Abstract
Apolipoprotein(a) [apo(a)] consists of a series of tandemly repeated modules known as kringles that are commonly found in many proteins involved in the fibrinolytic and coagulation cascades, such as plasminogen and thrombin, respectively. Specifically, apo(a) contains multiple tandem repeats of domains similar to plasminogen kringle IV (designated as KIV1 to KIV10) followed by sequences similar to the kringle V and protease domains of plasminogen. The KIV domains of apo(a) differ with respect to their ability to bind lysine or lysine analogs. KIV10 represents the high-affinity lysine-binding site (LBS) of apo(a); a weak LBS is predicted in each of KIV5–KIV8 and has been directly demonstrated in KIV7. The present study describes the first crystal structure of apo(a) KIV7, refined to a resolution of 1.45 Å, representing the highest resolution for a kringle structure determined to date. A critical substitution of Tyr-62 in KIV7 for the corresponding Phe-62 residue in KIV10, in conjunction with the presence of Arg-35 in KIV7, results in the formation of a unique network of hydrogen bonds and electrostatic interactions between key LBS residues (Arg-35, Tyr-62, Asp-54) and a peripheral tyrosine residue (Tyr-40). These interactions restrain the flexibility of key LBS residues (Arg-35, Asp-54) and, in turn, reduce their adaptability in accommodating lysine and its analogs. Steric hindrance involving Tyr-62, as well as the elimination of critical ligand-stabilizing interactions within the LBS are also consequences of this interaction network. Thus, these subtle yet critical structural features are responsible for the weak lysine-binding affinity exhibited by KIV7 relative to that of KIV10.
Keywords: Apolipoprotein(a), kringle, lysine binding, crystal structure
Since its discovery in 1963, lipoprotein(a) [Lp(a)]1 has become a major focus in the area of atherosclerotic disease (Berg 1963). Elevated plasma levels of Lp(a) (>20–30 mg/dL) have been correlated with an increased risk for the development of a variety of atherosclerotic disorders (for review, see Durrington 1995; Scanu and Edelstein 1995; Koschinsky and Marcovina 1997; Marcovina et al. 1999). Moreover, a marked inherited variability has been observed in plasma Lp(a) levels that vary from virtually undetectable to >100 mg/dL within the population (for review, see Marcovina and Koschinsky 1998).
Lp(a) resembles low-density lipoprotein (LDL) in both lipid composition as well as in the presence of apolipoprotein B-100 (apoB-100), yet is distinguished by the presence of a unique glycoprotein, apolipoprotein(a) [apo(a)]. This hydrophilic protein is covalently linked to the apoB-100 moiety of LDL by a single disulfide bond and is thought to bestow the unique structural and functional properties attributed to Lp(a) (Fless et al. 1986). Apo(a) exhibits high sequence identity to plasminogen (Pgn), a serine protease zymogen involved in the fibrinolytic system (McLean et al. 1987). The structure of apo(a) consists of multiple tandem repeats of domains resembling plasminogen kringle IV (KIV) (∼75–85% similarity) followed by single copies of sequences resembling Pgn KV and the protease domain (∼90% similarity) (McLean et al. 1987). The KIV domains of apo(a) can be classified into 10 types based on amino acid sequence (designated KIV1 to KIV10), all of which are present in single copy with the exception of KIV2 (Lackner et al. 1993; van der Hoek et al. 1993). The varying numbers of KIV2, ranging from <5 to >50, form the basis for the apo(a) isoform size heterogeneity observed within the population (Lackner et al. 1993; van der Hoek et al. 1993; Haibach et al. 1998; Marcovina and Koschinsky 1998; Utermann 1999).
Despite extensive research into understanding the structure and function of apo(a)/Lp(a), very little has been elucidated regarding its pathophysiological role. The unique structure of this lipoprotein has suggested both a proatherogenic role, because of its similarity to LDL, as well as a potential prothrombotic/antifibrinolytic role, as a result of its resemblance to plasminogen. Moreover, Lp(a) is localized in the arterial intima, preferentially accumulating at the sites of atherosclerotic lesions, presumably due to its ability to interact with various components of the extracellular matrix (ECM) (for review, see Marcovina and Koschinsky 1998). Numerous studies have demonstrated that, similar to plasminogen, apo(a) binds a variety of biological substrates (e.g., fibrin(ogen), cell surface receptors) through lysine-binding sites (LBS) present within some of its KIV domains. Of the apo(a) KIV domains, KIV10 bears the greatest sequence similarity to Pgn KIV and is the only domain that contains the critical residues implicated in the interactions of Pgn KIV with lysine and its analogs. Indeed, the only exception is the conservative substitution of Arg-35 for the lysine residue present in Pgn KIV (McLean et al. 1987; Guevara et al. 1993).
