The structure of melon necrotic spot virus is reported.
Keywords: Tombusviridae, tombusviruses, carmoviruses, coat proteins
Abstract
The structure of melon necrotic spot virus (MNSV) was determined at 2.8 Å resolution. Although MNSV is classified into the genus Carmovirus of the family Tombusviridae, the three-dimensional structure of MNSV showed a higher degree of similarity to tomato bushy stunt virus (TBSV), which belongs to the genus Tombusvirus, than to carnation mottle virus (CMtV), turnip crinkle virus (TCV) or cowpea mottle virus (CPMtV) from the genus Carmovirus. Thus, the classification of the family Tombusviridae at the genus level conflicts with the patterns of similarity among coat-protein structures. MNSV is one of the viruses belonging to the genera Tombusvirus or Carmovirus that are naturally transmitted in the soil by zoospores of fungal vectors. The X-ray structure of MNSV provides us with a representative structure of viruses transmitted by fungi.
1. Introduction
Melon necrotic spot virus (MNSV) is a small icosahedral virus (approximately 30 nm in diameter) that belongs to the genus Carmovirus of the family Tombusviridae. In this family, the virus particle consists of a protein shell, a single strand of genomic RNA and two short nongenomic RNAs. The protein shell is composed of 180 copies of chemically identical coat-protein subunits and is arranged in a T = 3 icosahedral lattice. The MNSV genome encodes five proteins, p29, p89, p7A, p7B and p42 (Genoves et al., 2006 ▶), but only p42 is included in the virus particle.
The structures of several viruses of the family Tombusviridae have already been reported. From the genus Tombusvirus, the structure of tomato bushy stunt virus (TBSV) has been determined at 2.9 Å (Olson et al., 1983 ▶; Harrison et al., 1978 ▶). From the genus Carmovirus, the capsid protein structure of carnation mottle virus (CMtV; 3.2 Å; Morgunova et al., 1994 ▶), the viral structure of turnip crinkle virus (TCV; 3.2 Å; Hogle et al., 1986 ▶) and the viral structure of cowpea mottle virus (CPMtV; 7.0 Å; Ke et al., 2004 ▶) are known. In the genus Necrovirus of the family Tombusviridae, the structure of tobacco necrosis virus (TNV) has also been determined (2.24 Å; Oda et al., 2000 ▶). The structure of TNV has no protruding domain (P-domain) and is obviously different from those of the genera Tombusvirus and Carmovirus.
In a recent study, characteristic structures of the coat proteins of the genera Tombusvirus and Carmovirus were precisely compared with each other (Ke et al., 2004 ▶). Despite their low sequence homology, they share highly homologous three-dimensional structures. The coat proteins of these viruses consist of the same three domains: the RNA-binding domain (R-domain), the surface domain (S-domain) and the P-domain. Ke and coworkers summarized the structural features of carmoviruses in comparison with tombusviruses as follows. (i) Carmoviruses have a different P-domain orientation relative to the S-domain to that found in tombusviruses. The relative angle between the S-domains and P-domains of carmoviruses is smaller by about 10° than that found in TBSV. (ii) Carmoviruses lack the nine C-terminal residues that interact with the adjacent P-domain around the twofold axis of TBSV. The total surface-contact areas between adjacent P-domains in carmoviruses are significantly smaller than those in TBSV. This appears to be because the carboxyl-terminus of TBSV, which forms an extensive contact, is longer than that of carmoviruses by nine residues. (iii) Carmoviruses have a deletion in the βC strand of the S-domain.
