Abstract
Glycopeptide dependence for growth in enterococci results from mutations in the ddl gene that inactivate the host D-Ala:D-Ala ligase. The strains require glycopeptides as inducers for synthesis of resistance proteins, which allows for the production of peptidoglycan precursors ending in D-Ala-D-Lac instead of D-Ala-D-Ala. The sequences of the ddl gene from nine glycopeptide-dependent Enterococcus faecium clinical isolates were determined. Each one had a mutation consisting either in a 5-bp insertion at position 41 leading to an early stop codon, an in-frame 6-bp deletion causing the loss of two residues (KDVA243-246 to KA), or single base-pair changes resulting in an amino acid substitution (E13 → G, G99 → R, V241 → D, D295 → G, P313 → L). The potential consequences of the deletion and point mutations on the 3-D structure of the enzyme were evaluated by comparative molecular modeling of the E. faecium enzyme, using the X-ray structure of the homologous Escherichia coli D-Ala:D-Ala ligase DdlB as a template. All mutated residues were found either to interact directly with one of the substrates of the enzymatic reaction (E13 and D295) or to stabilize the position of critical residues in the active site. Maintenance of the 3-D structure in the vicinity of these mutations in the active site appears critical for D-Ala:D-Ala ligase activity.
Keywords: D-Ala:D-Ala ligase, glycopeptide-dependence, DNA sequencing, comparative molecular modeling
D-Ala:D-Ala ligase catalyses the dimerization of D-Ala before its incorporation in late peptidoglycan precursors (Neuhaus 1960; Walsh 1989). Its key role in bacterial growth is indirectly demonstrated by the fact that mutants in this enzyme are unable to multiply unless they can use an alternative pathway for cell wall synthesis. This situation occurs in glycopeptide-dependent mutants of glycopeptide-resistant enterococci that are isolated in vitro or from patients treated for prolonged periods of time with vancomycin (for review, see Kirkpatrick et al. 1999). These bacteria possess an inactive D-Ala:D-Ala ligase (Baptista et al. 1997; Sifaoui and Gutmann 1997; Van Bambeke et al. 1999) and, thus, for cell wall synthesis, rely on the activity of a D-alanine:D-lactate (D-Ala:D-Lac) ligase encoded by the van operon that normally confers resistance to glycopeptides (Arthur et al. 1996a). Production of the enzyme is inducible by vancomycin or teicoplanin in VanA-type strains but only by vancomycin in VanB-type isolates (Arthur et al. 1996a). The ddl gene in Enterococcus faecalis vancomycin-dependent mutants (Van Bambeke et al. 1999) suffers mutations affecting an amino acid highly conserved in the D-Ala:D-Ala, D-Ala:D-Lac, and D-Ala:D-Serine ligases (Evers et al. 1996). The mutated residues are thought to play an important role in substrate binding or catalysis on the basis of the mutagenesis of the homologous Escherichia coli enzyme (Shi and Walsh 1995) and of the modeled 3-D structure of the E. faecalis enzyme (Prévost et al. 2000). Thus, mutations identified in E. faecalis, first, impair the interaction of the enzyme with Mg2+ (an essential cofactor in the reaction); second, perturb the binding of D-Ala2 (the second D-Ala entering the active site); or, third, alter the transfer of phosphate from ATP to D-Ala1. An Enterococcus avium mutant has a nonsense mutation leading to the production of a truncated, inactive enzyme (Sifaoui and Gutmann 1997). Similarly, an E. faecium strain has been described in which a frame-shift mutation at the very beginning of its ddl gene leads to an inactive enzyme (Casadewall and Courvalin 1999). This mutant does not require vancomycin for growth because its glycopeptide resistance proteins are constitutively expressed (Périchon et al. 1997).
We describe, at both phenotypic and genotypic levels, nine glycopeptide-dependent clinical isolates of E. faecium. We have determined the sequence of their ddl gene. We have analyzed by comparative molecular modeling the possible impact of the mutations on the structure of the active site of the D-Ala:D-Ala ligase to gain more insight into the structure–activity relationship of this key enzyme in bacterial metabolism.
Results
Characterization of the strains
Five of the vancomycin-dependent isolates, recovered from various specimens in four hospitals, were of the vanB genotype. The four remaining isolates were vancomycin-teicoplanin-dependent E. faecium of vanA genotype (Table 1). Strains BM4480 to BM4486 were isolated from patients receiving vancomycin, and strains BM4487 and BM4488 from patients receiving teicoplanin. The nine strains displayed different restriction patterns by pulse-field gel electrophoresis (Fig. 1 ▶) and by susceptibility to antibiotics other than glycopeptides (Table 1), except for isolates BM4482 and BM4483, which came from the same hospital. This suggests that the strains are independently derived except for the two latter ones, which correspond to the same clone.
Table 1.
