Abstract
Streptococcal pyrogenic exotoxin A (SpeA) is produced by Streptococcus pyogenes, and has been associated with severe infections such as scarlet fever and Streptococcal Toxic Shock Syndrome (STSS). In this study, the crystal structure of SpeA1 (the product of speA allele 1) in the presence of 2.5 mM zinc was determined at 2.8 Å resolution. The protein crystallizes in the orthorhombic space group P21212, with four molecules in the crystallographic asymmetric unit. The final structure has a crystallographic R-factor of 21.4% for 7,031 protein atoms, 143 water molecules, and 4 zinc atoms (one zinc atom per molecule). Four protein ligands—Glu 33, Asp 77, His 106, and His 110—form a zinc binding site that is similar to the one observed in a related superantigen, staphylococcoal enterotoxin C2. Mutant toxin forms substituting Ala for each of the zinc binding residues were generated. The affinity of these mutants for zinc ion confirms the composition of this metal binding site. The implications of zinc binding to SpeA1 for MHC class II recognition are explored using a molecular modeling approach. The results indicate that, despite their common overall architecture, superantigens appear to have multiple ways of complex formation with MHC class II molecules.
Keywords: Molecular recognition, X-ray crystallography, superantigen, pyrogenic exotoxin, zinc binding
Superantigens are potent T-cell mitogens that crosslink major histocompatibility complex (MHC) class II molecules of antigen presenting cells with T-cell receptors (TCRs) to stimulate large populations of T-cells in a Vβ-specific manner (Marrack and Kappler 1990). This stimulation leads to a massive cytokine release that may result in acute systemic illness and clinical shock (Herman et al. 1991). The superantigen family includes the staphylococcal enterotoxins (SEs) A, B, D, C1–3, D, E, G, H, I, toxic shock syndrome toxin-1 (TSST-1), the streptococcal pyrogenic exotoxins (Spes) A, C, G, and J, and the streptococcal superantigens SSA, SMEZ, and SMEZ2.
Streptococcal pyrogenic exotoxin A (SpeA) is produced by a majority of streptococcal strains isolated from patients with streptococcal toxic shock syndrome (STSS) (Musser et al. 1991; Chausee et al. 1996). STSS is characterized by rash, hypotension, multiorgan failure, and high mortality rate, and these symptoms are commonly associated with superantigen-mediated syndromes (Stevens 1995). There are at least four alleles (1–4) of the speA gene isolated so far. Crystal structure of one of these allelic forms, SpeA1, was determined previously (Papageorgiou et al. 1999; Earhart et al. 2000). The structure showed the conserved two-domain architecture (N- and C-terminal domains) and the presence of a long, solvent-accessible α-helix (common to all known members of the superantigen family) that spans the center of the molecule. Based on biochemical data and structural considerations, an MHC class II binding site at the N-terminal domain was identified. This site is referred to as the `generic site', and it is found in most of the known microbial superantigen structures (except in SEH, SpeC, SpeH, and SMEZ2) (for a recent review, see Papageorgiou and Acharya 2000).
Structure-based sequence alignment of SpeA1 with staphyloccocal enterotoxin C2 revealed a putative zinc binding site formed by residues Asp 77, His 106, and His 110. Biochemical and structural studies have shown that most members (with the exception of SEB and TSST-1) of the superantigen family possess a zinc binding site, and a bound zinc ion has been identified in the structures of SEA, SEC2, SED, SEH, SpeA, SpeC, SpeH, and SMEZ2. The proposed role of the zinc ion is to act as a bridge for MHC class II binding, thus providing an alternative mechanism for MHC class II binding. In SED, which possesses two zinc binding sites, the toxin is able to form homodimers (Sundström et al. 1996a). Moreover, zinc has also been found to play a role in the thermostability of certain members of the superantigen family (Cavallin et al. 2000). Thus, from the available data it has become clear that despite common structural and functional associations, each member of the superantigen family appears to have adopted a significantly different mode of forming the MHC class II–superantigen–TCR ternary complex. To further characterize the role of zinc ion on the superantigenic properties of SpeA1 we have determined the crystal structure of this toxin to 2.8 Å resolution in the presence of zinc.