A dominant role for KIV10 in the lysine-binding function of Lp(a) was first demonstrated by analyses involving Rhesus monkey Lp(a) in which a Trp70→Arg substitution in the KIV10 LBS (Tomlinson et al. 1989), also found in ∼2% of the human population (Scanu et al. 1994), was associated with a lysine-binding deficiency (Lys−) of the corresponding Lp(a) (Scanu et al. 1993, 1994; Scanu and Edelstein 1994). However, the ability of these KIV10-defective apo(a) species to form Lp(a) particles, a process mediated by lysine interactions, suggested the presence of additional LBS within the molecule (Edelstein et al. 1995). Moreover, the apo(a) species isolated from these Lys−Lp(a) particles maintained some degree of lysine-binding ability (Edelstein et al. 1995). Functional studies involving truncated apo(a) derivatives soon confirmed these claims by identifying an additional LBSs within the KIV5–KIV9 region of apo(a) that is essential to the process of Lp(a) assembly (Ernst et al. 1995; Gabel et al. 1996). Moreover, molecular modeling studies have predicted the presence of weak LBS in each of KIV5–KIV8, despite substitutions of some critical residues (Guevara et al. 1993). Thus, it is generally accepted that apo(a) KIV5–KIV8 each contain weak LBS that contribute to the lysine affinity of apo(a), but are masked in the context of Lp(a).
The existence of a weak LBS in KIV7 has been recently shown (M.N. Rahman, L. Becker, V. Petrounevitch, B.C. Hill, Z. Jia, and M.L. Koschinsky, unpubl. results). This study also represented the first attempt to characterize an LBS of apo(a) other than that of KIV10. The KIV7 domain was found to exhibit an affinity for lysine and its analogs 10-fold weaker (Kd = 230 ± 42 μM for ɛ-aminocaproic acid) in comparison with apo(a) KIV10 (Kd = 33 ± 4 μM for ɛ-aminocaproic acid), as well as differences in ligand specificity.
The ability of KIV7 to bind lysine supports the notion that this kringle may contribute to the lysine-binding ability of apo(a) and, therefore, to the overall function of the protein. Moreover, given the differences in ligand specificity relative to KIV10, the KIV7 domain may mediate unique interactions that contribute to the overall function of apo(a). Although these interactions may not be relevant in the context of Lp(a), the existence of both uncomplexed apo(a) and apo(a) fragments has been demonstrated (for review, see Scanu 1998), thus suggesting potential roles in free apo(a). In previous studies, we have shown that KIV7 contains a lysine- and proline-sensitive site capable of mediating interactions with plasmin-modified fibrinogen (M.N. Rahman, L. Becker, V. Petrounevitch, B.C. Hill, Z. Jia, and M.L. Koschinsky, unpubl. results), implying a potential contribution to the intimal trapping of apo(a) in areas of fibrin deposition. Furthermore, using truncated recombinant apo(a) derivatives, the KIV7 domain has been shown to contribute to noncovalent interactions that precede covalent Lp(a) assembly (Gabel and Koschinsky 1998; Trieu and McConathy 1998), and to mediate proline-sensitive cell surface binding to CHO cells (Trieu and McConathy 1998). More recently, in vitro binding experiments have demonstrated that apo(a) KIV7 has a higher binding affinity for certain lysine-containing peptides than apo(a) KIV10 (L. Becker and M.L. Koschinsky, unpubl. results).
In the present study, we report the high-resolution crystal structure of apo(a) KIV7 at 1.45 Å. Examination and comparison of the crystal structure with that of apo(a) KIV10, as well as those of plasminogen KI, KIV, and KV have allowed us to determine more definitively the structural basis underlying the properties of the apo(a) KIV7 LBS, specifically, the features contributing to its weaker lysine affinity relative to apo(a) KIV10. It is hoped that this information may provide further insight into the interactions that may be mediated by this kringle domain. Moreover, it may allow the development of specific inhibitors for KIV7-mediated interactions that enable the disruption of processes including Lp(a) assembly and intimal trapping of uncomplexed apo(a). This may prove to be a novel approach in lowering plasma Lp(a) levels or, perhaps, disrupt some of the pathogenic effects of apo(a)/Lp(a).