Properties of E. faecium clinical isolates
| MIC (μg/ml) | |||||
| Straina (genotype) | Phenotypeb | Vm | Te | Te in the presence of Vm (10 μg/ml) | Resistance to antibiotics in the presence of Vm (10 μg/ml)b |
| BM4147 (vanA) | VmR TeR | 256 | 256 | 256 | Em Gm Lm Sm Tc |
| BM4281 (vanB) | VmR TeS | 256 | 1 | 2 | Rif |
| BM4480 (vanB) | VmD | 1024 | NAc | 32 | Em Gm Lm Sm |
| BM4481 (vanB) | VmD | 512 | NA | 16 | Cm Em Rif Tc |
| BM4482 (vanB) | VmD | 512 | NA | 32 | Em Gm |
| BM4483 (vanB) | VmD | 512 | NA | 32 | Em Gm |
| BM4484 (vanA) | VmD TeD | 512 | 32 | 32 | Em Gm |
| BM4485 (vanB) | VmD | 1024 | NA | 64 | Em Gm Lm Sm |
| BM4486 (vanA) | VmD TeD | 512 | 32 | 32 | Em Gm Lm Sm |
| BM4487 (vanA) | VmD TeD | 512 | 32 | 32 | Em Lm Rif Sm |
| BM4488 (vanA) | VmD TeD | 512 | 32 | 32 | Em Lm Rif Sm Tc |
a Origin and/or reference: BM4147 (Leclercq et al. 1988); BM4281 (Quintiliani and Courvalin 1996); BM4480 and BM4486, Thomas Jefferson University Hospital, Philadelphia, PA; BM4481, Pittsburg, PA; BM4482 and BM4483, Hahnemann University Hospital, Philadelphia, PA; BM4484, Saint James's Hospital, Dublin, Ireland; BM4485, Veterans Affairs Medical Center, East Orange, NJ (Dever et al. 1995); BM4487, Saint George's Hospital, London, UK (Farrag et al. 1996); BM4488, Institut Gustave Roussy, Villejuif, France (Chachaty et al. 1998).
b D, dependent; R, resistant; S, susceptible; Cm, chloramphenicol; Em, erythromycin; Gm, gentamicin 500 μg/ml; Lm, lincomycin; Rif, rifampicin; Sm, streptomycin, 500 μg/ml; Te, teicoplanin; Tc, tetracycline; Vm, vancomycin.
c NA, not applicable.
Fig. 1.

DNA fingerprinting of Enterococcus faecium strains. Pulsed-field gel electrophoresis of total DNA from glycopeptide-dependent clinical isolates digested by SmaI.
Sequence of the ddl gene from BM4147 (wild-type strain)
The amino acid sequence deduced from that of the ddl gene from E. faecium BM4147 was aligned with that of the corresponding enzyme from E. faecalis and with the DdlB ligase of E. coli (Fig. 2 ▶). The E. faecium enzyme contained 358 amino acids; that is, 10 more than the E. faecalis enzyme. The sequence identity was very high (78%) between the two enterococcal enzymes, and both were similar to that of E. coli (36% amino acid identity for E. faecium and 37% for E. faecalis). All the residues suspected to play an important role in the enzymatic activity, based on the crystal structure of the DdlB E. coli enzyme, were conserved in the three proteins.
Fig. 2.

Deduced amino acid sequence of the D-Ala:D-Ala ligase from Enterococcus faecium aligned with those of Enterococcus faecalis and Escherichia coli. Numbering refers to the E. faecium sequence (GenBank accession no. AF294727). Conserved amino acids predicted to play a key role in enzymatic activity on the basis of the X-ray structure of the E. coli enzyme are shown in bold lettering. Mutated positions (Table 2) are indicated by arrows, and deleted residues are in brackets.
Sequence of the ddl gene from the dependent strains
Every glycopeptide-dependent strain had a mutation in its ddl gene compared to that of BM4147 (Table 2). The ddl genes in BM4480 and BM4481 had a 5-bp GAGCA repeat between codons 14 and 15, resulting in a frame-shift mutation. The ddl gene of BM4485 had an in-frame deletion of 6 bp at codons 244 and 245, leading to the loss of two residues (Asp 244 and Val 245). The six remaining strains showed point mutations in their ddl genes. Only two mutations, at positions 13 and 295, affected residues known to be involved in the binding of the substrates to the active site of the enzyme (Shi and Walsh 1995). The other mutations affected residues that are conserved among the ligases but not thought to play a key role in enzymatic activity. Residues at positions 99 and 313 are conserved in D-Ala:D-Ala, D-Ala:D-Lac, and D-Ala:D-Ser ligases, and Val 241 is conserved only in D-Ala:D-Ala ligases of various species of enterococci (Evers et al. 1996).
Table 2.
Mutations in the ddl gene of glycopeptide-dependent clinical isolates
| Strain | Mutationa | Amino acid change |
| BM4480 | codons 14–15 (CAT GAA → CAG AGC ATG AAG) | frameshift |
| BM4481 | codons 14–15 (CAT GAA → CAG AGC ATG AAG) | frameshift |
| BM4482 | codon 295 (GAT → GGT) | Asp → Gly |
| BM4483 | codon 295 (GAT → GGT) | Asp → Gly |
| BM4484 | codon 13 (GAG → GGG) | Glu → Gly |
| BM4485 | codons 243–246 (AAA GAC GTA GCA → AAA GCA) | Lys Asp Val Ala → Lys Ala |
| BM4486 | codon 99 (GGG → AGG) | Gly → Arg |
| BM4487 | codon 313 (CCA → CTA) | Pro → Leu |
| BM4488 | codon 241 (GTC → GAC) | Val → Asp |
a Mutations are shown in bold lettering.
