UlaG, the putative l-ascorbate-6-phosphate lactonase encoded by the ulaG gene from the utilization of l-ascorbate regulon in E. coli, has been cloned, overexpressed, purified using standard chromatographic techniques and crystallized in a monoclinic space group. Crystals were obtained by the sitting-drop vapour-diffusion method at 293 K. A data set diffracting to 3 Å resolution was collected from a single crystal at 100 K.
Keywords: UlaG, l-ascorbate metabolism, enterobacterial metabolism
Abstract
UlaG, the putative l-ascorbate-6-phosphate lactonase encoded by the ulaG gene from the utilization of l-ascorbate regulon in Escherichia coli, has been cloned, overexpressed, purified using standard chromatographic techniques and crystallized. Crystals were obtained by sitting-drop vapour diffusion at 293 K. Preliminary X-ray diffraction analysis revealed that the UlaG crystals belonged to the monoclinic space group C2, with unit-cell parameters a = 104.52, b = 180.69, c = 112.88 Å, β = 103.26°. The asymmetric unit is expected to contain six copies of UlaG, with a corresponding volume per protein weight of 2.16 Å3 Da−1 and a solvent content of 43%.
1. Introduction
Escherichia coli UlaG is a representative member of the so-called UlaG family, members of which have only been found in 11 species from three enterobacteria genera: Escherichia, Salmonella and Shigella. In E. coli, the gene encoding UlaG (Genbank P39300; ULAG_ECOLI), denoted ulaG, is located in one of the two divergently transcribed operons which constitute the utilization of l-ascorbate (ula) regulon (Campos et al., 2002 ▶). The ula regulon encodes most proteins involved in the anaerobic uptake and phosphorylation of l-ascorbate and the first steps of its catabolism, as well as a transcription repressor, UlaR. Although UlaG has been reported to be essential for cell viability under anaerobic conditions when l-ascorbate is the only available carbon source (Zhang et al., 2003 ▶), little is known about its exact catalytic activities. UlaG has been tentatively described as an l-ascorbate-6-phosphate lactonase (EC 3.1.1.–) and proposed to catalyze the first step in l-ascorbate-6-phosphate degradation: its transformation to 3-dehydro-l-gulonate-6-phosphate (Zhang et al., 2003 ▶). It also exhibits phosphodiesterase activity in vitro, hydrolyzing a phosphodiester bond in the artificial chromogenic substrate bis-p-nitrophenyl phosphate (bis-pNPP; Kuznetsova et al., 2005 ▶). In order to elucidate the role of UlaG in the anaerobic metabolism of l-ascorbate in prokaryotes, we have undertaken the determination of its three-dimensional structure. The structure of UlaG in complex with potential ligands and inhibitors should provide valuable information about substrate specificity.
In this work, we report the successful cloning, expression, purification and crystallization of UlaG from E. coli, as well as the preliminary characterization of UlaG crystals by X-ray crystallographic methods.
2. Methods and results
2.1. Cloning and recombinant expression
The 1062 bp gene encoding full-length UlaG (354 amino-acid residues, 40 061 Da) was amplified by PCR using genomic DNA from E. coli K12 strain ECL1 (Lin, 1976 ▶) as a template. The forward and reverse primers were 5′-CCGGAATTCATTAAAGAGGAGAAATTAACCATGGCGATGAGTAAAGTGAAAAG-3′ and 5′-GAA GATCTCAGGATGACTTGAACGGCAGATC-3′, respectively. The amplified fragment was digested with EcoRI and BglII (shown in bold) and ligated into similarly digested pQE60 (Qiagen) with a C-terminal hexahistidine tag in frame. The resultant plasmid, pQE60-UlaG, was verified by sequencing. For expression, pQE60-UlaG was transformed into Escherichia coli M15(pREP4) strain (Qiagen). Overnight starter cultures inoculated with freshly transformed cells (typically less than two weeks old) were diluted 50-fold in 1 l Superior Broth (SB, AthenaES) supplemented with 100 mg ml−1 ampicillin and 50 mg ml−1 kanamycin and grown at 300 K for 3–4 h at 250 rev min−1. On reaching an OD600 of ∼0.6, the culture was transferred to a 293 K shaker set at 220 rev min−1 and induction was brought about 1 h later with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG). Cells were harvested after 4 h by centrifugation at 5000g for 30 min at 277 K, flash-frozen in liquid nitrogen and stored at 193 K prior to further processing.