Results and Discussion
Quality of the structure
The crystallographic details for the SpeA1-zinc complex are shown in Table 1. The final model (four molecules in the asymmetric unit) contains 7,031 nonhydrogen protein atoms, 4 zinc ions, and 143 water molecules. The root mean square (r.m.s.) deviation between the zinc-bound structure and the native structure is 0.33 Å (mole 1), 0.35 Å (mole 2), 0.30 Å (mole 3), and 0.38 Å (mole 4). The regions that deviate most between the two structures include the flexible disulphide loop and the first six residues at the N-terminus. Exclusion of these areas from the calculation improves the r.m.s. deviation to 0.26 Å (mole 1), 0.25 Å (mole 2), 0.24 Å (mole 3), 0.33 Å (mole 4). The Ramachandran plot for all four molecules shows 85% of the residues in allowed regions and the rest in generously allowed regions. Residues 1 and 2 were not modeled due to poor density. Residues 5, 88, 112, 115, 179, and 180 in all four molecules and residues 91 and 92 in molecules 2, 3, and 4 were modeled as alanines due to insufficient density. Molecule 1 will be used throughout the discussion.
Table 1.
Crystallographic data processing and refinement statistics
| Cell dimensions (Å) | a = 126.9, b = 101.3, c = 82.0 |
| Space group | P212I2 4 mole/a.u. |
| Resolution (Å) | 40.0–2.8 |
| No. of measurements | 144123 |
| No. of unique reflections | 26185 |
| Completeness (%) | 95.8 (61.3)a |
| I/σ | 6.4 (5.3) |
| Rmerge (%)b | 10.8 (28.5) |
| Refinement | |
| Rcryst (%)c | 21.4 |
| Rfree (%)d | 28.0 |
| No. of protein atoms | 7031 |
| No. of water molecules | 143 |
| Temperature factors (Å2) | |
| Main chaine | 28.4, 30.1, 24.9, 43.5 |
| Side chain | 30.4, 33.2, 28.5, 45.1 |
| Zinc ligands | 29.3, 34.2, 23.2, 52.0 |
| Zinc ion (average) | 48.0 |
| RMSD in bond lengths (Å) | 0.007 |
| RMSD in bond angles (°) | 1.32 |
a Outermost shell 2.9–2.8 Å.
bRmerge = Σ(│Ij − <I>│)/Σ<I>, where Ij is the observed intensity of reflection j and <I> is the average intensity of multiple observations.
cRcryst = Σ││Fo│ − │Fc││/Σ│Fo│, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
d 5% of the data that were used for the calculation of Rfree were randomly excluded from the refinement.
e Temperature factors for individual molecules are quoted.
The zinc binding site
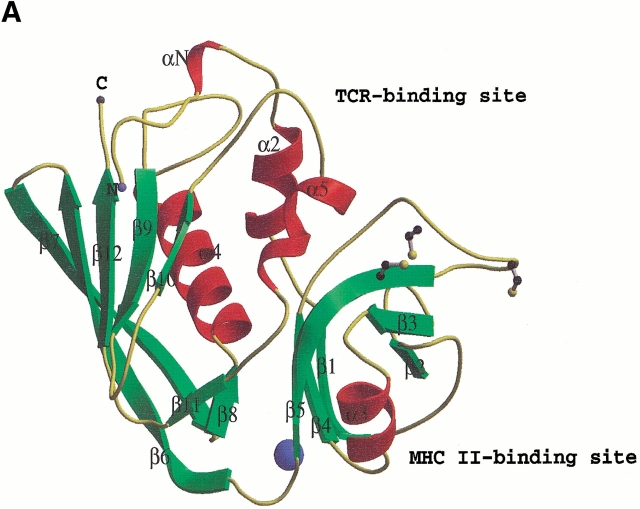
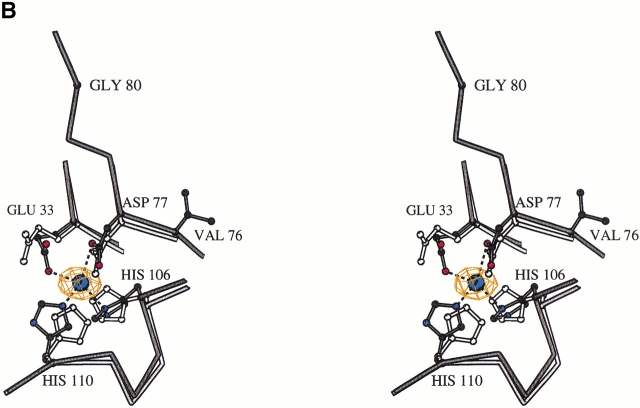
The crystal structure of the SpeA1–zinc complex revealed the presence of a zinc binding site at the interface between the N- and C-terminal domains of SpeA1 (Fig. 1A ▶). The zinc ligands are Glu 33, Asp 77, His 106, and His 110 (Fig. 1B ▶). In the presence of a zinc ion, slight movement of these residues surrounding the zinc ion was observed in the complex structure (Fig. 1B ▶) compared to the previously determined native structure of SpeA1 in the absence of zinc (Papageorgiou et al. 1999). The distances between the ligands and the zinc ion are Glu 33 OE1—1.98 Å, Asp 77 OD1—2.47 Å, His 106 ND1—2.06 Å and His 110 NE2—2.17 Å. These values are comparable to those from other zinc binding superantigens, and fall within the range of distances documented by Alberts et al. (1998) for tetrahedrally coordinated zinc ions in proteins. A similar study was recently conducted by Earhart et al. (2000). They reported an identical zinc binding site as presented here in addition to a cadmium binding site involving residues 87–98 from the disulfide loop. In an independent study presented here, we address the biological implications of MHC class II recognition by SpeA1 in the presence of zinc.
