistent with the values found for the other sites. This difference in behavior can be explained either by a difference of correlation times between xenon and protons or by a site occupancy that is lower than for the other sites.
Fig. 4.
Representation of the interaction sites of xenon with wheat ns-LTP as determined by the SPINOE experiments. On the protein backbone (blue tube) is superimposed a representation of residues for which polarization transfer between laser-polarized xenon and protons is observed. Xenon atoms are shown as small magenta spheres that are numbered accordingly to the site number I, II, IIb, III, and IV. The distance restraints used to build the model are displayed as magenta dashed lines. The longest distance restraint in model 2 is the one involving γCH3 of Val90 (8.1 Å), but the same distance = 4 Å using model 6.
Discussion
Comparison of the interaction sites
The comparison of the xenon sites in wheat ns-LTP determined by the solvation simulation, the analysis of proton chemical shifts under xenon pressure, and the SPINOE approach reveal similarities. i) Eight of the amino acids that were detected in SPINOE experiments exhibit variations of their proton chemical shifts >0.02 ppm under 5 bars of xenon. For the other eight amino acids, except Ala 38 and Pro 12 (for which accurate measurement is impossible), variations >0.01 ppm are observed. ii) The xenon sites found with the data derived from the SPINOE experiments are in excellent agreement with those detected by the solvation simulations. The two sites A and A′ found in all the 15 structures coincide with sites I and (II, IIb), respectively. The short distances between sites I and II, and I and IIb (6.2 Å and 4.7 Å, respectively), associated with the expected large dynamics of xenon in the cavity, imply that possibly only one xenon atom explores all three sites. A similar behavior has already been reported by X-ray crystallography studies (Quillin et al. 2000). Moreover, sites B and C, which are not always found by the solvation simulation, are nevertheless observed by the SPINOE approach (site III and IV, respectively).
These results prove that the xenon is able to bind inside the protein cavity without requiring large distortions of the ns-LTP structure. However, the observation of only one NMR signal for each spin species, and the partial sites occupancies deduced from the 1H chemical shift analysis under xenon pressure, are clear indications of complex dynamics, which may affect the protein on a large time domain as already observed on a mutant of lysozyme T4 (Mulder et al. 2000). Resorting to laser-polarized xenon proton cross-relaxation in combination with the analysis of the protein dynamics by classical relaxation approach would help to understand the mechanisms that are responsible for the binding. The study of the competitive complexation of the protein by a lipid and xenon would be interesting, but it is expected to be complicated by the fast relaxation of laser-polarized xenon with the aliphatic part of the lipid.
The behavior of xenon inside the cavity of this protein differs from those of small organic molecules of similar size (Liepinsh et al. 1999). Indeed, the only evidence of unambiguous interactions when using methane, ethane, or cyclopropane under pressures ranging from 5 bars to 140 bars, were with Leu 77 and Tyr 79 located at the entrance of the cavity. For larger molecules, such as cyclohexane or benzene, and when using saturated solutions at ambient pressure, intermolecular nOes have been observed with protons inside the cavity or at the surface. In particular for benzene, which exhibited the largest number of nOes, interactions with most of the hydrophobic protons of the cavity were observed, indicating no specific and preferential binding, in contrast to xenon, which seems to be located in well-defined sites. This tendency is corroborated by results obtained using X-ray crystallography under noble gas pressure (Quillin et al. 2000) and molecular simulation (Mann and Hermans 2000). It seems to arise from the large polarizability of xenon, which increases the binding energy. Moreover the cost in entropy is lower than for a nonspherical organic compound.
