A procedure for microseeding into nanolitre crystallization drops is described with selected successful examples.
Keywords: crystallization, crystal optimization, microseeding, additives
Abstract
A simple semi-automated microseeding procedure for nanolitre crystallization experiments is described. Firstly, a microseed stock solution is made from microcrystals using a Teflon bead. A dilution series of this microseed stock is then prepared and dispensed as 100 nl droplets into 96-well crystallization plates, facilitating the incorporation of seeding into high-throughput crystallization pipelines. This basic microseeding procedure has been modified to include additive-screening and cross-seeding methods. Five examples in which these techniques have been used successfully are described.
1. Introduction
X-ray crystallography remains the method of choice for determining the detailed three-dimensional structures of biological macromolecules in order to gain insight into their function and interactions. The method depends on the ability to grow well ordered single crystals. In the era of structural genomics/proteomics, major advances in protein crystallization have been made with the use of robotics, which has both automated the crystallization experiment and reduced the amount of protein required by an order of magnitude, improving the reproducibility of the experiments and allowing a larger number to be set up (Sulzenbacher et al., 2002 ▶; Brown et al., 2003 ▶; Hosfield et al., 2003 ▶; Walter et al., 2003 ▶; Berry et al., 2006 ▶). Standard screening and optimization procedures (Walter et al., 2005 ▶) on the protein of interest may yield only a microcrystalline precipitate or a bundle of small crystals that are unsuitable for X-ray diffraction analysis. However, these microcrystals may be used as seeds to produce single crystals that are suitable for structural studies (Bergfors, 2003 ▶). Here, we report a simple semi-automated microseeding protocol which has been integrated into our high-throughput platform and has facilitated a number of structural analyses. This technique has also been applied in combination with additive screening (Walter et al., 2005 ▶) and cross-seeding methods (Stura & Wilson, 1992 ▶). The procedures are semi-automated, as opposed to fully automated, because they require manual intervention during seed preparation and drop dispensing.
Crystallization is dependent on two processes: the formation of nuclei (for a discussion, see García-Ruiz, 2003 ▶) and the subsequent growth of an ordered crystalline lattice around the nuclei. However, the optimal conditions for nucleation may not be the same as for crystal growth: nucleation occurs when the solution is supersaturated, whereas ordered growth of crystals is optimal in a state of lower supersaturation, a ‘metastable’ solution. A complication is that the chemical components required for these two stages may also differ, as exemplified by the technique of ‘microseed matrix screening’ (Ireton & Stoddard, 2004 ▶; D’Arcy et al., 2007 ▶), in which crystals grown in one set of conditions are seeded into a second different screen of crystallization solutions. Indeed, seeding provides a general method of decoupling crystal nucleation from crystal growth (for a comprehensive review, see Bergfors, 2003 ▶). Usually, previously obtained crystals are used as seeds to eliminate the requirement for spontaneous nucleation in a new crystallization experiment and a lower concentration of protein is used, which in turn promotes the growth of better ordered crystals. Since macroseeding, which involves seeding with visible single crystals, is not generally amenable to automation unless with specialized equipment (e.g. Gerdts et al., 2006 ▶; Viola et al., 2007 ▶), we chose to use microseeding, in which the seeds, which are too small to be viewed directly, are dispensed as a dilution series. Here, we describe several seeding protocols developed to work in conjunction with our existing procedures for nanolitre crystallization (Walter et al., 2003 ▶, 2005 ▶).
2. Materials and methods
2.1. Protein preparation
Five proteins (four viral and one bacterial, as detailed in §3) were produced. Three of them [Murray Valley encephalitis virus (MVEV) helicase, MVEV NS2b-NS3 and transcriptional regulator NMB0838-1] were produced as soluble proteins in Escherichia coli using standard expression and purification protocols of the Oxford Protein Production Facility (OPPF) and Division of Structural Biology (Alzari et al., 2006 ▶; Au et al., 2006 ▶). CrmE was also expressed in E. coli, but was refolded. GP64 was produced as a glycoprotein in insect cells. Proteins were concentrated using centrifugal concentrators (Vivascience/Sartorius, Stonehouse, UK; Millipore, Massachusetts, USA) to an appropriate level according to a ‘Pre-Crystallization Test’ (PCT; Hampton Research, California, USA) in 20 mM Tris–HCl pH 7.5, 200 mM NaCl prior to crystallization.
