The high-resolution mass-spectrometric characterization, crystallization and X-ray diffraction studies of a recombinant IgE Fab fragment in complex with bovine β-lactoglobulin are reported.
Keywords: antibodies, IgE, food allergens, mass spectrometry
Abstract
A D1 Fab fragment containing the allergen-binding variable domains of the IgE antibody was characterized by ESI FT–ICR mass spectrometry and crystallized with bovine β-lactoglobulin (BLG) using the hanging-drop vapour-diffusion method at 293 K. X-ray data suitable for structure determination were collected to 2.8 Å resolution using synchrotron radiation. The crystal belonged to the orthorhombic space group P212121, with unit-cell parameters a = 67.0, b = 100.6, c = 168.1 Å. The three-dimensional structure of the D1 Fab fragment–BLG complex will provide the first insight into IgE antibody–allergen interactions at the molecular level.
1. Introduction
An allergy is an adverse immune response to normally harmless substances (allergens). It has been estimated that 20–30% of the population of developed countries suffer from allergy-related diseases, which include atopic dermatitis, rhinitis and asthma (Zuercher et al., 2006 ▶). IgEs are a class of antibodies that play a crucial role in the development of allergic symptoms. IgE exists at a low concentration in the serum of humans: compared with the normal IgG level (approximately 13.5 mg ml−1), even the most seriously allergic individuals rarely have an IgE level of greater than 1 µg ml−1 (Goldsby et al., 2003 ▶). Normally, IgE antibodies only respond as a defence against parasitic infections. The defects in IgE regulation suffered by genetically predisposed persons also invoke common environmental antigens to stimulate inappropriate IgE production. Circulating IgE sensitizes mast cells or basophiles through binding to the high-affinity receptor for IgE (Fc∊RI) located on their surface. Cross-linkage of receptor-bound IgE and multivalent allergens triggers blood basophiles and tissue mast cells to release pharmacologically active mediators such as histamine into the surrounding tissue, which causes allergic manifestations.
Food allergies are common among young children. Milk, egg, peanuts, wheat, soy, tree nuts, fish and shellfish have been noticed to be responsible for the majority of food-induced allergic reactions (Sicherer & Sampson, 2006 ▶). Food allergens are naturally existing proteins, but technological processing, digestion and heat treatment of food products can yield new allergic epitopes (Helm & Burks, 2000 ▶). Despite intensive investigations, the question of whether there are common protein features for protein allergenicity still remains unclear. However, pollen and food allergens have been found to belong to only very few of the thousands of protein families (Breiteneder & Radauer, 2004 ▶; Breiteneder & Mills, 2005 ▶; Radauer & Breiteneder, 2006 ▶).
Bovine β-lactoglobulin (BLG, Bos d 5), caseins and α-lactalbumin are the most allergenic proteins present in cow’s milk. BLG is a relatively small protein of 162 amino-acid residues that normally exists as a dimer. BLG variants A and B differ at position 64 (Asp/Gly) and 118 (Val/Ala). BLG belongs to the lipocalin family, a large group of structurally and functionally related proteins. The true physiological function of BLG is not clear, but its capability to bind retinol and fatty acids may indicate a role as a general transport protein (Flower, 1996 ▶). Much structural characterization has been performed with BLG, including the study of different crystal forms (Aschaffenburg et al., 1965 ▶) and detailed structure determinations (Brownlow et al., 1997 ▶; Oliveira et al., 2001 ▶).
The main purpose of the analysis of IgE antibody–allergen interactions is to discover important details relating to the molecular mechanisms of allergies. Here, we present the high-resolution mass-spectrometric analysis, crystallization and X-ray diffraction studies of bovine BLG in complex with a D1 Fab fragment that contains the allergen-binding variable domains from a recombinant IgE antibody isolated from a phage display library constructed from the lymphocytes of a milk-allergic patient.