Conclusions
In this article we have shown for the first time that a protein cavity can be probed using magnetization transfer from laser-polarized xenon. For wheat ns-LTP, the detection of unambiguous local interactions between protein protons and xenon has allowed the identification of xenon atoms at sites that are in remarkable agreement with the positions derived by solvation simulations and by using the analysis of proton chemical shift variations under xenon pressure. Interestingly, these sites exhibit similar features as those found in other proteins by X-ray diffraction under xenon pressure (Prangé et al. 1998), that is, regions rich in methyl groups. Amino acids at the surface of the protein, which present chemical shift variations in the presence of xenon, are not detected by the SPINOE approach. This indicates that either they result from small local structural modifications or that the residence time of xenon is too short to allow efficient heteronuclear cross-relaxation (Lühmer et al. 1999; Berthault et al. 2001). When compared to small organic molecules, xenon appears to be involved in more specific interactions. In addition, the use of ambient pressure, made possible by resorting to laser-polarized gas, brushes aside the obstacles associated with pressure-induced structural modifications. All these results lead us to believe that the use of laser-polarized xenon for characterizing protein hydrophobic cavities is a promising technique. However, it is important to keep in mind that the success of this method requires strong xenon magnetization (polarization >5%, pressure above 1 bar, or isotope-enriched gas; for discussion, see Desvaux et al. 2000). The present example of a protein in water is even more difficult than previous demonstrations using small cage molecules because the assignment of the 1D spectrum is difficult to achieve and may become completely impossible on larger protein, and the increase of the dipolar correlation times on complexation of the Xe in the cavity favors simultaneous self-relaxation of the proton and xenon magnetization to the detriment of heteronuclear cross-relaxation (Berthault et al. 2001). In this study, this increase, even in buried sites such as sites I and II, was not as dramatic as it could have been considering that the xenon–proton interactions are still observable.
The use of this approach based on the SPINOE for other proteins, for instance, to check the consistency with results obtained by X-ray diffraction under xenon pressure, requires a very careful choice of the experimental conditions (temperature, gas pressure, concentration of the protein) in the present stage of technical developments. In the future, higher xenon polarization associated with larger available quantities of laser-polarized gas may open the way to the detection of interactions with protein surfaces. Working with 'pseudo' steady-state xenon magnetization via the use of a system delivering laser-polarized xenon in continuous flow (Seydoux et al. 1999) would make the assignment easier and the increase of xenon self-relaxation relative to heteronuclear cross-relaxation less problematic. More precise distance restraints or dynamic investigation of the complex would then become accessible, and results in nondeuterated water (H2O) may be reachable. These future developments will become of high value for interpreting the role of hydrophobic pockets in biology, particularly in relation to their properties of adaptation and dynamics (Mulder et al. 2000; Quillin et al. 2000).
Materials and methods
Materials
To reduce the influence of the 1H water magnetization on the laser-polarized xenon magnetization, the protein was lyophilized in D2O several times and subsequently dissolved in D2O (99.97% from Eurisotop) to obtain a final concentration of 0.8 mM inside medium-sized, walled NMR tubes from Wilmad closed by J. Young valves (p2H = 5.4). Before each experiment these 5-mm tubes were carefully degassed by at least three freeze–pump–thaw cycles. The desired amount of gas (xenon, nitrogen, or helium) was then added above the solution. Xenon enriched in isotope 129 (96% from Eurisotop) was used for the SPINOE experiments.
NMR
NMR experiments were acquired on a Bruker DRX 600 spectrometer equipped with a three-axis gradients TBI probehead (1H, 15N, broadband). The 129Xe Larmor frequency was 166 MHz. Interactions between xenon and wheat ns-LTP and polarization transfer between laser-polarized xenon and protein protons were observed in the temperature interval 275K–308K. However, the results reported herein were acquired at 293K because it allowed us to obtain narrow NMR lines by minimizing temperature gradients during the SPINOE experiments. The proton peak assignment at 293K was deduced from the assignment previously published (308K; Simorre et al. 1991) on the basis of 2D TOCSY (Braunschweiler and Ernst 1983) and NOESY (Jeener et al. 1979) experiments, using the XEASY software (Bartels et al. 1995).
Study of the 1H chemical shift variations
Experimental evidence of interactions between wheat ns-LTP and xenon were obtained by following the 1H chemical shifts determined by 2D TOCSY experiments (matrices composed of 420 × 4096 real points, resolution in the direct dimension without zero filling of 2.8 Hz per point, i.e., 0.005 ppm) as a function of gas pressure using the XEASY software (Bartels et al. 1995). However, because even a moderate pressure can alter the protein structure, a reference gas was needed. During preliminary experiments, helium and nitrogen were considered and pressures up to 20 bars were explored. We finally decided to resort to nitrogen as a reference because its solubility in water is higher than that of helium (Clever 1979), and its pressure in the NMR tube could be more carefully defined because, in contrast to helium, it cannot diffuse through Teflon valves. Finally, the maximal pressure was reduced to 5 bars, which enabled the safe use of medium-sized walled NMR tubes instead of thick-walled tubes, thus leading to better resolution and signal-to-noise ratio. The same samples (D2O as solvent) as those used for SPINOE were used, so no exchangeable protons were detected. Because of the lower sensitivity of nonexchangeable protons to their environment (particularly methyl groups) the observed shifts were smaller than those observed for HN protons (Moindrot 2000), but presented a higher degree of confidence. Variations of chemical shifts are given relative to the degassed solution.