2.2. Crystallization and microseeding
2.2.1. Initial screening
Crystallization experiments were carried out in 96-well crystallization plates (CrystalQuick plate 609101, Greiner Bio-One Ltd, Stonehouse, UK) using nanolitre droplets dispensed using a Cartesian Honeybee X8 (previously termed Microsys MIC4000, Genomic Solutions, Huntingdon, UK) in the OPPF crystallization facility as described previously (Walter et al., 2003 ▶, 2005 ▶; Mayo et al., 2005 ▶). Initial screening was performed as sitting drop vapour-diffusion experiments with drops of 100 nl protein plus 100 nl reservoir equilibrated against 95 µl reservoir solution using commercially available screening solutions (Hampton Research, California, USA; Emerald BioStructures, deCode Genetics, Washington, USA and Molecular Dimensions, Newmarket, UK).
2.2.2. Microseed preparation
Microseeding experiments were initiated for projects where, even after optimization experiments, only microcrystals in the form of a crystalline precipitate or clusters of small crystals which diffracted poorly were obtained. These microcrystals were transferred to 50 µl of a stabilizing solution with an appropriately sized loop (usually some 100 µm in diameter), crushed by vortexing for 90 s with a Teflon bead (‘Seed Bead’, Hampton Research; Luft & DeTitta, 1999 ▶) and then used as microseed stock solution. The exact composition of the stabilizing solution was determined empirically as a solution which contained a high enough concentration of precipitant and protein such that the microcrystals did not dissolve. Typically, the stabilizing solution was made from 50% reservoir solution, 10% protein solution and 40% water. The microseed stock solution was serially diluted 1 in 4 with stabilizing solution to create a range of seeding solutions with sequentially reduced seed content: from undiluted seeds to a 4096-fold dilution of the seeds.
Microseeding crystallization experiments were carried out in Greiner crystallization plates with all nanolitre drops being dispensed by the Cartesian Honeybee instrument. Two changes were made to the standard screening protocol (Walter et al., 2003 ▶). Firstly, all reservoir wells contained the same precipitant, usually at 60% of the original crystal screen solution; secondly, seeding solution instead of, or in addition to, reservoir solution was added to the protein drop.
2.2.3. Basic microseeding
For the basic microseeding experiment, drops were dispensed as follows: 100 nl protein solution was dispensed to all 96 wells followed by 100 nl microseed solution in rows. For each row a successively diluted seeding solution was used, prepared as described above. Sealed plates were then placed into the automated imaging and storage system of the OPPF, allowing the progress of the crystallization experiments to be recorded and monitored externally (Mayo et al., 2005 ▶). Any remaining solutions of microseed stock and dilutions were frozen unaliquoted and stored at 253 K for later use (freezing of seed solutions is also described by D’Arcy et al., 2007 ▶). Storage in this way did not appear to reduce efficacy in subsequent experiments (seed solutions for MVEV helicase, for example, were still effective after three freeze–thaw cycles).
2.2.4. Additive and cross-seeding
Variations of the basic microseeding experiment were developed to make use of additive-screening and cross-seeding methods. In both types of experiments the same seed solution was used for all 96 drops. Results from the preceding basic microseeding experiment were used as guidance to decide which seed dilution would be most appropriate.
For additive microseeding, drops were dispensed as follows: 200 nl protein plus 100 nl reservoir plus 100 nl seed solution plus 100 nl additive screen (Hampton Research Additive Screen HR2-428 diluted fivefold with water as described by Walter et al., 2005 ▶). For volatile additives no additive was dispensed into the drops but 20 µl of undiluted additive was pipetted into the reservoir. The protein, reservoir and seed solutions were dispensed ‘on the fly’ while the additives were transferred as aliquots from a second plate.