Results and Discussion
Crystal structures from two different crystallization conditions
We have determined two structures of KIV7 from crystals grown under two different conditions (see Materials and Methods). In molecular replacement calculations, a strong and unambiguous solution gave rise to a correlation coefficient of 57% and initial R factor of 44%, after which refinement proceeded smoothly. The statistics of both reflection data and refinement are summarized in Table 1. Although the two forms are isomorphous, at rotating anode X-ray source crystals obtained in the presence of ammonium sulfate showed much better diffraction (∼1.85 Å) than those obtained in the presence of sodium chloride (∼2.9 Å). Thus, only the crystals from ammonium sulfate were characterized further at a synchrotron source. Subsequent electron density map revealed the presence of a sulfate ion trapped close to the LBS, which is likely the reason for the better crystal quality under these conditions. The sulfate ion is found within the cationic region of LBS, and is in contact with both Arg-35 and Tyr-62 (2.88 Å and 2.60 Å). Although of lower resolution (2.9 Å), the structure of the sodium chloride-derived crystal is essentially identical to that of the high-resolution structure (r.m.s.d. 0.27 Å) with little difference observed in the LBS. Thus, sulfate binding does not appear to alter the kringle structure to any appreciable extent. Consequently, we will only refer to the high-resolution structure in our discussions below.
Table 1.
Data collection and refinement statistics
| Crystal parameters and data statistics | Sulfate complex | Apo kringle |
| Space group | P212121 | P212121 |
| Cell dimension a (Å) | 35.11 | 32.76 |
| Cell dimension b (Å) | 40.49 | 39.87 |
| Cell dimension c (Å) | 65.46 | 62.56 |
| Solvent content (%) | 23.1 | 23.27 |
| No. of molecules in asymmetric unit | 1 | 1 |
| Resolution range (Å) | 25.00–1.45 | 25.0–2.90 |
| Total no. of reflection | 48859 | 7031 |
| No. of unique reflections | 15774 | 1789 |
| Rsym | 0.069 | 0.092 |
| Completeness (%) | 91.8 | 88.0 |
| Refinement | Sulfate complex | Apo kringle |
| Resolution range (Å) | 6.0–1.45 | 6.0–2.90 |
| R (%) | 17.1 | 18.9 |
| Rfree (%) (10% data) | 19.6 | 26.7 |
| No. of protein atoms | 683 | 683 |
| No. of water atoms | 215 | 0 |
| No. of hetero atoms | 5 | 0 |
| R.m.s.d. of bond lengths (Å) | 0.044 | 0.007 |
| R.m.s.d. of bond angles (°) | 1.42 | 1.17 |
Overall structure of KIV7 and comparison with other kringles
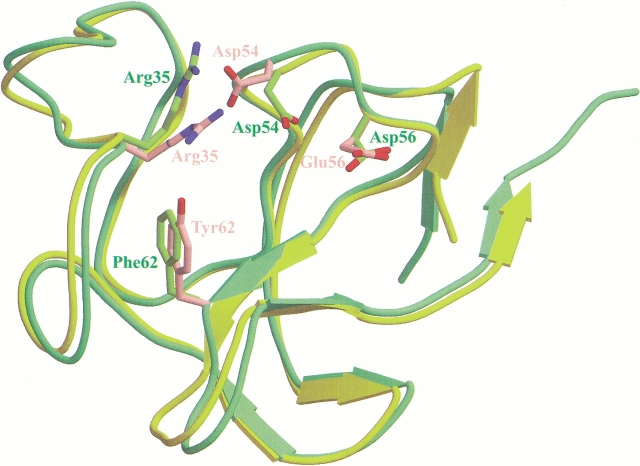
The KIV7 structure is well defined with the exception of the region encompassing residues 83–103 of the carboxyl-terminal region in which no electron density was observed. As expected, the overall structure of KIV7 is very similar to the X-ray structures of other kringles previously determined, including Pgn kringle I (KI) (Wu et al. 1994), Pgn KIV (Mulichak et al. 1991), Pgn KV (Chang et al. 1998), and apo(a) KIV10 (Mikol et al. 1996) (Fig. 1 ▶). Although superposition of the Cα atom traces of the plasminogen KI, KIV, KV, and apo(a) KIV10 structures with that of KIV7 showed no significant differences (r.m.s.d. 0.86 Å, 0.40 Å, 0.96 Å, and 0.50 Å, respectively), a few exceptions were observed including the Cα distances between His-33 of KIV7 and Pro-33 of Pgn KI (1.92 Å), between Gln-34 of KIV7 and Arg-34 of Pgn KI (2.47 Å), between His-33 of KIV7 and Ser-34 of Pgn KV (3.93 Å), and between Gln-34 of KIV7 and Ile-35 of Pgn KV (2.18 Å). Because of our high-resolution data, strong difference density was apparent at the carboxyl-terminal region of the kringle. Consequently, we were able to add four more residues, namely Pro-79, Val-80, Met-81, and Glu-82, at the carboxyl terminus, which, after refinement, fit into the map very well.