MNSV was classified into the genus Carmovirus based on its genome organization and probable translation strategy (Riviere & Rochon, 1990 ▶; Lommel et al., 2005 ▶). However, a sequence-alignment study of coat proteins has shown that MNSV has higher similarity to the genus Tombusvirus than to the genus Carmovirus (Riviere et al., 1989 ▶). In contrast to the viruses of known three-dimensional structures from the genera Carmovirus and Tombusvirus, MNSV is transmitted by a fungal vector. Of the 14 recognized species of the genus Carmovirus, only three viruses [MNSV, cucumber soil-borne virus (CSBV) and squash necrosis virus (SqNV)] are transmitted by fungal vectors. Of the 13 recognized species in the genus Tombusvirus, only cucumber necrosis virus (CNV) is transmitted by a fungal vector (Rochon et al., 2004 ▶). Recently, the mechanism of infection by zoospores has been studied for CNV. The infection process in the fungal vector is mediated by an interaction between a coat protein and a specific zoospore receptor (Robbins et al., 1997 ▶). It has been suggested that CNV infects the zoospore through a specific glycoprotein receptor (Kakani et al., 2003 ▶). Based on the TBSV structure, the predicted structure of CNV suggests that the receptor-binding site of CNV is located near the viral quasi-threefold axis (Kakani et al., 2001 ▶). MNSV and CNV are transmitted by the same fungal vector (Olpidium bornovanus) and MNSV can competitively inhibit CNV infection (Kakani et al., 2001 ▶), suggesting that MNSV shares a zoospore receptor with CNV.
In this study, we determined the structure of MNSV at 2.8 Å resolution in order to make comparisons with members of the genera Carmovirus and Tombusvirus and to elucidate the structural features of MNSV as a representative structure of viruses of the genera Tombusvirus and Carmovirus that are transmitted by fungal vectors.
2. Materials and methods
2.1. Purification of MNSV
MNSV KS strain (Kubo et al., 2005 ▶) was purified from melon cotyledons (Cucumis melo L.) inoculated with the viral strain. Frozen materials were homogenized in 0.05 M sodium phosphate buffer pH 7.0 including 0.02 M ascorbic acid and chloroform using a Waring blender. The material was clarified by centrifugation at 8500g for 10 min and the supernatant was adjusted to pH 5.0 by the addition of HCl. After further centrifugation, the supernatant was ultracentrifuged to precipitate viral particles. The precipitant was suspended in 0.05 M sodium phosphate buffer pH 7.0 and purification of the viral particle was achieved by sucrose-density gradient centrifugation on 10–40%(w/v) sucrose. To obtain more highly purified viral particles, the viral fraction was subjected to sucrose-density gradient centrifugation on 25–55%(w/v) sucrose. Finally, the virus fraction was concentrated by ultracentrifugation and stored at 277 K. Purified particles were negatively stained with 2% uranyl acetate and verified by electron microscopy.
2.2. Crystallization
Crystallization was carried out at 277 K using the hanging-drop vapour-diffusion method. The concentration of purified MNSV was adjusted to 14.3 mg ml−1 in 10 mM sodium phosphate buffer pH 7.0. The crystals most suitable for X-ray analysis were obtained when a 2 µl drop containing 10%(w/v) virus, 100 mM sodium phosphate buffer pH 7.0 and 4.5%(w/v) PEG 2000 was equilibrated with a reservoir solution containing 100 mM sodium phosphate buffer pH 7.0 and 9%(w/v) PEG 2000. Crystals grew to dimensions of 0.5 × 0.5 × 0.3 mm within a week. The crystals belong to the cubic space group I23, with unit-cell parameters a = b = c = 375 Å.
2.3. X-ray diffraction experiments
X-ray diffraction data were collected from native crystals using a DIP-6040 imaging-plate detector (MacScience/Bruker AXS, Yokohama, Japan) on beamline BL44XU at SPring-8. The X-rays were monochromated with an Si(111) monochrometer. The wavelength of the X-rays was tuned to 0.90 Å and collimated to a size of 50 × 50 µm. The camera distance was 600 mm in order to separate the diffraction spots. All data sets were collected at 100 K. The intensity data were processed and integrated using the MOSFLM program (Leslie, 1992 ▶) and scaled using the SCALA program (Collaborative Computational Project, Number 4, 1994 ▶). Intensity data statistics are summarized in Table 1 ▶.
Table 1. Summary of diffraction data and refinement statistics.