3-D structure of the E. faecium D-Ala:D-Ala ligase by comparative molecular modeling
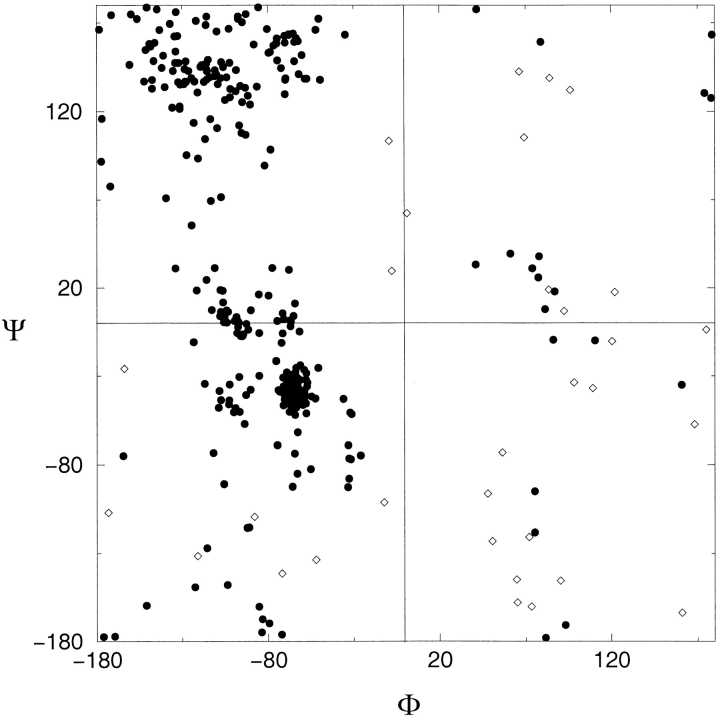
A 3-D model of the E. faecium D-Ala:D-Ala ligase was constructed using as a template the three-dimensional structure of the corresponding enzyme from E. coli (Fig. 3 ▶), as their sequences exhibit significant amino acid identity (36%; see Fig. 2 ▶). The degree of accuracy in various stereochemical features of the model was comparable to that of the X-ray template structure. These criteria (Ramachandran plot, peptide bonds planarity, nonbonded interactions, hydrogen bonds, closeness of side chain dihedral angles to ideal values) were consistent with crystal structures determined at a resolution of 2–2.7 Å. For example, the percentage of nonglycine and nonproline residues in most favored regions of the Ramachandran plot was 88% for the E. coli X-ray structure and 72% for the E. faecium model (Fig. 4 ▶). The regions of the protein model with the poorest stereochemistry were located between residues 30 and 90, as well as in the last C-terminal 14 residues for which there are no equivalent residues in the template structure and contribute toward 11% of the poorer stereochemistry of the E. faecium model (Fig. 4 ▶). These portions, therefore, had to be modeled ab initio, a procedure that is known to yield relatively inaccurate results for insertions larger than seven to eight residues (Sanchez and Sali 1997).
Fig. 3.

Structural comparison, shown in stereo, of Enterococcus faecium and Escherichia coli D-Ala:D-Ala ligases complexed with ADP and the phosphinophosphate inhibitor. Top: 3-D model of the E. faecium D-Ala:D-Ala ligase computed with the Modeller software (Sali and Blundell 1993) from the alignment in Figure 2 ▶. Bottom: Backbone representation of the X-ray structure of E. coli D-Ala:D-Ala ligase determined at 2.3-Å resolution. Each structure houses ADP and the inhibitor, depicted as orange thick sticks, and two Mg2+ ions, shown as spherical balls. Backbones of amino acids are drawn in dark blue.
Fig. 4.
Ramachadran plot of the Enterococcus faecium 3-D model. The (φ, ψ) values are represented by filled circles. Nonglycine residues whose (φ, ψ) angle values fall in generously allowed and disallowed regions, as defined in PROCHECK (Laskowski et al. 1993), are shown as open diamonds. Most of those residues are located in the 30–90-protein fragment as well as in the last C-terminal 14 residues for which there are no equivalent residues in the Escherichia coli structure.