Fig. 1.
Superposition of KIV7 and KIV10 backbone structures, highlighting the four key LBS side chains. KIV7 backbone is in green and the side chains are in pink. KIV10 backbone is in yellow and the side chains are in green. In the KIV7 crystal structure reported here, we were able to determine one more amino-terminal and four more carboxy-terminal residues than, for example, the KIV10 crystal structure.
LBS and comparison with other kringles
In general, the KIV7 LBS is similar to those previously reported for KIV10, Pgn KI, and KIV. The LBS of KIV7 is bordered by Trp-32–Arg-35, Asp-54–Glu-56, Trp-60–Tyr-62, and Arg-69–Tyr-72. Three key elements are involved in ligand interaction: a cationic center (Arg-35, Arg-69), an anionic center (Asp-54, Glu-56), and a hydrophobic center (Trp-60, Trp-70, Tyr-62, Tyr-72). There are a number of interactions observed among these residues. In addition to a number of critical interactions that will be discussed in detail later, the guanidinium group of Arg-69 forms two hydrogen bonds with the backbone carbonyl group of Trp-32 (2.83 Å and 2.86 Å) and Glu-56 forms a hydrogen bond with Tyr-72 (2.62 Å).
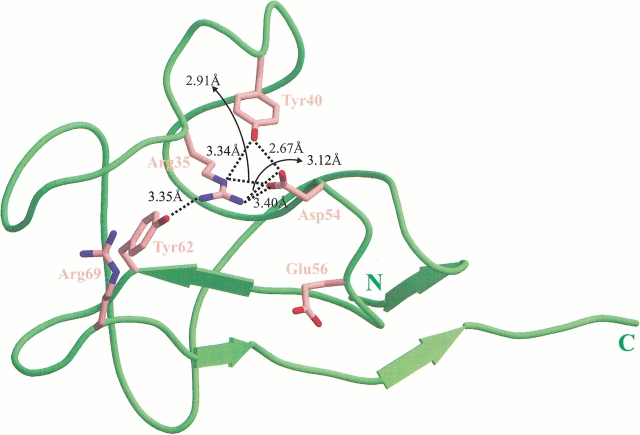
Despite the general similarity in LBS between KIV7 and KIV10, there are a number of seemingly subtle but critical differences. Specifically, two substitutions occur within the binding site: at residue 62 where Phe in KIV10 is replaced by Tyr in KIV7, and at residue 56 where Asp in KIV10 is replaced by Glu in KIV7 (Fig. 1 ▶). The Phe-62→Tyr-62 change leads to at least three main structural consequences. First, Tyr-62, which is located in the center of the cationic region, forms a hydrogen bond with the side chain of Arg-35 (Fig. 2 ▶). Molecular modeling demonstrates that the extra hydroxyl group of Tyr-62 (cf. Phe in KIV10) would result in steric hindrance with the ligand if it were to bind in a similar manner as in KIV10. Second, because of this hydrogen bond interaction with Tyr-62, the position of Arg-35 is more centrally located within the LBS in comparison with its corresponding position in KIV10 (Fig. 2 ▶). Within the KIV10 LBS, Arg-35 appears to play an important role in stabilizing the carboxylate group of lysine or lysine analogs (Mochalkin et al. 1999). In KIV7, however, the side chain of Arg-35 is restrained by its hydrogen bond interaction with Tyr-62; this was also observed in the sulfate-free, low-resolution structure of KIV7. Not only would the position of the Arg-35 guanidinium group in KIV7 reduce the accessibility of the LBS, the reduction in the flexibility of both Tyr-62 and Arg-35 would also greatly impair the adaptability in accommodating the incoming ligand. Hydrogen bond formation between Tyr-40 and the Arg-35 NE atom acts to further restrict the rotational freedom of Arg-35. In comparison, the guanidinium group of Arg-35 in KIV10 is positioned away from the binding pocket and, naturally, has no such contact with Phe-62. Therefore, KIV10 contains a more open cationic region with a more accessible LBS and a greater potential for adaptability. Interestingly, although two other kringles, Pgn KI and KV, both contain a Tyr residue at the equivalent position of Tyr-62 in KIV7, they lack the corresponding interacting partner at the equivalent position of Arg-35. Third, the position of Asp-54 in KIV7 is different from that