2. Materials and methods
2.1. Purification of D1 Fab fragment
The BLG-specific D1 antibody was isolated from a combinatorial IgE scFv phage display library constructed from the lymphocytes of a milk-allergic patient that contains the IgE VH gene pool combined with either the κ or λ VL pool. Detailed description of the isolation and characterization of the D1 Fab fragment will be published elsewhere (Jylhä et al., manuscript in preparation). The D1 Fab fragment contains the variable domains selected from the IgE antibody phage display library with the Cκ and CH1 region of human IgG1. The Fd chain was designed to contain the human IgG1 constant region since our previous expression experiments have shown that recombinant IgE Fab fragments are produced in lower amounts in Escherichia coli (Laukkanen et al., 2003 ▶). The D1 Fab fragment was produced in the E. coli RV308 strain (ATCC 31608) by a fed-batch fermentation (4.5 l) in a Bio-Flow IV fermentor (New Brunswick) and purified by metal-affinity chromatography essentially as described by Nevanen et al. (2001 ▶). The Fab fragment was purified from the culture supernatant (4.5 l) using a Cu2+ Chelating Sepharose Streamline column (Amersham-Pharmacia), with a yield of 95 mg. The purified D1 Fab fragment was analyzed on a 15% Coomassie-stained SDS–PAGE gel under nonreducing conditions. The purity was ∼95% and the interchain disulfide bond between the light and heavy chain was formed (data not shown). The equilibrium dissociation constant (K d = 1.4 × 10−9 M) and kinetic parameters (k on = 1.4 × 106, k off = 1.54 × 10−3) determined by BIAcore indicate strong binding of the D1 Fab fragment to BLG, which is typical for IgE antibodies. The amino-acid sequences of the heavy (Fd) and light chain of the D1 Fab fragment are shown in Fig. 1 ▶.
Figure 1.
Deduced amino-acid sequences of the heavy (Fd) and light chain of the D1 Fab fragment. The complementarity-determining regions (CDRs) are underlined, the constant regions are in bold and the hexahistidine tag at the C-terminus of the heavy chain is in italics.
2.2. Mass spectrometry
All mass-spectrometric experiments were performed with a 4.7 T Bruker BioAPEX-II ESI FT–ICR mass spectrometer (Bruker Daltonics, Billerica, Massachusetts, USA), as described in detail previously (Jänis et al., 2004 ▶). Protein samples were desalted using PD-10 columns (Amersham Biosciences, Bucks, UK) and the concentrations were determined by absorbance at 280 nm using extinction coefficients of 61 590 M −1 cm−1 (D1 Fab) and 16 500 M −1 cm−1 (BLG). The samples were directly electrosprayed at a flow rate of 1.5 µl min−1; N2 (69 kPa) was used as a drying and a nebulizing gas. In order to avoid dissociation of noncovalent protein complexes, the following parameter settings were carefully optimized. The drying-gas temperature was set to 353 K, the capillary exit potential was adjusted to 70 V and the ions were accumulated in the hexapole for 250 ms. The data sets (256–1000 co-added time-domain transients, 256 or 512 kword each) were fast Fourier transformed prior to magnitude calculation and external frequency-to-m/z calibration with respect to the ions of an ES Tuning Mix (Agilent Technologies, Santa Clara, California, USA).