Structure-loss of function relationships in glycopeptide-dependent mutants
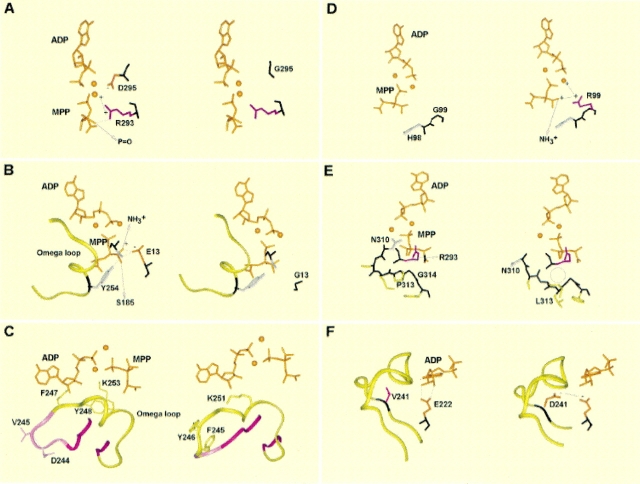
We used the resulting model to identify structural features that would explain the lack of D-Ala:D-Ala ligase activity in the mutants of E. faecium. We first validated our approach by examining the models constructed for the wild-type enzyme and two of the mutant proteins affected by a single amino acid change (Asp 295 to Gly and Glu 13 to Gly) previously known to be part of the active site and involved in catalytic activity (Shi and Walsh 1995). As shown in Figure 5A ▶, Asp 295 occupies a crucial location in the active site. It interacts directly with one of the two Mg2+ ions assisting the phosphate group transfer from ATP and forms a H bond with Arg 293. The latter residue has an important catalytic role in the E. coli ligase. Asp 295 has been proposed to interact with the carboxylate group of D-Ala1 substrate, as it makes contact with the inhibitor P–O group (Shi and Walsh 1995). The replacement of Asp 295 by Gly in BM4482 and BM4483 should grossly perturb the phosphate group transfer because of the loss of a negative charge. Moreover, glycine, because of its lack of side chain, generates larger backbone mobility that could give rise to a local structural rearrangement of the polypeptidic chain. Figure 5B ▶ shows that Glu 13 could play a critical role in stabilizing D-Ala1 binding, as it is within hydrogen bond distance to the amino group of the inhibitor. Recent pKa calculations of the E. coli enzyme have indicated that the corresponding E. coli residue, Glu 15, is one of the most probable participants in the deprotonation step of D-Ala2 NH3+ group (Carlson et al. 1999). In addition to this role, a H-bond triad is formed involving Tyr 254, Ser 185, and Glu 13 side chains, whose function, as in the E. coli enzyme (Fan et al. 1994), could be to secure the mobile loop 246–258 on the top of the active site so as to position Lys 253 toward the γ-phosphate of ATP. Given its location in the active site, its interaction with other active site residues, and its conservation in ligase sequences, the mutation of Glu 13 into Gly is expected to strongly impair the enzyme activity by removing a net charge and a number of H-bond partners.
Fig. 5.
Modeled structure of the active site of Enterococcus faecium D-Ala:D-Ala ligases. The wild-type active site is shown on the left, and that of the mutant with impaired enzymatic activity is on the right. ADP and methylphosphinophosphate (MPP) are depicted as orange sticks, and the two Mg2+ ions are shown as orange spherical balls. H bonds are shown as dashed lines. (A) Asp 295 → Gly mutation. In the wild-type enzyme, Asp 295 negatively charged (−) side chain (in orange) makes favorable interactions (solid lines) in the wild-type enzyme with one Mg2+ and the Arg 293 (in pink) positively charged side chain (+), which itself H bonds with the MPP P−O group. The Gly substitution results in the loss of these interactions and may affect the conformation of Arg 293 side chain because of its proximity to one Mg2+, shown as a solid line, that is no longer counterbalanced by Asp 295. (B) Glu 13 → Gly mutation. The Glu 13 negatively charged side chain (−) makes a favorable interaction with the MPP NH3+ group and is involved in a H-bond triad with Ser 185 and Tyr 254 side chains; the latter being part of the ω-loop illustrated by a light-green ribbon. The Gly substitution results in the loss of these interactions and may affect the conformation of the Ser 185 side chain and, in turn, that of Tyr 254 side chain (depicted in light gray). (C) DV244–245 deletion. The ribbon illustrates the backbone conformation of the 238–260 region. The pink ribbon corresponds to the extended segments that precede and follow the ω-loop (residues 246–258), shown in green. The side chains of Phe 247, Tyr 248, and Lys 253 (Phe 245, Tyr 246, and Lys 251 in the mutant), which are strongly affected by the deletion, are also drawn as green sticks. (D) Gly 99 → Arg mutation. The substitution is likely to induce unfavorable interactions of the positively charged (+) Arg side chain (in pink) with the MPP NH3+ group and one Mg2+, illustrated as solid lines. (E) Pro 313 → Leu mutation. The substitution results in the conformational change 310–315 portion of the backbone (in black). In particular, the Asn 310 side chain (in light gray), which is important for the stabilization of one Mg2+, is strongly affected. The circle indicates the empty space formed between Arg 293 (in pink) and the 310–315 fragment resulting from the substitution. (F) Val 241 → Asp mutation. In the mutant structure, the negatively charged (−) Asp241 (in orange) may affect the conformation of another negatively charged (−) side chain, E222 (also in orange), which stabilizes the ribose of ADP via the formation of two hydrogen bonds.
Residues 244–245, which are deleted in BM4485, are very close to Phe 247 and Tyr 248, residues in direct contact with the inhibitor molecule (Fig. 5C ▶). The latter residues are in an ω-loop composed of residues 246–258 that is thought to swing over the active site after the substrates bind (Parsons et al. 1988; Healy et al. 2000). Figure 5C ▶ shows that in the wild-type protein, residues 243–246 form a turn following an extended segment and preceding the ω-loop, which adopts a helical conformation. These residues may be important for the hinge-bending motion that will bring the ω-loop over the active site to occur. The model (Fig. 5C ▶) shows that the deletion grossly affects the position of the ω-loop, including Phe and Tyr residues, which in turn, could result in a deficiency in the substrate binding. Most importantly, the conformation of Lys 253 (251 in the mutant), a conserved residue that is likely to be involved in the stabilization of transfer of the phosphate group of ATP, is also perturbed by this deletion, suggesting an impairment of activity for the mutant enzyme.