of KIV10. As observed in other kringle structures, an acidic residue at this position is critical to the LBS in the stabilization of the cationic end of zwitterionic ligands such as lysine. In KIV7, Asp-54 is involved in three electrostatic interactions with Arg-35 (NH1, NH2, and NE), as well as a further hydrogen bond interaction with Tyr-40, the result of which is the positioning of Asp-54 away from the binding pocket and toward the solvent space (Fig. 2 ▶). Furthermore, these interactions would potentially restrict the flexibility of Asp-54. Therefore, it is likely that Asp-54 may not be able to interact with and stabilize the amino group of the ligand. In contrast, the corresponding Asp-54 residue in KIV10 does not interact with Arg-35 or Tyr-40, and is located closer to the binding pocket than Asp-54 of KIV7. Clearly Asp-54 of KIV10 has a greater rotational flexibility to allow it to interact with and stabilize the amino group of ligands. Thus, it appears that the difference in the positioning of Asp-54 in the KIV7 LBS is due to the positioning of Arg-35, which, in turn, is dependent on the presence of Tyr-62.
Fig. 2.
The interaction network of KIV7. Dashed lines represent the interaction network "triggered" by Tyr-62 in KIV7; the distances are indicated (3.4 Å cut-off distance used).
The KIV7 domain has been demonstrated to play a role in the initial noncovalent interactions between apo(a) and apoB-100 that precede specific disulfide bond formation in the process of Lp(a) assembly (Gabel and Koschinsky 1998; Trieu and McConathy 1998). It has further been shown that this initial noncovalent step can be inhibited by lysine, lysine analogs, arginine, phenylalanine, and proline (Edelstein et al. 1995; Frank et al. 1995; Gabel and Koschinsky 1998; Koschinsky et al. 1997). Interestingly, the KIV7 domain is capable of mediating lysine- and proline-sensitive interactions with plasmin-modified fibrinogen (M.N. Rahman, L. Becker, V. Petrounevitch, B.C. Hill, Z. Jia, and M.L. Koschinsky, unpubl. results) and mediating proline-sensitive binding to cell surfaces (Trieu and McConathy 1998). Proline contains no structural homology to lysine and lysine analogs; it lacks positive and negative segments separated by a sufficient distance required to interact with the anionic and cationic centers of LBS. However, although proline is too short to interact with both charged centers of the binding pocket simultaneously, we were able to model the binding of this amino acid to the KIV7 LBS in a manner that involves only the cationic center. Proline can be oriented in a manner such that its carboxyl group forms an electrostatic interaction with Arg-69, whereas the ring nitrogen atom forms two hydrogen bonds with Tyr-62 and Arg-35. Alternatively, the carboxyl group of proline can be oriented to form a salt bridge with Arg-35, whereas the ring nitrogen atom forms a hydrogen bond with Arg-69. The cyclic structure of proline would putatively restrict the flexibility of the nitrogen atom, making it more favorable to form stabilizing interactions. The substitution of Tyr-62 with Phe-62 in both KIV10 and Pgn KIV, and the substitution of Arg-35 by Ile-35 and Phe-36 in Pgn KI and KV would make it impossible to establish the aforementioned interactions, which may explain why proline binds to KIV7 more favorably than to other kringles.