NMR experiments with laser-polarized xenon
In our experimental setup, which is described in detail in Desvaux et al. (2000), xenon was polarized by optical pumping through the spin-exchange method (Herman 1965; Walker and Happer 1997); that is, the intermediate step was the polarization of the electronic spins of an alkali metal. A cell heated at 368°K, that contained a mixture of xenon and nitrogen (1 : 3 for a mean pressure of 210 mbar) and a few droplets of rubidium was irradiated by a titanium : sapphire laser beam at a wavelength of 794.7 nm. After ∼10 mn of optical pumping, the polarized xenon was condensed and separated from nitrogen. Frozen laser-polarized xenon was displaced from the optical pumping room to the fringe field of an NMR superconducting magnet. Here it was introduced into an NMR tube by condensation using a home-built extruded rotor that was filled with liquid nitrogen. This tube contained the liquid protein solution. A pressure of ∼1 bar per pumping cycle and an average useful polarization for the SPINOE experiment of ∼16% were obtained.
The detection of polarization transfer between the laser-polarized xenon and the protein protons was obtained by the SPINOE pulse sequence (Song et al. 1997) modified to reach a level of stability better than 0.1% (Desvaux et al. 2000; Berthault et al. 2001), thus >10 times lower than ΔMIk/MIk. This was made possible because magnetic susceptibility shifts caused by the presence of the polarized gas were taken into account by applying two inversion pulses on the xenon magnetization every other FID. Radiation-damping effects of the xenon magnetization were suppressed by using a CHIRP pulse (Böhlen et al. 1989) combined with a pulsed-field gradient as inversion pulse (Berthault et al. 1999). The initial conditions of the proton spin system corresponded to complete saturation obtained by a series of 32-proton, 90° hard pulses, each followed by a pulsed-field gradient of random amplitude. The recycling delay (100 ms + 450 ms of acquisition) was then reduced as much as possible. Obviously, considering that the self-relaxation time of the dissolved xenon was relatively short (∼2 mn), the ideal number of accumulations of the SPINOE sequence to obtain the best signal-to-noise ratio depended on the mixing time τm and the initial xenon magnetization. Consequently, all spectra were stored separately and were added in the processing, thus defining the optimal number, which finally ranged between 16 and 64 accumulations. Before each SPINOE experiment, the NMR tube was vigorously shaken to refresh dissolved laser-polarized xenon by exchanging it with the gaseous reservoir. About 30 s after reinsertion of the tube into the magnet, the xenon magnetization was monitored by a small read pulse (∼2°), and the SPINOE experiment, which started by four dummy scans, was acquired. In each run, corresponding to the result of one optical pumping cycle, an average of 15 different SPINOE experiments (representing about 45 min of experiment time) could be acquired before the xenon signal became too small, and no polarization transfer could be safely detected. The SPINOE spectra of Fig. 3 ▶ corresponds to a composition of six different runs, all of which were acquired with positive xenon-spin temperature. Because of the xenon magnetization decay during the experiment, and, hence, the loss of proton signal, collection of 2D SPINOE spectra were prohibited. Consequently, the peak assignment of the 1D SPINOE spectra was obtained by comparison with 2D TOCSY experiments that were acquired under exactly the same conditions (temperature, sample, spectrometer, and pressure of xenon). This assignment procedure was obviously difficult because of spectral overlap, low signal-to-noise ratio, and possible artifacts. However, the measurement of a large number of spectra at different mixing times reduces the risk of a misinterpretation and helps the assignment.
Solvation simulation and molecular modeling
Molecular modeling was conducted with the SYBYL 6.5 package (TRIPOS Associate Inc.) on SGI workstations, using the 15 refined solution structures of wheat ns-LTP deposited at the PDB (code 1GH1).
The solvation simulation was performed using the xenon parameters defined in SYBYL, considering two layers of solvent around the protein which was kept rigid during all the procedure. The program tried to locate inside and around the protein the largest number of xenon atoms considered as rigid spheres whose diameter is the van der Waals one. The atoms that were external to the cavity were removed.