Gly 99 does not interact directly with the inhibitor in the wild-type protein (Fig. 5D ▶); however, the adjacent His 98 makes contact with D-Ala1 side chain. Both residues are invariant in ligases (Evers et al. 1996). The model shows that the positively charged side chain of Arg present at position 99 in the mutant enzyme of BM4486 is close to the NH3+ group of the inhibitor and to one of the two Mg2+ ions. This spatial arrangement could lead to a reduced affinity of D-Ala1 and of the Mg2+ ion for the active site pocket and will likely impair enzyme activity.
Residues Pro 313 (replaced by Leu in strain BM4487) and Gly 314 are invariant in ligases (Evers et al. 1996). As shown in Figure 5E ▶, Pro 313 does not directly interact with the inhibitor, but the NH backbone of Gly 314 forms a H bond with the P–O group. Pro 313 also makes hydrophobic contacts with the side chain of Arg 293, an important catalytic residue. The model shown in Figure 5E ▶ for the Pro to Leu mutant located the Leu side chain on the opposite side of the backbone relative to Arg 293, leaving the latter less constrained. In addition, the side chain of Asn 310, a conserved residue (Evers et al. 1996) that stabilizes one of the Mg2+ ions via its carbonyl side chain group, is moved away from this ion. This may have a detrimental effect on the phosphate transfer step from ATP to the D-ala substrate. It thus seems that the Pro 313 to Leu mutation produces a protein with a higher local flexibility that affects the enzyme activity.
Finally, the Val 241 residue (mutated into Asp in E. faecium BM4488) is located in a region close to the active site pocket but that exhibits only a weak similarity with the E. coli sequence (see Fig. 2 ▶). As correct sequence alignments are crucial for the production of reliable 3-D models constructed by comparative modeling, the quality of the model in this protein region is uncertain. Nevertheless, in the mutant enzyme, Asp can possibly repel the Glu 222 side chain, which establishes H bonds with the ribose of ADP in the wild-type enzyme (Fig. 5F ▶), and therefore, Asp can affect the binding of one of the substrates to the enzyme. Moreover, the absence of positive charge to stabilize the Asp residue could perturb the correct position of the ω-loop, as in the 244–245 deletion.
Discussion
This study analyses all of the clinical isolates of glycopeptide-dependent E. faecium, an organism that causes more concern than other enterococcal species with respect to glycopeptide resistance (Moellering 2000). These strains show high resistance to vancomycin and a lower level of resistance to teicoplanin (in the presence of vancomycin as inducer for VanB-type strains, or of vancomycin or teicoplanin as inducer for VanA-type strains). Both glycopeptides appear to be equipotent inducers of the production of resistance proteins by VanA-type dependent mutants. The relatively low level of resistance to teicoplanin in all dependent strains (Table 1) would appear to indicate a low production of resistance proteins (Arthur et al. 1996b). This was experimentally confirmed in vancomycin-dependent E. faecalis mutants and attributed to the exclusive synthesis, by these strains, of precursors ending in D-Ala-D-Lac, making the production of the resistance enzymes removing D-Ala-D-Ala-containing precursors superfluous (Van Bambeke et al. 1999).
Considering the molecular mechanism involved in glycopeptide dependence, there is growing evidence that the appearance of this phenotype follows the generation of mutations in the gene encoding the D-Ala:D-Ala ligase that lead to enzyme inactivation (Baptista et al. 1997; Fraimow et al. 1994; Van Bambeke et al. 1999). Glycopeptide-dependent mutants may, therefore, be considered as useful tools to refine structure–activity relationships for this enzyme, which may represent an attractive target for new antibiotics. First described by Neuhaus in 1960, D-Ala:D-Ala ligase plays a key role in cell wall biosynthesis (Walsh 1989). Yet it has only been purified from a few bacterial species (Neuhaus 1962; Daub et al. 1988; Zawadzke et al. 1991), and its 3-D structure has only been established for the E. coli enzyme (Fan et al. 1994). In a previous study (Prévost et al. 2000), we showed that molecular modeling applied to D-Ala:D-Ala ligase E. faecalis mutants (Baptista et al. 1997; Van Bambeke et al. 1999) could help in understanding changes in a 3-D enzyme structure that lead to its inactivation. In the absence of X-ray data, we have applied this approach to the D-Ala:D-Ala ligase of E. faecium. The deduced sequence of the ddl gene in E. faecium showed high sequence similarity with the corresponding E. faecalis enzyme, as well as with that of E. coli. In particular, and as described for the E. faecalis enzyme (Prévost et al. 2000), all the residues involved in the ATP-dependent ligation of the carboxyl group of one substrate to the amino function of another substrate, which is the common feature of the enzymes belonging to the ATPgrasp family (Galperin and Koonin 1997), are also conserved in the E. faecium enzyme. Likewise, the E. faecium, E. faecalis, and E. coli enzymes possess an organization of their ATP-binding site similar to that of cAMP-dependent protein kinase, a member of an unrelated family of protein kinases (Denessiouk et al. 1998). The mutated amino acids in the glycopeptide-dependent mutants are all conserved in D-Ala:D-Ala ligases (Evers et al. 1996); interestingly, however, they were not necessarily predicted to be directly involved in substrate recognition (Fig. 2 ▶). Although the degree of enzyme impairment was not measured biochemically, the lack of growth of the mutants in the absence of vancomycin (which induces expression of the structural gene for the D-Ala:D-Lac ligase) strongly suggests a complete lack of D-Ala:D-Ala ligase activity. This is indeed what has been found previously in vancomycin-dependent mutants of E. faecalis, which produced 0% of