In conclusion, the overall structure of KIV7 is very similar to that shared by other characterized kringles and also contains a well-defined LBS consisting of cationic, anionic, and hydrophobic centers similar to that found in apo(a) KIV10, as well as in Pgn KI and KIV. However, substitution of Tyr-62 in KIV7 for the corresponding Phe-62 residue in KIV10, results in the formation of a unique interaction network in KIV7. The pairing of Tyr-62 and Arg-35 is the key to this cascade, or sequential formation, of the interaction network; other kringles have only one of the two residues rather than both simultaneously. The rigidity of LBS residues is increased as the result of the networked interaction. The high resolution X-ray structure of KIV7 provides us with the confidence with which we are able to provide some insights into the lysine-binding properties of KIV7. Furthermore, the structure has enabled us to observe these subtle but critical structural features that are apparently responsible for the important functional difference between KIV7 and other kringles including KIV10.
Materials and methods
Protein crystallization
Recombinant KIV7 protein was overexpressed using a pET16b vector in Escherichia coli BL21 (DE3) cells and purified as published elsewhere (Rahman et al. 2001). Hexagonal and cubical shaped crystals of KIV7 protein were obtained using two different conditions: (1) 21–23% ammonium sulfate, 0.1 M MES at pH 5.5–6.0, (2) 2.0 M NaCl, 0.1 M MES at pH 6.5, 0.1 M KH2PO4, and 0.1 M NaH2PO4. Crystals were grown at room temperature by vapor diffusion (hanging drop method). Crystal growth was very slow; typical time required for growing diffraction-quality crystals was 3–4 mo.
Data collection and processing
Crystals were initially tested using the in-house X-ray facility, which consists of a 30-cm MarResearch imaging plate, and Rigaku rotating anode X-ray generator operated at 50 kV, 100 mA. Further diffraction data were collected on a synchrotron source at CHESS A1 station using ADSC Quantum-4 CCD detector. For all data collection, a single crystal was transferred from cryoprotection solution that contained 20% glycerol and crystallization buffer. Crystals were flash-frozen in liquid propane cooled by liquid nitrogen, and subsequently placed under a stream of nitrogen gas at 100K. Data were processed and scaled with Denzo and Scalepack (Otwinowski and Minor 1997).
Structure solution, refinement, and substrates modeling of KIV7
Phase determination was accomplished by molecular replacement using the EPMR program (Kissinger et al. 1997). The diffraction data used for molecular replacement was a lower resolution data set obtained from rotating anode X-ray source, whereas the probing model was the crystal structure of human plasminogen kringle 4 (PDB accession code 1PK4; Mulichak et al. 1991). Refinement was carried out using CNS package (Brünger et al. 1999). The coordinates of the final refined structure have been deposited in PDB (accession code 1I71). Modeling was performed using Sybyl (version 5.3, Tripos Inc.).
Acknowledgments
This work is supported by Canada Institutes of Health Research. Z.J. holds a Canada Research Chair in Structure Biology. We thank CHESS staff for assistance in data collection.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Lp(a), lipoprotein(a)
LDL, low-density lipoprotein
apoB-100, apolipoprotein B-100
apo(a), apolipoprotein(a)
Pgn, plasminogen
K, kringle
ECM, extracellular matrix
LBS, lysine-binding site(s)
Lys−, lysine-binding deficiency
Kd, dissociation constant
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.01701.