To locate the xenon interaction sites from the results of the SPINOE experiments, we started from a solution structure (code 1GH1, model 2) that was kept rigid in the following steps, and we added the xenon atoms. The 26 SPINOE cross-peaks were converted into distance restraints (2.3 Å < r < 7.5 Å) using double weighting for the 13 restraints observed at short mixing times with a high signal-to-noise ratio. When methyl groups were involved, pseudoatoms located at the carbon coordinates were used (explaining the upper range of distance restraints). The locations of xenon were finally obtained by minimization in the TRIPOS force field using the 26 distances restraints. We confirmed consistency of the results by simultaneously minimizing the protein structure and the xenon locations without experimental restraints and observing the stability of the formed complex, by considering other 1GH1 models and observing similar binding sites (they are relatively well-defined by nOes in various directions), and by searching for missing nOes. After checking, all protons close to xenon sites (distances <6 Å) exhibit nOes, even if they were not previously detected because of spectral overlap or low signal-to-noise ratio. For example, for the expected nOes with methyl groups of Leu 51 and Leu 77, ambiguous peaks were observed in the SPINOE spectra at the associated chemical shifts.
Acknowledgments
We greatly acknowledge S. Moindrot for her involvement in the first steps of this work. We thank L. Bull, J.C. Gabriel, and D. Sy for helpful comments. The ns-LTP samples were provided by D. Marion (INRA Nantes).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
FID, Free Induction Decay
Ns-LTP, nonspecific lipid transfer protein
nOe, nuclear Overhauser effect
NOESY, nuclear Overhauser effect spectroscopy
TOCSY, total correlation spectroscopy
SPINOE, spin polarization-induced nuclear Overhauser effect
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.47001.
Supplemental material: See www.proteinscience.org.
References
- Bartels, C., Xia, T.E., Billeter, M., Güntert, P., and Wüthrich, K. 1995. The program XEASY for computer-supported NMR spectra analysis of biological macromolecules. J. Biomol. NMR 5 1–10. [DOI] [PubMed] [Google Scholar]
- Berthault, P., Desvaux, H., Le Goff, G., and Pétro, M. 1999. A simple way to properly invert intense nuclear magnetization: Application to laser-polarized xenon. Chem. Phys. Lett. 314 52–56. [Google Scholar]
- Berthault, P., Landon, C., Vovelle, F., and Desvaux, H. 2001. Première détection d'un transfert d′aimantation entre du xénon polarisé par laser et des protons d'une protéine. C. R. Acad. Sci. Paris Sér. IV, in press.
- Böhlen, J.-M., Rey, M., and Bodenhausen, G. 1989. Refocusing with chirped pulses for broadband excitation without phase dispersion. J. Magn. Reson. 84 191–197. [Google Scholar]
- Bowers, C.R., Storhaug, V., Webster, C.E., Bharatam, J., Cottone, III, A., Gianna, R., Betsey, K., and Gaffney, B.J. 1999. Exploring surfaces and cavities in lipoxygenase and other proteins by hyperpolarized xenon-129 NMR. J. Am. Chem. Soc. 121 9370–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweiler, L. and Ernst, R.R. 1983. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53 521–528. [Google Scholar]
- Charvolin, D., Douliez, J.P., Marion, D., Cohen-Addad, C., and Pebay-Peyroula, E. 1999. The crystal structure of a wheat non-specific lipid transfer protein (nsLTP1) complexed with two phospholid molecules at 2.1 Å resolution. Eur. J. Biochem. 264 562–568. [DOI] [PubMed] [Google Scholar]
- Clever, H.L. 1979. IUPAC solubility data series. Pergamon Press, Oxford, UK.