precursors ending in D-Ala-D-Ala (Van Bambeke et al. 1999). The consequences of two mutations, Asp 295 → Gly (in strains BM4482 and BM4483) and Glu 13 → Gly (in strain BM4484), can be explained on the basis of homology with the E. coli enzyme (Fan et al. 1994; Shi and Walsh 1995) because they affect residues directly involved in the binding of the substrate (see Fig. 5A,B ▶). This confirms the importance of Asp 295 in catalysis, observed for point mutants of E. faecalis (Asp → Val [Van Bambeke et al. 1999]) and of E. coli (Asp → Asn [Shi and Walsh 1995]). We also demonstrate the critical role of Glu 13, as was surmised from the studies with E. coli (where a mutation of the corresponding Glu [at position 15] to Gln causes a 100-fold decrease in activity [Shi and Walsh 1995]). In addition, we suggest that the H bond established between Glu 13 and the amino group of the inhibitor (and, therefore, most likely with D-Ala1) is critical for binding. The other point mutations, Gly 99 → Arg, Pro 313 → Leu, and Val 241 → Asp, affect residues that do not interact directly with the substrates and were, therefore, only rationalized by molecular modeling. As discussed in the study of the E. faecalis enzyme (Prévost et al. 2000), comparative modeling provides accurate models in regions homologous with those of the E. coli template. This was clearly the case in two of the three regions studied corresponding to mutations at positions 99 (strain BM4486; Fig. 5D ▶) and 313 (strain BM4487; Fig. 5E ▶). The regions of lowest structural confidence were those corresponding to substitution at position 241 (strain BM4488; Fig. 5F ▶) and the deletion at positions 244–245 (in strain BM4485; Fig. 5C ▶), as the sequence homology in these regions with the E. coli template is weak (Sanchez and Sali 1997; Prévost et al. 2000). The latter deletion is, however, of interest because the deleted residues did not appear to enter in direct contact with the substrate. The model showed that these four mutations may indirectly affect the conformation of the active site. This type of mutant is rarely studied by site-directed mutagenesis, as opposed to active site residues for which functional groups are spatially proximal to the bound substrate or to the reaction intermediates. An important aspect of the active site is that invariance extends to a number of amino acids that do not themselves make contact with the substrate but that, rather, provide substructure or scaffold on which amino acids contacting the substrate are supported. The present data bear some similarity to those obtained with the ketosteroid isomerase of Pseudomonas putida and with the Influenza virus neuraminidase (Varghese et al. 1992), which showed that a H-bond network provides the structural organization needed by the enzyme to maintain the active site geometry optimized for both function and stability (Kim et al. 2000). Other studies have also pointed to the involvement of flexible surface loops, such as that affected in strain BM4485 (Fig. 5C ▶), in the mechanism of enzyme-catalyzed reactions (First and Fersht 1993). Certain residues located in these loops may not be directly involved in the enzyme catalytic mechanism but seem essential for the movement of the loop that brings the active site residues close to the substrate.
Finally, several mutations (Asp 295 → Gly, Gly 99 → Arg, Glu 13 → Gly, Val 241 → Asp) are accompanied by the gain or the loss of a charged residue or by a modification in the orientation of a charged residue (in the mutant having a deletion of residues 244 and 245). The subsequent perturbation of electrostatic interactions could restrict or even possibly prevent the access to the active site or the correct positioning in the active site of one of the substrates.
The present data indicate that point mutations offer a larger potential for enzyme inactivation than could be deduced from the examination of the substrate binding site itself. Maintenance of the geometry of the catalytic pocket thus appears critical for enzymatic activity. These conclusions may open interesting perspectives for the understanding of the mechanism of action of enzymes belonging to the same family as well as for the design of new antibiotics.
Materials and methods
Bacterial strains and growth conditions
Certain properties of the clinical isolates studied are summarized in Table 1. Glycopeptide-dependent strains were recovered from patients undergoing vancomycin therapy, with the exception of strains BM4487 and BM4488, which were isolated from patients receiving teicoplanin. The clinical isolates were grown in brain heart infusion (BHI, Difco Laboratories) supplemented with 10 μg/mL vancomycin. The nondependent strains were grown in nonsupplemented BHI.
Phenotypic and genotypic characterization of the strains
The MICs of vancomycin and teicoplanin were determined after 48 h of incubation by the method of Steers et al. (1959) with 105 CFU per spot on BHI agar or BHI agar supplemented with 10 μg/mL vancomycin (to determine the MICs of teicoplanin for the vancomycin-dependent strains). Susceptibility to antibiotics other than glycopeptides was determined by disk agar diffusion on BHI supplemented with 10 μg/mL vancomycin. The species of enterococci and the glycopeptide resistance genotype were determined by PCR (Dutka-Malen et al. 1995) with DNA from E. faecium BM4147 (vanA) and E. faecalis BM4281 (vanB) as positive controls (Leclercq et al. 1988; Quintiliani and Courvalin 1996). All strains were found to be E. faecium.
Pulse-field gel electrophoresis of genomic DNA
Glycopeptide-dependent strains were analyzed by pulse-field gel electrophoresis. Genomic DNA embedded in agarose plugs (Miranda et al. 1991) was digested overnight at 30°C with 40 U of SmaI endonuclease. The restriction fragments were separated by using a clamped homogenous electric field with a CHEF-DR II system (Bio-Rad Laboratories) with a pulse ranging from 2 to 22 s for 16 h.