References
- Berg, K. 1963. A new serum type system in man—the Lp system. Acta Pathol. Microbiol. Scand. 59 369–382. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grossekunstleve, R.W., Jiang, J-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1999. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Chang, Y., Mochalkin, I., McCance, S.G., Cheng, B., Tulinsky, A., and Castellino, F.J. 1998. Structure and ligand binding determinants of the recombinant kringle 5 domain of human plasminogen. Biochemistry 37 3258–3271. [DOI] [PubMed] [Google Scholar]
- De Serrano, V.S. and Castellino, F.J. 1993. Specific anionic residues of the recombinant kringle 2 domain of tissue-type plasminogen activator that are responsible for stabilization of its interaction with omega-amino acid ligand. Biochemistry 32 3540–3548. [DOI] [PubMed] [Google Scholar]
- Durrington, P.N. 1995. Lipoprotein(a). Bailliéres Clin. Endocrinol. Metab. 9 773–795. [DOI] [PubMed] [Google Scholar]
- Edelstein, C., Mandala, M., Pfaffinger, D., and Scanu, A.M. 1995. Determinants of lipoprotein(a) assembly: A study of wild-type and mutant apolipoprotein(a) phenotypes isolated from human and rhesus monkey lipoprotein(a) under mild reductive conditions. Biochemistry 34 16483–16492. [DOI] [PubMed] [Google Scholar]
- Ernst, A., Helmhold, M., Brunner, C., Pethö-Schramm, A., Armstrong, V.W., and Müller, H-J. 1995. Identification of two functionally distinct lysine-binding sites in kringle 37 and in kringles 32–36 of human apolipoprotein(a). J. Biol. Chem. 270 6227–6234. [DOI] [PubMed] [Google Scholar]
- Fless, G.M., ZumMallen, M.E., and Scanu, A.M. 1986. Physicochemical properties of apolipoprotein(a) and lipoprotein(a-) derived from the dissociation of human plasma lipoprotein(a). J. Biol. Chem. 261 8712–8718. [PubMed] [Google Scholar]
- Frank, S., Durovic, S., Kostner, K., and Kostner, G.M. 1995. Inhibitors for the in vitro assembly of Lp(a). Arterioscler. Thromb. Vasc. Biol. 15 1774–1780. [DOI] [PubMed] [Google Scholar]
- Gabel, B.R. and Koschinsky, M.L. 1998. Sequences within apolipoprotein(a) kringle IV types 6–8 bind directly to low-density lipoprotein and mediate noncovalent association of apolipoprotein(a) with apolipoprotein B-100. Biochemistry 37 7892–7898. [DOI] [PubMed] [Google Scholar]
- Gabel, B.R., May, L.F., Marcovina, S.M., and Koschinsky, M.L. 1996. Lipoprotein(a) assembly: quantitative assessment of the role of apo(a) kringle IV types 2–10 in particle formation. Arterioscler. Thromb. Vasc. Biol. 16 1559–1567. [DOI] [PubMed] [Google Scholar]
- Guevara, J. Jr., Jan, A.Y., Knapp, R., Tulinsky, A., and Morrisett, J.D. 1993. Comparison of ligand-binding sites of modeled apo(a) kringle-like sequences in human lipoprotein(a). Arteriosclerosis and Thrombosis 13 758–770. [DOI] [PubMed] [Google Scholar]
- Haibach, C., Kraft, H.G., Köchl, S., Abe, A., and Utermann, G. 1998. The number of kringle IV types 3–10 is invariable in the human apo(a) gene. Gene 208 253–258. [DOI] [PubMed] [Google Scholar]
- Kissinger, C.R. Gehlhaar, D.K., and Fogel, D.B. 1999. Rapid automated molecular replacement by evolutionary search. Acta Cryst. D55 484–491. [DOI] [PubMed] [Google Scholar]
- Klezovitch, O., Edelstein, C., and Scanu, A.M. 1996. Evidence that the fibrinogen-binding domain of apo(a) is outside the lysine-binding site of kringle IV-10. J. Clin. Invest. 98 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschinsky, M.L. and Marcovina, S.M. 1997. Lipoprotein(a): structural implications for pathophysiology. Int. J. Clin. Lab. Res. 27 14–23. [DOI] [PubMed] [Google Scholar]
- Koschinsky, M.L., Marcovina, S.M., May, L.F., and Gabel, B.R. 1997. Analysis of the mechanism of lipoprotein(a) assembly. Clin. Genet. 52 338–346. [DOI] [PubMed] [Google Scholar]
- Lackner, C., Cohen, J.C., and Hobbs, H.H. 1993. Molecular definition of the extreme size polymorphism in apolipoprotein(a). Hum. Mol. Genet. 2 933–940. [DOI] [PubMed] [Google Scholar]