For cross-seeding, the seed solution was made from the original underivatized protein and this solution was then dispensed into the drops of the derivatized protein. Otherwise, this procedure was identical to basic microseeding as described above, except that the same seed solution was used for all the drops.
3. Results
The layout and results of a typical basic microseeding experiment are shown in Fig. 1 ▶ (in this case for MVEV helicase, see below). The seed solution was successively fourfold diluted with stabilizing solution from the original undiluted seed stock solution in row G to a 4096-fold dilution in row A. Fewer crystals were obtained as the seed solution was diluted and these crystals took longer to grow. In wells containing higher seed concentrations a few crystals had already appeared after 1 d and continued to grow over the following days. In some of these wells additional crystals appeared in subsequent days. These wells are marked 10 in Fig. 1 ▶ as the total number of crystals present could not be established unambiguously owing to their small size. No crystals were observed in the absence of seeds (row H), in which only the stabilizing solution was added to the drops.
Figure 1.
General layout of the basic microseeding experiment in a 96-well crystallization plate [drops shown are of Murray Valley encephalitis virus (MVEV) helicase]. In row G the seed stock solution was used undiluted and in row H no seeds were used. In rows F to A the seed solution was diluted each time by factor of four starting with the seed stock solution to yield solutions with decreasing seed content as shown on the right. For the seeding experiment of MVEV helicase the number of crystals observed in each well after ∼5 d are listed. Wells containing no crystals are left blank and drops containing ten or more crystals are marked 10.
3.1. Examples
The basic microseeding and microseeding with additive screening protocols have been used successfully for several proteins.
3.1.1. Murray Valley encephalitis virus (MVEV) helicase
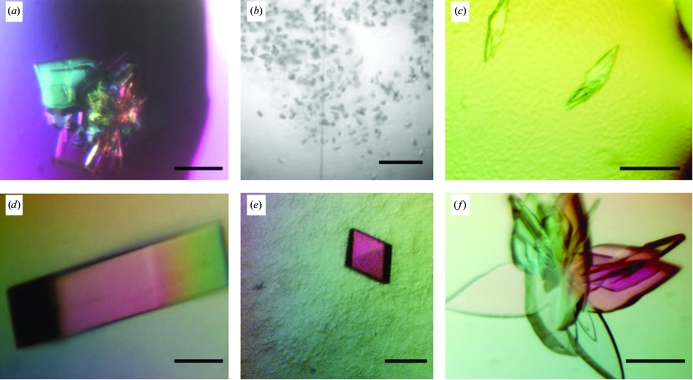
MVEV helicase, the C-terminal domain of nonstructural protein 3 (NS3), crystallized within 2 d in 5% PEG 6000, 0.1 M HEPES pH 7.0, forming bundles of stacked plates (Fig. 2 ▶ a). These bundles showed disordered anisotropic diffraction to 3.5 Å resolution when tested at the UK MRC beamline BM14 at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). The basic microseeding experiment produced optically superior crystals of a rod-like morphology (Fig. 2 ▶ d) within a few days, which diffracted to 1.8 Å resolution when exposed at the ESRF beamline ID23.1 and allowed the structure to be determined (Mancini et al., 2007 ▶; PDB code 2v8o). The space group and unit-cell parameters for crystals before and after seeding did not change.
Figure 2.
Examples of crystal improvement before and after microseeding: (a), (d) Murray Valley encephalitis virus (MVEV) helicase; (b), (e) baculovirus GP64; (c), (f) full-length MVEV NS2b-NS3. The solid bar represents 0.1 mm in all images.