Amplification, cloning, and sequencing of the ddl gene
Purified total DNA was used as a template for amplification by PCR using pfu polymerase and primers 5′-GAGTAAATCACTGAACGATT and 5′-GGTTACGCAATCACTCCAGCCT designed from the sequence of the ddl gene from E. faecium BM4339 (Casadewall and Courvalin 1999). Amplification was carried out in a 100–-μL volume containing 50 pmol of each primer, 50 nmol of each deoxynucleotide, reaction buffer, 100 ng of DNA, and 10 U of pfu DNA polymerase. A total of 30 cycles were run; each cycle consisted of 1 min denaturation at 94°C, 1 min annealing at 54°C, and 2 min elongation at 72°C. The PCR products were purified from agarose gels (Sephaglas kit, Pharmacia), cloned into pCR-Blunt (Invitrogen Leek), and sequenced by the dideoxychain termination method (Sanger et al. 1977) using T7 DNA polymerase (Sequenase kit, U.S. Biochemical) and [α-35S]dATP (Amersham Radiochemical Center). Two independent PCR products were sequenced for each strain studied.
Comparative molecular modeling
The procedure followed was that used for building the model of the E. faecalis enzyme (Prévost et al. 2000). In brief, the sequence of the target enzyme was aligned with that of the DdlB of E. coli, which displays ∼36% identity with the E. faecium enzyme (Evers et al. 1996; this work). The model was produced for the entire enzyme in the presence of ADP and methylphosphinophosphate in the active site. The latter molecule is an irreversible inhibitor of the enzyme replacing D-Ala-D-Ala in the active site of the E. coli enzyme (Parsons et al. 1988; Fan et al. 1994). Using Modeller4 software (Sali and Blundell 1993), the 3-D structure was built based on the satisfaction of spatial restraints extracted from the alignment and on the optimization of the interaction energy of the molecule. The model obtained was then validated by appropriate programs (Modeller; Procheck [Laskowski et al. 1993]), as described previously for the E. faecalis model (Prévost et al. 2000).
Acknowledgments
M.P. and F.V.B. are Chercheurs Qualifiés of the Belgian Fonds National de la Recherche Scientifique. This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases and by the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires from the Ministére de l'Education Nationale de la Recherche et de la Technologie. We thank H.S. Fraimow, E. Chachaty, and L.L. Dever for the gift of strains.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
D-Ala, D-Alanine
D-Ala:D-Ala ligase, D-Alanine:D-Alanine ligase
3-D, three-dimensional
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.39101.
References
- Arthur, M., Reynolds, P.E., and Courvalin, P. 1996a. Glycopeptide resistance in enterococci. Trends Microbiol. 4 401–407. [DOI] [PubMed] [Google Scholar]
- Arthur, M., Depardieu, F., Reynolds, P.E., and Courvalin, P. 1996b. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21 33–44. [DOI] [PubMed] [Google Scholar]
- Baptista, M., Depardieu, F., Reynolds, P.E., Courvalin, P., and Arthur, M. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25 93–105. [DOI] [PubMed] [Google Scholar]
- Carlson, H.E., Briggs, J.M., and McCammon, J.A. 1999. Calculation of the pKa values for the ligands and side chains of Escherichia coli D-alanine:D-alanine ligase. J. Med. Chem. 42 109–117. [DOI] [PubMed] [Google Scholar]
- Casadewall, B. and Courvalin, P. 1999. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 181 3644–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachaty, E., Ciupek, C., Schoepfer, C., Hartmann, O., and Tancrede, C. 1998. Glycopeptide-dependent vanA Enterococcus faecium bacteriemia after prolonged therapy with teicoplanin. In: Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, San Diego, CA, Abstract C-87
- Daub, E., Zawadzke, L.E., Botstein, D., and Walsh, C.T. 1988. Isolation, cloning, and sequencing of the Salmonella typhimurium ddlA gene with purification and characterization of its product, D-alanine:D-alanine ligase. Biochemistry 27 3701–3708. [DOI] [PubMed] [Google Scholar]
- Denessiouk, K.A., Lehtonen, J.V., Korpela, T., and Johnson, MS. 1998. Two "unrelated" families of ATP-dependent enzymes share extensive structural similarities about their cofactor binding sites. Protein Sci. 7 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever, L.L., Smith, S.M., Handwerger, S., and Eng, R.H.K. 1995. Vancomycin-dependent Enterococcus faecium isolated from stool following oral vancomycin therapy. J. Clin. Microbiol. 33 2770–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka-Malen, S., Evers, S., and Courvalin, P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers, S., Casadewall, B., Charles, M., Dutka-Malen, S., Galimand, M., and Courvalin, P. 1996. Evolution of structure and substrate specificity in D-alanine:D-alanine ligases and related enzymes. J. Mol. Evol. 42 706–712. [DOI] [PubMed] [Google Scholar]
- Fan, C., Moews, P.C., Walsh, C.T., and Knox, J.R. 1994. Vancomycin-resistance: structure of D-alanine:D-alanine ligase at 2.3 Å resolution. Science 266 439–443. [DOI] [PubMed] [Google Scholar]