Values in parentheses are for the outer shell.
| Data-collection parameters | |
| X-ray source | BL44XU, SPring-8 |
| Wavelength () | 0.9 |
| Resolution range () | 152.502.79 (2.942.79) |
| No. of observed reflections | 2846632 (229963) |
| No. of unique reflections | 208594 (24692) |
| Space group | I23 |
| Unit-cell parameters () | a = b = c = 375 |
| R merge † (%) | 16.4 (60.8) |
| Completeness (%) | 99.2 (80.2) |
| Refinement statistics | |
| No. of protein atoms | 35255 |
| No. of water molecules | 39 |
| No. of calcium ions | 20 |
| R factor (%) | 20.84 |
| Free R factor (%) | 22.38 |
| Mean B value (2) | 32.5 |
| R.m.s.d.s from ideal values | |
| Bond lengths () | 0.01 |
| Bond angles () | 1.3 |
| Ramachandran plot | |
| Residues in allowed regions (%) | 100 |
| Residues in disallowed region (%) | 0 |
R
merge = 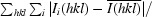
 , where I
i(hkl) is the intensity of the measured reflection i and
, where I
i(hkl) is the intensity of the measured reflection i and  is the averaged I(hkl) of equivalent reflections.
is the averaged I(hkl) of equivalent reflections.
2.4. Structure determination and refinement
The three-dimensional structure of MNSV was determined by the molecular-replacement (MR) method using the X-ray structure of TBSV (PDB code 2tbv). The initial σA-weighted electron-density map was improved by solvent flattening, histogram matching and the noncrystallographic symmetry (NCS) averaging technique using the DM program (Cowtan & Main, 1996 ▶) from the CCP4 suite. The modified electron-density map revealed the main chain and most of the side chains. The structural model was built using the Coot program (Emsley & Cowtan, 2004 ▶) and refined using REFMAC5 (Collaborative Computational Project, Number 4, 1994 ▶) under the restraint of noncrystallographic fivefold symmetry. The structural refinement was monitored by an R factor evaluated for reflections used in refinement and a free R evaluated for reflections excluded from refinement. The quality of the final model was assessed using PROCHECK (Laskowski et al., 1993 ▶). CCP4mg (Potterton et al., 2004 ▶) and UCSF Chimera (Pettersen, 2004 ▶) were used to generate the figures. The contacting surface area within the P-domain dimer interface was calculated using the AREAIMOL program (Lee & Richards, 1971 ▶) in CCP4. Structure-based sequence alignment was performed using the DALI server (Holm & Park, 2000 ▶).
3. Results and discussion
3.1. Quality of the structure
The crystallographic asymmetric unit of MNSV contained five icosahedral asymmetric units, each comprising one-twelfth of the whole virus particle and composed of 180 chemically identical protein subunits arranged in a T = 3 lattice. Three subunits (A, B and C molecules) are present in each icosahedral asymmetric unit. The electron density refined by the noncrystallographic symmetry averaging method coupled with the density-modification method was interpreted in terms of an atomic model. Of the 390 amino-acid residues of each subunit, 296 residues of the A subunit, 296 residues of the B subunit and 331 residues of the C subunit were unambiguously located in the electron-density map. A total of 7051 protein atoms, four calcium ions and 41 water molecules were included in the structural model. The structure was successfully refined to an R factor of 20.8% and a free R factor of 22.4% at 2.8 Å resolution. The Ramachandran plots for the final structural model indicated that 88.1% of nonglycine residues were in the most favoured region, 11.8% were in additional favoured regions and no residues were in disallowed regions.
The A, B and C subunits exhibited slightly different conformations from their counterparts in other T = 3 viruses. The C subunit had additional ordered structure in the N-terminal region in comparison with the A and B subunits (Fig. 1 ▶). Of the 390 residues in each subunit, residues 1–94 of subunits A and B and 1–59 of subunit C were disordered in the inner part of the capsid. No electron density was assigned to RNA. Large peaks clearly visible in the electron-density map at subunit-contact regions were assigned to calcium ions after structural refinement, based on their temperature factors and coordination geometries. MNSV had four calcium ions in each icosahedral asymmetric unit and their binding sites are conserved in TBSV (Fig. 2 ▶; Olson et al., 1983 ▶). Most T = 3 plant viruses contain calcium ions, which promote virus assembly and stabilize the virus particle (Robinson & Harrison, 1982 ▶). As in other viruses, the calcium ions in MNSV may stabilize the viral structure.
Figure 1.