3.1.2. Baculovirus GP64
The deglycosylated form of a baculovirus fusion protein GP64 produced a microcrystalline shower in 10% PEG 6000, 0.1 M citrate pH 5.0 (Fig. 2 ▶ b). This material was considered to be too small to be tested for diffraction. The basic microseeding protocol produced single crystals which diffracted to a resolution of 3.5 Å in space group C2 when exposed at station ID14.1, ESRF. Further microseeding experiments with the additive screen resulted in crystals in space group H32 with over half of the additives. These crystals were used for heavy-atom soaking experiments and of the 30 crystals tested for diffraction after soaking, two diffracted to 2.9 Å resolution (Fig. 2 ▶ e) at BM14, ESRF (Kadlec et al., work submitted).
3.1.3. MVEV NS2b-NS3
The full-length MVEV NS3 consists of the helicase and a serine-protease domain which requires a peptide from the NS2b protein for correct folding and functionality. Small multiple crystals grew in 20% PEG 6000, 0.1 M MES pH 6.0 with either 0.2 M MgCl2 or CaCl2 within a few days (Fig. 2 ▶ c). Diffraction images from these crystals, tested using an in-house rotating-anode X-ray source, gave data to 4.0 Å resolution and revealed the presence of multiple lattices. Basic microseeding and optimization experiments did not lead to better crystals; however, by microseeding with an additive screen a cluster of large blade-shaped crystals could be obtained (Fig. 2 ▶ f). These yielded data to 2.6 Å resolution at station ID29, ESRF and the structure determination is in progress (Assenberg et al., in preparation). The space group for crystals before and after seeding was P21, with similar unit-cell parameters.
3.2. Cross-seeding
The method of cross-seeding, in which microcrystals of one protein are used as seeds to grow crystals of a derivative protein, is well established (Stura & Wilson, 1992 ▶; Bottomley et al., 1994 ▶). We have transferred this method to microseeding and describe two examples in more detail below.
3.2.1. Vaccinia virus CrmE
Unlabelled (native) CrmE, an extracellular protein from Vaccinia virus that mimics the human TNFα receptor, crystallized readily in 0.2 M KH2PO4, 20% PEG 3350. However, the selenomethionine (SeMet) labelled protein was refractory to crystallization under these conditions. Using the native crystals for microseeding facilitated the routine growth of SeMet-labelled crystals, which enabled the further refinement of the crystallization and cryoprotection conditions that proved essential for the structure solution. The structure has recently been reported (Graham et al., 2007 ▶; PDB code 2uwi).
3.2.2. Neisseria transcriptional regulator NMB0838-1
The transcriptional regulator NMB0838-1 of N. meningitidis strain MC58 (serogroup B) crystallized in space group P41212 (or P41212) in 2 M sodium/potassium phosphate and yielded a data set to 2.7 Å resolution at station ID14.2, ESRF. Attempts to solve the structure using molecular-replacement methods were unsuccessful and so SeMet derivatized protein was produced. Crystals of the derivative could only be obtained by seeding with native crystals and diffracted to 2.5 Å resolution at BM14, ESRF. The unit-cell parameters and space group of the SeMet-derivatized crystals were similar to those for the native protein and structure determination is in progress.
4. Discussion
The desirability of establishing effective procedures for every stage of the structure-determination pipeline has prompted us to design semi-automated microseeding methods which go beyond the usual protocols for optimizing crystallization conditions. We have developed a straightforward protocol to perform microseeding experiments on our high-throughput platform which can easily be transferred to other systems and offers several benefits.
(i) Using 100 nl drops in seeding experiments has the advantage of requiring less protein material and/or being able to set up more drops.
(ii) Seeding solutions are dispensed by pipetting robots, which are faster and more reproducible than manual pipetting.
(iii) Integrating the microseeding experiment into the existing crystallization pipeline provides the benefits of plate tracking and crystal imaging (Mayo et al., 2005 ▶).
These protocols have allowed us to grow well ordered crystals of large enough size that we have been able to collect data sufficient to solve the structures of five proteins that had proved recalcitrant to crystallographic analysis. The protocols have therefore been incorporated into our crystallization platform as a generic rescue tool.