2.3. Crystallization
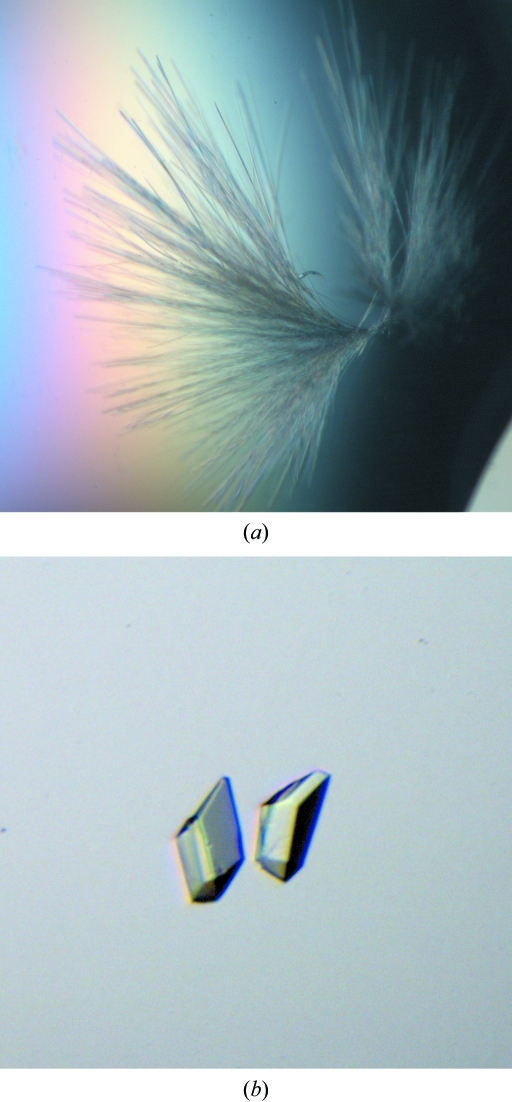
Crystallization experiments were performed using the hanging-drop vapour-diffusion method at 293 K. Crystallization droplets were prepared by mixing 2 µl D1 Fab solution (1.4 mg ml−1 in 20 mM phosphate buffer pH 7.0), 1 µl BLG (Sigma, L3908) solution (2 mg ml−1 in pure water) and 2 µl reservoir solution. The droplets were allowed to equilibrate against 500 µl reservoir solution. Initial crystallization conditions were found using the modified crystal screen PEG 3350 series for antibodies (Valjakka et al., 2000 ▶). Needle-like crystals (Fig. 2 ▶ a) appeared in the droplet containing 15%(w/v) PEG 3350, 0.1 M BTP {1,3-bis[Tris(hydroxymethyl)methylamino]propane} buffer at pH 5.5. Attempts to improve the crystal quality by varying the precipitant concentration, pH and buffer pH proved unsuccessful. In contrast, addition of detergent to the crystallization droplet had a remarkable effect on the outer morphology of the crystals. Crystals with approximate dimensions of 0.07 × 0.05 × 0.05 mm were obtained after one week by mixing 2 µl D1 Fab solution and 1 µl BLG solution with 2.5 µl reservoir solution containing 14%(w/v) PEG 3350, 0.1 M BTP pH 5.0 with the addition of 0.5 µl n-dodecyl-β-d-maltoside detergent from the Hampton Research Detergent Screening Kit 1 (Fig. 2 ▶ b).
Figure 2.
D1 Fab–BLG crystals grown (a) in the absence of detergent and (b) in the presence of detergent.
2.4. Data collection and processing
For data collection, crystals were transferred into a cryoprotectant solution [35%(w/v) PEG 3350, 0.1 M BTP pH 5.0] and mounted in nylon loops. After soaking, crystals were frozen by plunging them directly into liquid nitrogen. X-ray diffraction data were collected from the frozen crystal at 100 K using synchrotron radiation on beamline ID29 at the ESRF (Grenoble). The wavelength was 1.000 Å and the reflections were recorded with an ADSC Q315R detector. The crystal was exposed for 1 s with a 1.0° oscillation angle per frame at a crystal-to-detector distance of 400 mm. The data set was processed using XDS (Kabsch, 1993 ▶) and the true space group was determined with XPREP (SHELXTL software package). The data-collection statistics are presented in Table 1 ▶.