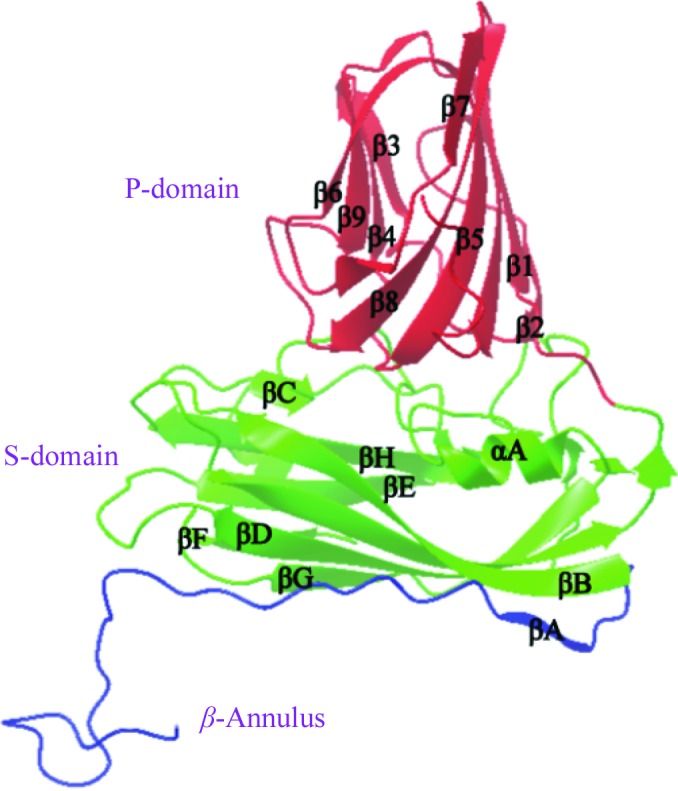
The coat-protein structure of MNSV: a ribbon drawing of the C subunit of MNSV with β-sheets and α-helices labelled. It consists of the P-domain (red) and S-domain (green) and a β-annulus structure at the N-terminus (blue).
Figure 2.
Calcium ions in subunit interfaces. (a) A coat-protein trimer consisting of A, B and C subunits drawn using a ribbon-and-wire model; calcium ions between two subunits are represented by grey balls. The coordination structures of the calcium ions are depicted in (b), (c) and (d). Calcium ions are shown as grey balls and residues coordinating to calcium ions are shown. Each coordination bond is drawn by a broken line and labelled with the interatomic distance. (b) Coordination structure of the calcium ion between the A and B subunits. (c) Coordination structure of those between the B and C subunits. (d) Coordination structure of that between the C and A subunits. The coordination-bond distances around each calcium ion are in the range 2.2–2.9 Å.
3.2. The coat-protein structure of MNSV
These results were consistent with the identity ratios of primary structure. Each MNSV coat protein exhibited the β-barrel jelly-roll motif (Rossmann et al., 1983 ▶) common in T = 3 plant viruses, which include viruses from the genera Tombusvirus and Carmovirus. Each of the three subunits A, B and C contained two domains: the S-domain (167 residues from Ser95 to Pro261) and the P-domain (129 residues from Ile262 to Ala390). These domains are connected by a flexible hinge region (four residues from Ile262 to Met265). As observed in TBSV and other T = 3 plant viruses, only the C subunit had a β-annulus (Asn60–Gly94; Fig. 1 ▶); this interacted with other β-annuli related by the threefold axis. The R-domain (Met1–Gly94) was disordered and is thought to interact with RNA inside the viral particle. The S-domain consisted of nine antiparellel β-sheets and two α-helices. Each of the secondary structures was nearly identical to that of TBSV, with an identical length (167 amino acids). The MNSV P-domain also had the jelly-roll structure typical of other viruses of the genera Tombusvirus and Carmovirus. The structure of the P-domain consisted of two antiparallel β-sheets and no α-helix.
To compare the tertiary structures of the two proteins, one structure was superposed on the other by least-squares fitting of corresponding Cα atoms using the DALI server. The structural difference between two molecules was evaluated using the root-mean-square deviation (r.m.s.d.) of Cα atoms. The r.m.s.d.s of the S-domains were 1.0 Å for MNSV/TBSV, 1.4 Å for MNSV/CMtV and 1.3 Å for TBSV/CMtV. Those of the P-domain were 2.0 Å for MNSV/TBSV, 2.3 Å for MNSV/CMtV and 2.2 Å for TBSV/CMtV. The structures of the S-domains were well conserved, consistent with the fact that the sequence identity for the S-domains is 10–30% higher than that for the P-domains.