2.2. Protein purification
The cell pellet was resuspended in four times the cell wet weight (∼20 ml per litre of culture) in lysis buffer containing 50 mM Tris pH 8, 500 mM NaCl, 20 mM imidazole, 30 µg ml−1 lysozyme, 20 µg ml−1 DNase I, 10 mM MgSO4, one tablet of EDTA-free Complete protease-inhibitor cocktail (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 2 mM β-mercaptoethanol. After 20 min incubation at 298 K, the cell suspension was sonicated for ten cycles (each cycle consisting of 45 s continuous operation followed by a 15 s pause interval). The crude lysate was clarified by centrifugation twice at 60 000g for 30 min at 278 K and filtration through a 0.45 µm membrane. UlaG was purified by metal-affinity chromatography using a 5 ml HisTrap column charged with Ni2+ (GE Healthcare) followed by size-exclusion chromatography (Superdex 200 10/300 GL column, GE Healthcare). A peak containing pure UlaG eluted with a molecular weight of ∼250 kDa, which corresponds to a highly oligomeric species, most likely a hexamer. Peak fractions containing pure UlaG, as judged from Coomassie-stained SDS–PAGE gels, were pooled and dialyzed against a buffer containing 20 mM Tris pH 8, 500 mM NaCl and 2 mM DTT. Dynamic light scattering using a Zetasizer Nano ZS instrument (Malvern) also indicated that the most likely quaternary structure of UlaG in solution was a hexamer, with an estimated molecular weight of 256 kDa and a polydispersity index of 23%. The protein concentration was determined from the absorbance at 280 nm.
2.3. Crystallization
Prior to setting up crystallization trials, UlaG was concentrated to 7.5 mg ml−1 in storage buffer (20 mM HEPES pH 7.4, 500 mM NaCl, 2 mM β-mercaptoethanol) and assayed with a pre-crystallization test (Hampton Research) in 24-well plates. Briefly, drops consisting of 1 µl UlaG at 7.5 mg ml−1 and 1 µl of one of four crystallization conditions [A1, 100 mM Tris pH 8.5, 2 M ammonium sulfate; B1, 100 mM Tris pH 8.5, 1 M ammonium sulfate; A2, 100 mM Tris pH 8.5, 0.2 M MgCl2.6H2O, 30%(w/v) polyethylene glycol 4000; B2, 100 mM Tris pH 8.5, 0.2 M MgCl2.6H2O, 15%(w/v) polyethylene glycol 4000] were arranged in a hanging-drop vapour-diffusion experiment over 500 µl reservoir solution. Within 1 h fine granular precipitates had developed in condition A2; they also appeared in the other conditions in 1 d. This suggested that the initial concentration was suitable for crystallization screenings. Crystals were readily obtained in various crystallization conditions from a coarse initial screening of ∼100 conditions performed in 24-well plates. The initial crystallization conditions were improved by optimizing the sample and precipitant concentrations as well as the pH, additive reagents and sample/reservoir solution volumes. Alternative reservoir solutions (1, 1.5 or 2 M NaCl) were also tried (but did not provide any improvement). The most suitable crystals for X-ray analysis were obtained with the sitting-drop vapour-diffusion method by mixing 1 µl sample solution (with a concentration of 7.5 mg ml−1) and 1 µl reservoir solution consisting of 20%(w/v) polyethylene glycol 3350, 200 mM lithium citrate at 293 K. The mixed sample was equilibrated against 500 µl reservoir solution. The crystals reached maximum dimensions of ∼80 × 50 × 25 µm within 3 d (Fig. 1 ▶).
Figure 1.
A representative native crystal of E. coli UlaG.
2.4. X-ray diffraction data collection and processing
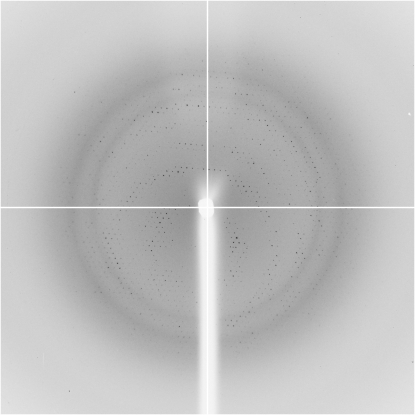
Prior to diffraction data collection, the crystals were cryoprotected by gently adding sterile-filtered glycerol directly to the crystallization drop to a final concentration of 20%(v/v) and letting it equilibrate for at least 24 h before flash-cooling in liquid nitrogen. A complete native X-ray diffraction data set was collected to ∼3 Å resolution from a single crystal using cryocooled (100 K) crystals at ID14-3, ESRF (Grenoble, France) (Fig. 2 ▶). The diffraction data were collected using 0.5° oscillations with 60 s exposure and no attenuation, with a crystal-to-detector distance of 237.27 mm (equivalent to 2.5 Å maximum resolution at the edge of the detector). The data were indexed, processed and scaled with XDS (Kabsch, 1993 ▶). The data-collection statistics are summarized in Table 1 ▶. The crystals belong to the monoclinic space group C2, with unit-cell parameters a = 104.52, b = 180.69, c = 103.26 Å, β = 103.26°.
Figure 2.
X-ray diffraction image from a native UlaG crystal. The edge of the detector corresponds to a resolution of 2.5 Å.