- Marcovina, S.M. and Koschinsky, M.L. 1998. Lipoprotein(a) as a risk factor for coronary artery disease. Am. J. Cardiol. 82 52–66. [DOI] [PubMed] [Google Scholar]
- Marcovina, S.M., Hegele, R.A., and Koschinsky, M.L. 1999. Lipoprotein(a) and coronary heart disease risk. Curr. Cardiol. Rep. 1 105–111. [DOI] [PubMed] [Google Scholar]
- McCance, S.G., Mengart, N., and Castellino, F.J. 1994. Amino acid residues of the kringle-4 and kringle-5 domains of human plasminogen that stabilize their interaction with omega-amino acid ligands. J. Biol. Chem. 269 32405–32410. [PubMed] [Google Scholar]
- McLean, J.W., Tomlinson, J.E., Kuang, W-J., Eaton, D.L., Chen, E.Y., Fless, G.M., Scanu, A.M., and Lawn, R.M. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature 330 132–137. [DOI] [PubMed] [Google Scholar]
- Mikol, V., LoGrasso, P.V., and Boettcher, B.R. 1996. Crystal structures of apolipoprotein(a) kringle IV37 free and complexed with 6-aminohexanoic acid and with p-aminomethylbenzoic acid: Existence of novel and expected binding modes. J. Mol. Biol. 256 751–761. [DOI] [PubMed] [Google Scholar]
- Mochalkin, I., Cheng, B., Klezovitch, O., Scanu, A.M., and Tulinsky, A. 1999. Recombinant kringle IV-10 modules of human apolipoprotein(a): Structure, ligand binding modes, and biological relevance. Biochemistry 38 1990–1998. [DOI] [PubMed] [Google Scholar]
- Mulichak, A.M., Tulinsky, A., and Ravichandran, K.G. 1991. Crystal and molecular structure of human plasminogen kringle 4 refined at 1.9-Å resolution. Biochemistry 30 10576–10588. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Rahman, M.N., Petrounevitch, V., Jia, Z., and Koschinsky M.L. 2001. Antifibrinolytic effect of single Apo(a) kringle domains: Relationship to fibrinogen binding. Protein Eng. (in press). [DOI] [PubMed]
- Scanu, A.M. 1998. Proteolytic modifications of lipoprotein(a): potential relevance to its postulated atherothrombogenic role. J. Investigative Med. 46 359–363. [PubMed] [Google Scholar]
- Scanu, A.M. and Edelstein, C. 1994. Apolipoprotein(a): Structural and functional consequences of mutations in kringle type 10 (or kringle 4–37). Clin. Genet. 46 42–45. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Kringle-dependent structural and functional polymorphism of apolipoprotein(a). Biochim. Biophys. Acta 1256 1–12. [DOI] [PubMed] [Google Scholar]
- Scanu, A.M., Miles, L.A, Fless, G.M., Pfaffinger, D., Eisenbart, J., Jackson, E., Hoover-Plow, J.L., Brunck, T., and Plow, E.F. 1993. Rhesus monkey lipoprotein(a) binds to lysine Sepharose and U937 monocytoid cells less efficiently than human lipoprotein(a): Evidence for the dominant role of kringle 437. J. Clin. Invest. 91 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu, A.M., Pfaffinger, D., Lee, J.C., and Hinman J. 1994. A single point mutation (Trp72→Arg) in human apo(a) kringle 4–37 associated with a lysine binding defect in Lp(a). Biochim. Biophys. Acta 1227 41–45. [DOI] [PubMed] [Google Scholar]
- Tomlinson, J.E., McLean, J.W., and Lawn, R.M. 1989. Rhesus monkey apolipoprotein(a): Sequence, evolution, and sites of synthesis. J. Biol. Chem. 264 5957–5965. [PubMed] [Google Scholar]
- Trieu, V.N. and McConathy, W.J. 1998. Functional characterization of T7 and T8 of human apolipoprotein(a). Biochem. Biophys. Res. Comm. 251 356–359. [DOI] [PubMed] [Google Scholar]
- Utermann, G. 1999. Genetic architecture and evolution of the lipoprotein(a) trait. Curr. Opin. Lipidol. 10 133–141. [DOI] [PubMed] [Google Scholar]
- van der Hoek, Y.Y., Wittekoek, M.E., Beisiegel, U., Kastelein, J.J.P., and Koschinsky, M.L. 1993. The apolipoprotein(a) kringle IV repeats which differ from the major repeat kringle are present in variably-sized isoforms. Hum. Mol. Genet. 2 361–366. [DOI] [PubMed] [Google Scholar]
- Wu, T-P., Padmanabhan, K.P., and Tulinsky, A. 1994. Structure of recombinant plasminogen kringle 1 and the fibrin binding site. Blood Coagulation Fibrinolysis 5 157–166. [DOI] [PubMed] [Google Scholar]