Three β-annuli from the C subunits formed three antiparallel β-sheets that stabilized interaction around the threefold axis. Compared with that of TBSV, the β-annulus of MNSV had one more residue (Asn60) included in a β-sheet. As in other plant viruses, the structure of the β-annulus around the threefold axis may play an important role in virus assembly (Sorger et al., 1986 ▶; Satheshkumar et al., 2005 ▶).
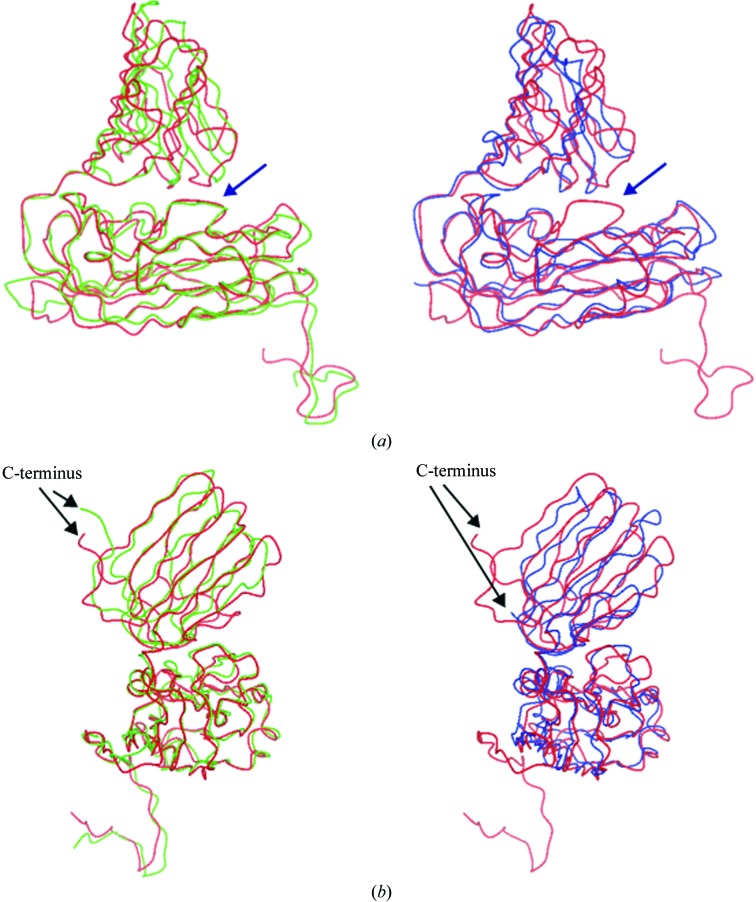
3.3. Comparison with other carmoviruses
The overall structure of MNSV showed that the angle of the P-domain relative to the S-domain was almost the same as for the tombusvirus TBSV and about 10° larger than that of carmoviruses (Fig. 3 ▶ a). MNSV has nine C-terminal residues that extend vertically from the capsid surface, just as in TBSV (Fig. 3 ▶ b). These residues are included in the P-domain dimeric interface, as in TBSV. The total surface-contact area of a P-domain with the adjacent P-domain in MNSV is 1470 Å2, which is comparable to that of TBSV (1570 Å2) and significantly larger than that of another carmovirus, CMtV (866 Å2). MNSV had almost the same βC-strand length as that of TBSV; this strand is deleted in all members of the genus Carmovirus except for MNSV. Consequently, the three-dimensional structure of MNSV was more similar to that of a tombusvirus than that of a carmovirus. These results are consistent with the sequence-alignment study, which also showed MNSV to be more similar to a tombusvirus than to a carmovirus. Thus, the classification of the family Tombusviridae at the genus level conflicts with the similarity of the primary and tertiary structures of the coat proteins.
Figure 3.
Comparison of some of the structural features of the capsid-protein structure. Shown here are images of the C subunit of the TBSV (a tombusvirus) and CMtV (a carmovirus) aligned with that of MNSV shown as Cα backbones coloured green, blue and red, respectively. (a) Comparison of βC insertions and hinge angles. Blue arrows represent the insertion at βC. (b) The C-termini of the three viruses. In MNSV and TBSV, the C-terminal residues make extensive interactions with the adjacent P-domains.