- Desvaux, H., Gautier, T., Le Goff, G., Pétro, M., and Berthault, P. 2000. Magnetization transfer between laser-polarized xenon and protons of a cage-molecule in water. Eur. Phys. J. D. 12 289–296. [Google Scholar]
- Ernst, J.A., Clubb, R.T., Zhou, H.X., Gronenborn, A.M., and Clore, G.M. 1995. Use of NMR to detect water within nonpolar protein cavities response. Science 270 1848–1849. [DOI] [PubMed] [Google Scholar]
- Gincel, E., Simorre, J.-P., Caille, A., Marion, D., Ptak, M., and Vovelle, F. 1994. Three-dimensional structure in solution of a wheat lipid transfer protein from multidimensional 1H-NMR data. Eur. J. Biochem. 226 413–422. [DOI] [PubMed] [Google Scholar]
- Gomar, J., Petit, M.-C., Sodano, P., Sy, D., Marion, D., Kader, J.-C., Vovelle, F., and Ptak, M. 1996. Solution structure and lipid binding of a non-specific lipid transfer protein extracted from maize seeds. Protein Sci. 5 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomar, J., Sodano, P., Sy, D., Shin, D.H., Lee, J.Y., Suh, S.W., Marion, D., Vovelle, F., and Ptak, M. 1998. Comparison of solution and crystal structures of maize nonspecific lipid transfer protein: A model for a potential in vivo lipid carrier protein. Proteins 31 160–171. [DOI] [PubMed] [Google Scholar]
- Heinemann, B., Vilbour, A.K., Reinholt, N.P., Molskov, B.L., and Poulsen, F.M. 1996. Structure in solution of a four-helix lipid binding protein. Protein Sci. 5 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, R.M. 1965. Theory of spin-exchange between optically pumped rubidium and foreign gas nuclei. Phys. Rev. 137A 1062–1065. [Google Scholar]
- Inoue, K., Yamada, H., Imoto, T., and Akasaka, K. 1998. High pressure NMR study of a small protein, gurmarin. J. Biomol. NMR 12 535–541. [DOI] [PubMed] [Google Scholar]
- Jeener, J., Meier, B.H., Bachmann, P., and Ernst, R.R. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71 4546–4553. [Google Scholar]
- Kader, J.-C. 1996. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. 47 627–654. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Min, K., Cha, H., Shin, D.H., Hwang, K.Y., and Suh, S.W. 1998. Rice non-specific lipid transfer protein: the 1.6 Å crystal structure in the unliganded state reveals a small hydrophobic cavity. J. Mol. Biol. 276 437–448. [DOI] [PubMed] [Google Scholar]
- Lerche, M.H., Kragelund, B.B., Belch, L.M., and Poulsen, F.M. 1997. Barley lipid-transfer protein complexed with palmitoyl CoA: The structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure 5 291–306. [DOI] [PubMed] [Google Scholar]
- Lerche, M.H. and Poulsen, F.M. 1998. Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer protein. Protein Sci. 7 2490–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepinsh, E., Sodano, P., Tassin, S., Marion, D., Vovelle, F., and Otting, G. 1999. Solvation study of the non-specific lipid transfer protein from wheat by intermolecular NOEs with water and small organic molecules. J. Biomol. NMR 15 213–225. [DOI] [PubMed] [Google Scholar]
- Lühmer, M., Goodson, B.M., Song, Y.-Q., Laws, D.D., Kaiser, L., Cyrier, M.C., and Pines, A. 1999. Study of xenon binding in cryptophane-A using laser-induced NMR polarization enhancement. J. Am. Chem. Soc. 121 3502–3512. [Google Scholar]
- Mann, G. and Hermans, J. 2000. Modeling protein-small molecule interactions: structure and thermodynamics of noble gases binding in a cavity in a mutant phage T4 lysozyme. J. Mol. Biol. 302 979–989. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W., Morton, A.G., and Dahlquist, F.W. 1995. Use of NMR to detect water within nonpolar protein cavities. Science 270 1847–1848. [DOI] [PubMed] [Google Scholar]
- Moindrot, S. 2000. Etacles structurales par RMN et moélisation moléculaire d'une protèine antimicrobienne d'origine végétale et d'un complexe entre une proteine végétale de transfert de lipides (LTP) et la prostaglandine Be. PhD thesis, University of Orléans, France.