Acknowledgments
The Oxford Protein Production Facility is funded by the Medical Research Council with additional finance from the Biotechnology and Biological Science Research Council and is part of the Structural Proteomics IN Europe consortia (SPINE, contract No. QLG2-CT-2002-00988, and SPINE2-Complexes, contract No. LSHG-CT-2006-031220) and VIZIER initiative (contract No. LSHG-CT-2004-511960). We thank the staff at BM14 and the ESRF for help with data collection. EJM and JMG are supported by the Royal Society. SCG is supported by the Nuffield Dominions Trust. DIS is a Medical Research Council Research Professor.
References
- Alzari, P. M. et al. (2006). Acta Cryst. D62, 1103–1113.
- Au, K. et al. (2006). Acta Cryst. D62, 1267–1275. [DOI] [PubMed]
- Bergfors, T. (2003). J. Struct. Biol. 142, 66–76. [DOI] [PubMed]
- Berry, I. M., Dym, O., Esnouf, R. M., Harlos, K., Meged, R., Perrakis, A., Sussman, J. L., Walter, T. S., Wilson, J. & Messerschmidt, A. (2006). Acta Cryst. D62, 1137–1149. [DOI] [PubMed]
- Bottomley, M. J., Robinson, R. C., Driscoll, P. C., Harlos, K., Stuart, D. I., Aplin, R. T., Clements, J. M., Jones, E. Y. & Dudgeon, T. J. (1994). J. Mol. Biol.244, 464–468. [DOI] [PubMed]
- Brown, J. et al. (2003). J. Appl. Cryst.36, 315–318.
- D’Arcy, A., Villard, F. & Marsh, M. (2007). Acta Cryst. D63, 550–554. [DOI] [PubMed]
- García-Ruiz, J. M. (2003). J. Struct. Biol.142, 22–31. [DOI] [PubMed]
- Gerdts, C. J., Tereshko, V., Yadav, M. K., Dementieva, I., Collart, F., Joachimiak, A., Stevens, R. C., Kuhn, P., Kossiakoff, A. & Ismagilov, R. F. (2006). Angew. Chem. Int. Ed.45, 8156–8160. [DOI] [PMC free article] [PubMed]
- Graham, S. C., Bahar, M. W., Abrescia, N. G. A., Smith, G. L., Stuart, D. I. & Grimes, J. M. (2007). J. Mol. Biol.372, 660–671. [DOI] [PubMed]
- Hosfield, D., Palan, J., Hilgers, M., Scheibe, D., McRee, D. E. & Stevens, R. C. (2003). J. Struct. Biol.142, 207–217. [DOI] [PubMed]
- Ireton, G. C. & Stoddard, B. L. (2004). Acta Cryst. D60, 801. [DOI] [PubMed]
- Luft, J. R. & DeTitta, G. T. (1999). Acta Cryst. D55, 988–993. [DOI] [PubMed]
- Mancini, E. J., Assenberg, R., Verma, A., Walter, T. S., Tuma, R., Grimes, J. M., Owens, R. J. & Stuart, D. I. (2007). Protein Sci.16, 2284–2300. [DOI] [PMC free article] [PubMed]
- Mayo, C. J., Diprose, J. M., Walter, T. S., Berry, I. M., Wilson, J., Owens, R. J., Jones, E. Y., Harlos, K., Stuart, D. I. & Esnouf, R. M. (2005). Structure, 13, 175–182. [DOI] [PubMed]
- Stura, E. A. & Wilson, I. A. (1992). Crystallization of Nucleic Acids and Proteins: A Practical Approach, edited by A. Ducruix & R. Giegé, pp. 99–126. Oxford University Press.
- Sulzenbacher, G. et al. (2002). Acta Cryst. D58, 2109–2115. [DOI] [PubMed]
- Viola, R., Carman, P., Walsh, J., Miller, E., Benning, M., Frankel, D., McPherson, A., Cudney, B. & Rupp, B. (2007). J. Appl. Cryst.40, 539–545. [DOI] [PMC free article] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]
- Walter, T. S., Diprose, J., Brown, J., Pickford, M., Owens, R. J., Stuart, D. I. & Harlos, K. (2003). J. Appl. Cryst.36, 308–314.