3.4. Structural comparison of MNSV with TBSV
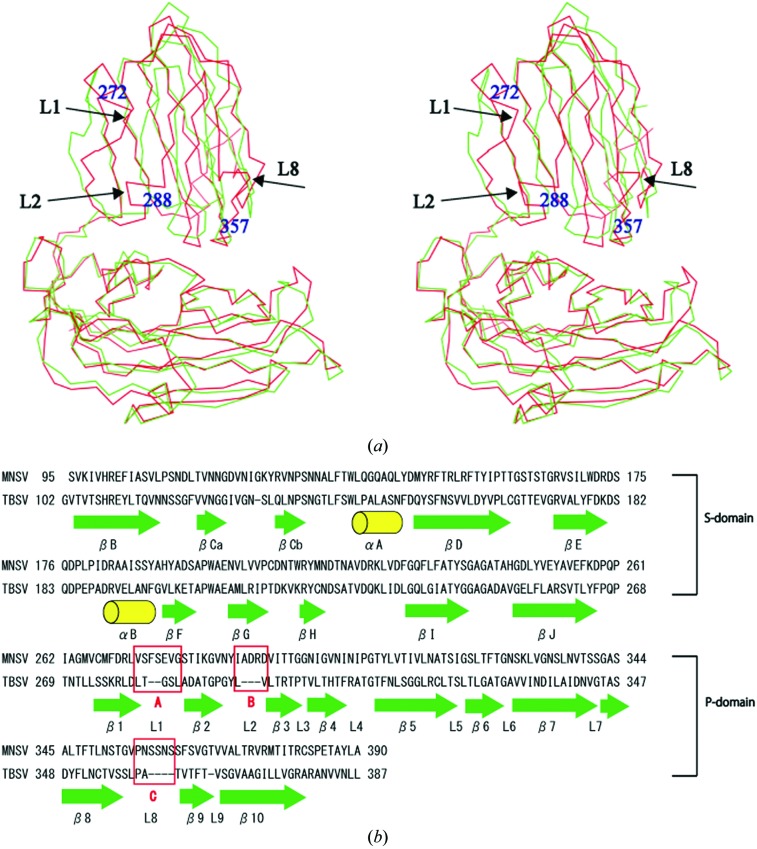
The r.m.s.d. of Cα atoms between the MNSV and TBSV proteins was 1.8 Å. When the superpositions were performed separately for the S-domain and P-domain, the r.m.s.d.s for the S-domain and P-domain were 1.0 and 2.0 Å, respectively. The sequences of MNSV and TBSV were aligned taking their tertiary structural homology into account. The sequence identity between the MNSV protein and the TBSV protein was 29% for S-domains and P-domains together, 42% for the S-domain alone and 16% for the P-domain alone; i.e. the S-domains of the two viruses were more conserved in both primary and tertiary structure than the P-domains. The P-domain of MNSV, with 129 amino acids, is ten residues longer than that of TBSV. The difference comprises insertions in several loop regions of MNSV: two residues in loop L1 (272–278), three residues in loop L2 (288–291) and four residues in loop L8 (357–362) (Figs. 4 ▶ a and 4 ▶ b).
Figure 4.
Comparison of the coat-protein structures of MNSV and TBSV. (a) Shown here are stereo images of the A subunits of MNSV (red) and TBSV (green). Arrows represent insertion sequences. (b) Structure-based alignment of the coat proteins of MNSV and TBSV. Green arrows represent β-sheet structure and yellow cylinders represent α-helices. The red letters A (272–278), B (288–291) and C (357–362) represent insertions.
MNSV is transmitted by the same fungal vector as CNV (Rochon et al., 2004 ▶), while TBSV is not transmitted by a fungus. In the case of CNV, fungal vector transmission is mediated by molecular interaction between the coat protein and a fungal receptor (Kakani et al., 2001 ▶, 2003 ▶). However, structural comparison between MNSV and TBSV has not successfully elucidated any specific residue that interacts with the fungal receptor at present.