- Montet, Y., Amara, P., Volbeda, A., Vernede, X., Hatchikian, E.C., Field, M.J., Frey, M., and Fontecilla-Camps, J.-C. 1997. Gas access to the active site of Ni-Fe hydrogenases probed by X-ray crystallography and molecular dynamics. Nature Struct. Biol. 4 523–526. [DOI] [PubMed] [Google Scholar]
- Mulder, F.A.A., Hon, B., Muhandiram, D.R., Dahlquist, F.W., and Kay, L.E. 2000. Flexibility and ligand exchange in a buried cavity mutant of T4 lysozyme studied by multinuclear NMR. Biochemistry 39 12614–12622. [DOI] [PubMed] [Google Scholar]
- Otting, G., Liepinsh, E., Halle, B., and Frey, U. 1997. NMR identification of hydrophobic cavities with low water occupancies in protein structures using small gas molecules. Nature Struct. Biol. 4 396–404. [DOI] [PubMed] [Google Scholar]
- Otting, G., Liepinsh, E., and Wüthrich, K. 1991. Protein hydration in aqueous solution. Science 254 974–979. [DOI] [PubMed] [Google Scholar]
- Poznanski, J., Sodano, P., Suh, S.W., Lee, J.Y., Ptak, M., and Vovelle, F. 1999. Solution structure of a lipid transfer protein extracted from rice seeds. Eur. J. Biochem. 259 692–708. [DOI] [PubMed] [Google Scholar]
- Prangé, T., Schiltz, M., Pernot, L., Colloc'h, N., Longhi, S., Bourguet, W., and Fourme, R. 1998. Exploring hydrophobic sites in proteins with xenon or krypton. Prot. Struct. Funct. Genet. 30 61–73. [PubMed] [Google Scholar]
- Quillin, M.L., Breyer, W.A., Grisworld, I.J., and Matthews, B.W. 2000. Size versus polarizability in protein-ligand interactions: Binding of noble gases within engineered cavities in phage T4 lysozyme. J. Mol. Biol. 302 955–977. [DOI] [PubMed] [Google Scholar]
- Rubin, S.M., Spence, M.M., Goodson, B.M., Wemmer, D.E., and Pines, A. 2000. Evidence of nonspecific surface interactions between laser-polarized xenon and myoglobin in solution. Proc. Natl. Acad. Sci. 97 9472–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux, R., Pines, A., Haake, M., and Reimer, J.A. 1999. NMR with a continuously circulating flow of laser-polarized 129Xe. J. Phys. Chem. B. 103 4629–4637. [Google Scholar]
- Shin, D.H., Lee, J.Y., Hwang, K.Y., Kim, K.K., and Suh, S.W. 1995. High resolution crystal structure of the non-specific lipid transfer protein from maize seedlings. Structure 3 189–199. [DOI] [PubMed] [Google Scholar]
- Simorre, J.-P., Caille, A., Marion, D., Marion, D., and Ptak, M. 1991. Twoand three-dimensional 1H NMR studies of a wheat phospholipid transfer protein: sequential resonance assignments and secondary structure. Biochemistry 30 11600–11608. [DOI] [PubMed] [Google Scholar]
- Sodano, P., Caille, A., Sy, D., de Person, G., Marion, D, and Ptak, M. 1997. 1H NMR and fluorescence studies of the complexation of DMPG by wheat non-specific lipid transfer protein. Global fold of the complex. FEBS Lett. 416 130–134. [DOI] [PubMed] [Google Scholar]
- Solomon, I. 1955. Relaxation processes in a system of two spins. Phys. Rev. 99 559–565. [Google Scholar]
- Song, Y.-Q., Goodson, B.M., Taylor, R.E., Laws, D.D., Navon, G., and Pines, A. 1997. Selective enhancement of NMR signals for α-cyclodextrin with laser-polarised xenon. Angew. Chem. Int. Ed. Engl. 36 2368–2370. [Google Scholar]
- Sterk, P., Booji, H., Schellekens, G.A., Van Kammen, A., and De Vries, S.C. 1991. Cell specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell. 3 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stith, A., Hitchens, T.K., Hinton, D.P., Berr, S.S., Driehuys, B., Brookeman, J.R., and Bryant, R.G. 1999. Consequences of 129Xe-1H cross-relaxation in aqueous solutions. J. Magn. Reson. 139 225–231. [DOI] [PubMed] [Google Scholar]
- Tassin-Moindrot, S., Caille, A., Douliez, J.P., Marion, D., and Vovelle, F. 1999. The wide binding properties of a wheat non-specific lipid transfer proteins. Solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 4 1117–1124. [DOI] [PubMed] [Google Scholar]
- Terras, F.R.G., Goderis, I.J., Van Leuven, F., Vanderleyden, J., Cammue, B.P.A., and Broekaert, W.F. 1992. In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to non-specific lipid transfer proteins. Plant Physiol. 100 1055–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, T.G. and Happer, W. 1997. Spin-exchange optical pumping of noble-gas nuclei. Rev. Mod. Phys. 69 629–642. [Google Scholar]
- Wolfenden, R. and Radzicka, A. 1994. On the probability of finding a water molecule in a nonpolar cavity. Science 265 936–937. [DOI] [PubMed] [Google Scholar]