Table 1. X-ray data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 67.0, b = 100.6, c = 168.1 |
| Resolution (Å) | 20–2.8 (2.9–2.8) |
| No. of unique reflections | 28674 (2799) |
| Redundancy | 7.2 |
| Average I/σ(I) | 13.1 (3.5) |
| Completeness (%) | 99.8 (100) |
| Rmerge (%) | 15.9 (55.2) |
3. Results and discussion
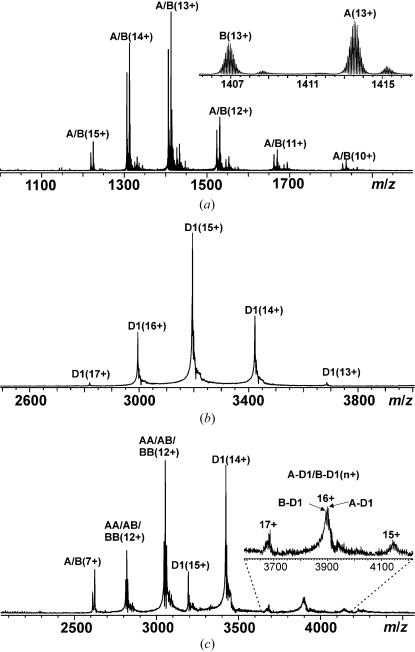
Mass spectrometry was performed to obtain information about the purity and homogeneity of the protein samples as well as to detect possible immunocomplex formation in solution. The isotopically resolved ESI FT–ICR mass spectrum of a BLG sample obtained in denaturing solution conditions [50:50(v:v) CH3CN:H2O] had two major peaks for each charge state, which corresponded to the two known BLG variants A and B (Fig. 3 ▶ a). The most abundant isotopic masses were determined to be 18 362.56 ± 0.08 Da for A (theoretical mass 18 362.45 Da) and 18 276.51 ± 0.05 Da for B (theoretical mass 18 276.41 Da). The A:B abundance ratio was ∼57:43. In native-like solution conditions (10 mM ammonium acetate buffer pH 5.0), BLG mainly existed in its dimeric form. However, at the lowest concentrations tested for BLG, peaks representing free monomers were also present, consistent with the reported K d value (∼7.0 µM) for the B variant (Sawyer & Kontopidis, 2000 ▶). The mass spectrum of the D1 Fab fragment measured in a 10 mM ammonium acetate buffer exhibited a distribution of charge states of a single protein form (Fig. 3 ▶ b), with a determined average mass of 47 911.7 ± 0.8 Da (theoretical mass 47 787.4 Da). The reason for the difference between the determined and the theoretical masses (124 Da) remained unclear. As the charge-state distribution of the D1 Fab fragment appeared in the m/z range 2800–3700, the experimentally determined masses are hereafter reported as average masses, since isotopic resolution cannot be achieved for ∼50 kDa proteins at these m/z values. The complex formation between BLG and the D1 Fab fragment was examined by measuring the mass spectrum of a nearly equimolar mixture of the two proteins in 10 mM ammonium acetate buffer (Fig. 3 ▶ c). In addition to the peaks for monomeric and dimeric BLG and the D1 Fab fragment, new peaks appeared at m/z 3600–4200, which corresponded to the charge states 15+, 16+ and 17+ of a noncovalent D1 Fab–BLG (monomer) immunocomplex. Since BLG exists as a mixture of A and B variants, two D1 Fab–BLG immunocomplexes (abbreviated A-D1 and B-D1) were detected (experimental masses 66 273.4 and 66 186.3 Da and calculated masses 66 274.0 and 66 187.9 Da for A-D1 and B-D1, respectively) with nearly the same abundance ratio as for free A and B.
Figure 3.
ESI FT–ICR mass spectra of (a) 5.0 µM BLG (variants A and B) in 50:50(v:v) CH3CN:H2O, (b) 9.4 µM D1 Fab fragment in 10 mM ammonium acetate buffer pH 5.0 and (c) a mixture of 9.4 µM D1 Fab fragment and 11.4 µM BLG in the same buffer. Numbers in parentheses (z+) denote different protein ion charge states as [m + zH]z+. The inset in (a) shows the isotopically resolved charge state 13+ for the A and B variants. The inset in (c) shows the charge states 15+, 16+ and 17+ of noncovalent D1 Fab–BLG immunocomplexes.