- Farrag, N., Eltringham, I., and Liddy, H. 1996. Vancomycin-dependent Enterococcus faecalis. Lancet 348 1581–1582. [DOI] [PubMed] [Google Scholar]
- First, E.A. and Fersht, A.R. 1993. Mutational and kinetic analysis of a mobile loop in tyrosyl-tRNA synthetase. Biochemistry 32 13658–13663. [DOI] [PubMed] [Google Scholar]
- Fraimow, H.S., Jundkind, D.L., Lander, D.W., Delso, D.R., and Dean, J.L. 1994. Urinary tract infection with an Enterococcus faecalis isolate that requires vancomycin for growth. Ann. Intern. Med. 121 22–26. [DOI] [PubMed] [Google Scholar]
- Galperin, M.Y. and Koonin, EV. 1997. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol activity. Protein Sci. 6 2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy, V.L., Lessard, I.A.D., Roper, D.I., Knox, J.R., and Walsh, C.T. 2000. Vancomycin-resistance in enterococci: Reprogramming of the D-Ala:D-Ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 7 R109–R119. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Jang, D.S., Nam, G.H., Choi, G., Kim, J.S., Ha, N.C., Kim, M.S., Oh, B.H., and Choi, K.Y. 2000. Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B. Biochemistry 39 4581–4589. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, B.D., Harrington, S.M., Smith, D., Marcellus, D., Miller, C., Dick, J., Karanfil, L., and Perl, T.M. 1999. An outbreak of vancomycin-dependent Enterococcus faecium in a bone marrow transplant unit. Clin. Infect. Dis. 29 1268–1273. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., McArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291 [Google Scholar]
- Leclercq, R., Derlot, E., Duval, J., and Courvalin, P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319 157–161. [DOI] [PubMed] [Google Scholar]
- Miranda, A.G., Singh, K.V., and Murray, B.E. 1991. DNA fingerprinting of Enterococcus faecium by pulse-field gel electrophoresis may be a useful epidemiologic tool. J. Clin. Microbiol. 29 2752–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering, R.C. 2000. Enterococcus species, Streptococcus bovis, and Leuconostoc species. In Principles and Practice of Infectious Diseases, 5th ed. (eds. G.L. Mandell, et al.), pp. 2147–2156. Churchill Livingstone, Philadelphia, PA.
- Neuhaus, F.C. 1960. The enzymatic synthesis of D-alanyl-D-alanine. Biochem. Biophys. Res. Com. 3 401–405. [DOI] [PubMed] [Google Scholar]
- ———. 1962. The enzymatic synthesis of D-alanyl-D-alanine. I. Purification and properties of the D-alanyl-D-alanine synthetase. J. Biol. Chem. 237 778–786. [PubMed] [Google Scholar]
- Parsons, W.H., Patchett, A.A., Bull, H.G., Schoen, W.R., Taub, D., Davidson, J., Combs, P.L., Springer, J.P., and Busch, R.D. 1988. Phosphinic acid inhibitors of D-alanyl-D-alanine ligase. J. Med. Chem. 31 1772–1778. [DOI] [PubMed] [Google Scholar]
- Périchon, B., Reynolds, P.E., and Courvalin, P. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41 2016–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost, M., Van Belle, D., Tulkens, P.M., Courvalin, P., and Van Bambeke, F. 2000. Modeling of the Enterococcus faecalis D-alanine:D-alanine ligase: Structure-based study of the active site in the wild-type enzyme and in glycopeptide-dependent mutants. J. Mol. Microbiol. Biotechnol. 2 321–330. [PubMed] [Google Scholar]
- Quintiliani, Jr., R. and Courvalin, P. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172 1–8. [DOI] [PubMed] [Google Scholar]
- Sali, A. and Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spacial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Sanchez, R. and Sali, A. 1997. Advances in comparative protein-structure modeling. Curr. Opin. Struct. Biol. 7 206–214. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. and Walsh, C.T. 1995. Active site mapping of Escherichia coli D-Ala:D-Ala ligase by structure-based mutagenesis. Biochemistry 34 2768–2776. [DOI] [PubMed] [Google Scholar]
- Sifaoui, F. and Gutmann, L. 1997. Vancomycin dependence in a VanA-producing Enterococcus avium strain with a nonsense mutation in the natural D-Ala:D-Ala ligase gene. Antimicrob. Agents Chemother. 41 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steers, E., Foltz, E.L., Graves, B.S., and Riden, J. 1959. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot. Chemother. 9 307–311. [PubMed] [Google Scholar]
- Van Bambeke, F., Chauvel, M., Reynolds, P.E., Fraimow, H.S., and Courvalin, P. 1999. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob. Agents Chemother. 43 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese, J.N., McKimm-Breschkin, J.L., Caldwell, J.B., Kortt, A.A., Colman, P.M. 1992. The structure of the complex between Influenza virus neuraminidase and sialic acid, the viral receptor. Proteins 14 327–332. [DOI] [PubMed] [Google Scholar]
- Walsh, C.T. 1989. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem. 264 2393–2396. [PubMed] [Google Scholar]
- Zawadzke, L.E., Bugg, T.D.H., and Walsh, C.T. 1991. Existence of two D-alanine:D-alanine ligases in Escherichia coli: Cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry 30 1673–1682. [DOI] [PubMed] [Google Scholar]