4. Conclusions
In this study, we determined the structure of MNSV at 2.8 Å resolution and found it to be more similar to that of a tombusvirus than to that of a carmovirus, even though MNSV is classified as a carmovirus based on its genome organization, probable translation strategy and protein sequence similarity (Riviere & Rochon, 1990 ▶). Our findings suggest that the structural features of the coat proteins must be considered as one of the criteria for the classification of viruses. The 2.8 Å structure of MNSV will be useful as a representative structure of the viruses of the genera Carmovirus and Tombusvirus that are transmitted by fungal vectors.
Supplementary Material
PDB reference: melon necrotic spot virus, 2zah, r2zahsf
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cowtan, K. D. & Main, P. (1996). Acta Cryst. D52, 43–48. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Genoves, A., Navarro, J. A. & Pallas, V. (2006). J. Gen. Virol. 87, 2371–2380. [DOI] [PubMed]
- Harrison, S. C., Olson, A. J., Schutt, C. E. & Winsker, F. K. (1978). Nature (London), 276, 368–373. [DOI] [PubMed]
- Hogle, J. M., Maeda, A. & Harrison, S. C. (1986). J. Mol. Biol. 191, 625–638. [DOI] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Kakani, K., Robbins, M. & Rochon, D. (2003). J. Virol. 77, 3922–3928. [DOI] [PMC free article] [PubMed]
- Kakani, K., Sgro, J. Y. & Rochon, D. M. (2001). J. Virol. 75, 5576–5583. [DOI] [PMC free article] [PubMed]
- Ke, J., Schmidt, T., Chase, E., Bozarth, R. F. & Smith, T. J. (2004). Virology, 321, 349–358. [DOI] [PubMed]
- Kubo, C., Nakazono-Nagaoka, E., Hagiwara, K., Kajihara, H., Takeuchi, S., Matsuo, K., Ichiki, T. U. & Omura, T. (2005). Plant Pathol. 54, 615–620.
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lee, B. & Richards, M. (1971). J. Mol. Biol. 55, 379–400. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Lommel, S. A., Martelli, G. P., Rubino, L. & Russo, M. (2005). Eighth Report of the International Committee on Taxonomy of Viruses, edited by C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger & L. A. Ball, pp. 907–936. New York: Academic Press.
- Morgunova, E. Y., Dauter, Z., Fry, E., Stuart, D. I., Stelmashchuk, V. Y., Mikhailovy, A. M., Wilson, K. S. & Vainshtein, B. K. (1994). FEBS Lett. 338, 267–271. [DOI] [PubMed]
- Oda, Y., Saeki, K., Takahashi, Y., Maeda, T., Naitow, H., Tsukihara, T. & Fukuyama, K. (2000). J. Mol. Biol. 300, 153–169. [DOI] [PubMed]
- Olson, A. J., Bricogne, G. & Harrison, S. C. (1983). J. Mol. Biol. 171, 61–93. [DOI] [PubMed]
- Pettersen, E. F. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed]
- Riviere, C. J., Pot, J. & Tremaine, J. H. (1989). J. Gen. Virol. 70, 3033–3042. [DOI] [PubMed]
- Riviere, C. J. & Rochon, D. M. (1990). J. Gen. Virol. 71, 1887–1896. [DOI] [PubMed]
- Robbins, M. A., Reade, D. & Rochon, D. M. (1997). Virology, 234, 138–146. [DOI] [PubMed]
- Robinson, I. K. & Harrison, S. C. (1982). Nature (London), 297, 563–568.
- Rochon, D., Kakani, K., Robbins, M. & Reade, R. (2004). Annu. Rev. Phytopathol. 42, 211–241. [DOI] [PubMed]
- Rossmann, M. G., Abad-Zapatero, C., Murthy, M. R., Liljas, L., Jones, T. A. & Standberg, B. (1983). J. Mol. Biol. 165, 711–736. [DOI] [PubMed]
- Satheshkumar, P. S., Lokesh, G. L., Murthy, M. R. N. & Savithri, H. S. (2005). J. Mol. Biol. 353, 447–458. [DOI] [PubMed]
- Sorger, P. K., Stockley, P. G. & Harrison, S. C. (1986). J. Mol. Biol. 191, 639–658. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: melon necrotic spot virus, 2zah, r2zahsf