Table 1. X-ray data-collection and processing statistics.
Values in parentheses are for the highest resolution shell (3.15–3.07 Å).
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 104.52, b = 180.69, c = 112.88, α = 90, β = 103.826, γ = 90 |
| Wavelength (Å) | 0.931 |
| Resolution range (Å) | 46.8–3.07 (3.15–3.07) |
| Measured reflections | 164017 |
| Unique reflections | 40591 |
| Completeness (%) | 96.7 (92.0) |
| Mean I/σ(I) | 9.7 (3.5) |
| Rmerge† (%) | 14.4 (53.2) |
R
merge = 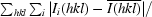
 , where Ii(hkl) is the ith intensity measurement of reflection h including symmetry-related reflections and
, where Ii(hkl) is the ith intensity measurement of reflection h including symmetry-related reflections and 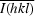 denotes the average.
denotes the average.
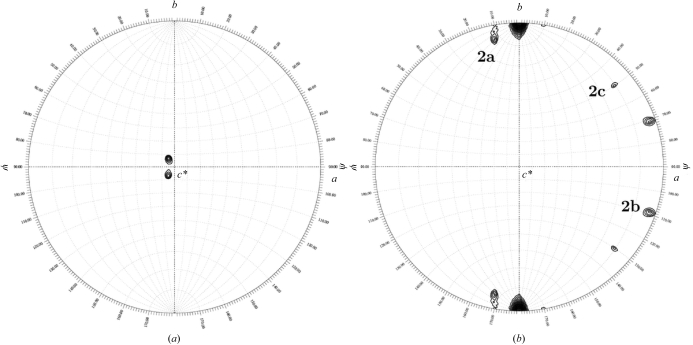
Cell-content analysis with Phaser (Storoni et al., 2004 ▶) reveals that the asymmetric unit is most likely to contain six copies of UlaG, with a crystal volume per protein weight of 2.16 Å3 Da−1 and a corresponding solvent content of 43%. Self-rotation calculations with GLRF (Tong & Rossmann, 1997 ▶) show a strong peak in the κ = 120° section (contoured from 7σ) at (ϕ, ψ) = (95°, 97°), indicating the presence of a noncrystallographic threefold axis (Fig. 3 ▶ a). A self-rotation function calculation with AMoRe (Navaza, 1994 ▶) with identical parameters shows that the threefold local symmetry axis is tilted 8.7° with respect to the normal to the xy plane. In addition, the κ = 180° section reveals three twofold axes perpendicular to the local threefold axis at (ϕ, ψ) = (152°, 12°), (2°, 109°) and (11°, 50°), consistent with a molecular 32 point group (Fig. 3 ▶ b). These observations are compatible with the existence in the asymmetric unit of a trimer of UlaG dimers in a pseudo-hexameric arrangement, with the local symmetry axis tilted 8.7° with respect to the plane normal to the crystallographic twofold axis. The asymmetric unit contains one biological unit in solution as shown by size-exclusion chromatography and dynamic light scattering.
Figure 3.
Self-rotation function calculated using UlaG crystals. A self-rotation search with κ = 120° was used to identify the noncrystallographic threefold axis of symmetry (a). The presence of two sets of three twofold axes in the xy plane clearly apparent at κ = 180° (labelled 2a, 2b and 2c) indicates that the UlaG oligomer has 32 point-group symmetry (b). Calculations and plots were performed and produced using GLRF using a Patterson integration radius of 38 Å and data between 15 and 4 Å resolution.
Attempts to solve the structure of E. coli UlaG by either molecular-replacement procedures or by anomalous diffraction using selenomethionine-labelled UlaG are in progress.
Acknowledgments
The authors are grateful to Alexander Popov and Dominique Bourgeois for assistance during data collection at ID14-3 (ESRF). This work was supported by grants from the Spanish Ministry of Science and Education (MEC): BFU 2006-15573 to MCV, BFU 2004-03586/BMC to JB and GEN2003-20642 to MC. FG was the recipient of a predoctoral fellowship from the Generalitat de Catalunya. FJF was supported by an I3P postdoctoral contract from MEC.
References
- Campos, E., Aguilar, J., Baldoma, L. & Badia, J. (2002). J. Bacteriol.184, 6065–6068. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kuznetsova, E., Proudfoot, M., Sanders, S. A., Reinking, J., Savchenko, A., Arrowsmith, C. H., Edwards, A. M. & Yakunin, A. F. (2005). FEMS Microbiol. Rev.29, 263–279. [DOI] [PubMed]
- Lin, E. C. (1976). Annu. Rev. Microbiol.30, 535–578. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed]
- Zhang, Z., Aboulwafa, M., Smith, M. H. & Saier, M. H. (2003). J. Bacteriol.185, 2243–2250. [DOI] [PMC free article] [PubMed]