Crystallographic data were collected from a single crystal that diffracted to a resolution of 2.8 Å. We used the signal-to-noise ratio (3.0) to define the resolution cutoff. The crystal belonged to space group P212121, with unit-cell parameters a = 67.0, b = 100.6, c = 168.1 Å, α = β = γ = 90°. The phasing problem was solved by the molecular-replacement method with MOLREP (Vagin & Teplyakov, 1997 ▶). The human monoclonal antibody 3D6 (He et al., 1992 ▶; PDB code 1dfb) and bovine β-lactoglobulin (Oliveira et al., 2001 ▶; PDB code 1b8e) structures were used as the search models for the initial rotation-function and translation-function calculations. A preliminary molecular-replacement solution indicated the presence of one D1 Fab–allergen immunocomplex with an approximate total molecular weight of 136 kDa (two D1 Fab fragments and one BLG dimer) in an asymmetric unit. The Matthews coefficient (Matthews, 1968 ▶) calculated for one antibody–allergen immunocomplex in this space group was 2.1 Å3 Da−1, corresponding to a solvent content of 40%. The determined complex structure is expected to provide significant information on allergenic BLG epitopes in addition to special features of IgE–allergen interactions.
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron-radiation facilities and would like to thank the staff of beamline ID29 for help in the collection of data. We are also grateful to Reetta Kallio-Ratilainen and Anja Pallas for their skilful technical assistance. This work was supported by the Sigrid Jusélius Foundation, the ISB graduate school and the Academy of Finland (grants 108533 and 208920).
References
- Aschaffenburg, R., Green, D. W. & Simmons, R. M. (1965). J. Mol. Biol.13, 194–201. [DOI] [PubMed]
- Breiteneder, H. & Mills, E. N. C. (2005). J. Allergy Clin. Immunol.115, 14–23. [DOI] [PubMed]
- Breiteneder, H. & Radauer, C. (2004). J. Allergy Clin. Immunol.113, 821–830. [DOI] [PubMed]
- Brownlow, S., Morais Gabral, J. H., Cooper, R., Flower, D. R., Yewdall, S. J., Polikarpov, I., North, A. C. T. & Sawyer, L. (1997). Structure, 5, 481–495. [DOI] [PubMed]
- Flower, D. R. (1996). Biochem. J.318, 1–14. [DOI] [PMC free article] [PubMed]
- Goldsby, R. A., Kindt, T. J., Osborne, B. A. & Kuby, J. (2003). Immunology, 5th ed. New York: W. H. Freeman & Co.
- He, X. M., Ruker, F., Casale, E. & Carter, D. C. (1992). Proc. Natl Acad. Sci. USA, 89, 7154–7158. [DOI] [PMC free article] [PubMed]
- Helm, R. M. & Burks, A. W. (2000). Curr. Opin. Immunol.12, 647–653. [DOI] [PubMed]
- Jänis, J., Turunen, O., Leisola, M., Derrick, P. J., Rouvinen, J. & Vainiotalo, P. (2004). Biochemistry, 43, 9556–9566. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Laukkanen, M.-L., Mäkinen-Kiljunen, S., Isoherranen, K., Haahtela, T., Söderlund, H. & Takkinen, K. (2003). J. Immunol. Methods, 278, 271–281. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Nevanen, T. K., Söderholm, L., Kukkonen, K., Suortti, T., Teerinen, T., Linder, M., Söderlund, H. & Teeri, T. T. (2001). J. Chromatogr. A, 925, 89–97. [DOI] [PubMed]
- Oliveira, K. M. G., Valente-Mesquita, V. L., Botelho, M. M., Sawyer, L., Ferreira, S. T. & Polikarpov, I. (2001). Eur. J. Biochem.268, 477–483. [DOI] [PubMed]
- Radauer, C. & Breiteneder, H. (2006). J. Allergy Clin. Immunol.117, 141–147. [DOI] [PubMed]
- Sawyer, L. & Kontopidis, G. (2000). Biochim. Biophys. Acta, 1482, 136–148. [DOI] [PubMed]
- Sicherer, S. H. & Sampson, H. A. (2006). J. Allergy Clin. Immunol.117, S470–S475. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Valjakka, J., Hemminki, A., Teerinen, T., Takkinen, K. & Rouvinen, J. (2000). Acta Cryst. D56, 218–221. [DOI] [PubMed]
- Zuercher, A. W., Fritsché, R., Corthésy, B. & Mercenier, A. (2006). Curr. Opin. Biotechnol.17, 198–203. [DOI] [PubMed]