Fig. 1.
(A) The structure of SpeA1–zinc complex. The bound zinc ion is shown as a blue sphere. The disulfide bridge is shown in ball-and-stick representation. The third cysteine residue (free cysteine), which is part of the disulfide loop, is also shown. The figure was created with the program BOBSCRIPT (Esnouf 1997). (B) Comparison of the zinc binding site in the native SpeA1 (white bonds) and in the zinc complex (black bonds). The difference electron density for the bound zinc ion is also shown.
The presence of a zinc binding site in SpeA1 was first proposed by Papageorgiou et al. (1999) on the basis of sequence comparison and the crystal structure of SEC2. The residues Asp 77, His 106, and His 110 of the SpeA1–zinc binding site are structurally equivalent to the residues Asp 83, His 118, and His 122 in SEC2 with the fourth zinc ligand in SEC2 being formed by Asp 9 of a symmetry related molecule in the crystal lattice. The SEA molecule contains two zinc binding sites—one on each N- and C-terminal domains (Schad et al. 1995; Sundström et al. 1996b). The SEC2-like zinc binding site of SEA (located at the C-terminal domain) also has three ligands structurally equivalent to SpeA1: Glu 39 (Glu 33 in SpeA1), Asp 83 (Asp 77), and His 118 (His 106), with a water molecule in SEA replacing the second histidine residue (His 110) found in SpeA1 (Schad et al. 1997). Different and structurally nonequivalent zinc binding sites have been identified in SEA (Schad et al. 1995; Sundström et al. 1996b), SED (Sundström et al. 1996a), SEH (Häkansson et al. 2000), and SpeC (Roussell et al. 1997). As a result, several putative roles for zinc in the structure and function of the superantigen family have been proposed: the zinc ion has been shown to have a role in the formation of (a) stable homodimers (for SED and SEH); (b) a second high-affinity MHC class II binding site (for SEA, SEC, SED, and SEH); and (c) in the thermostability of superantigens.
Zinc site mutants have decreased affinity for zinc
To confirm that the residues Glu 33, Asp 77, His 106, and His 110 comprise a zinc binding site, mutant forms of SpeA1 with Ala substituted at each of these positions were generated. The affinity of each mutant form for 65Zn2+ was determined by equilibrium dialysis (Table 2). The Kd of SpeA1 for zinc was found to be 2.3 μM, which is approximately 10-fold higher than the Kd of the staphylococcal superantigen SEA–zinc interaction (0.3 μM) (Sundström et al. 1996b). The Kd of the mutant form SpeA1–Asp77Ala for zinc was 60 μM, of SpeA1–His106Ala for zinc was 120 μM, and of SpeA1–H110Ala for zinc was 80 μM, indicating that the affinity of each of these toxins for zinc was significantly decreased when compared to the affinity of wild-type toxin for zinc. The SpeA1–Glu33Ala–zinc interaction had a Kd of 5.8 μM, which represents only a slight decrease in affinity. No zinc binding was observed with the double mutant SpeA1–Asp77Ala, His106Ala when 150 μM toxin was used in the experiment, indicating that the Kd of zinc for this mutant was much greater than 150 μM. Thus, the residues Asp 77, His 106, and His 110 are clearly necessary for zinc binding, while Glu 33 has a lesser role.
Table 2.
Affinity of SpeAl and mutant forms for 65Zn2+
| Protein | Kd, μM |
| SpeAl | 2.3 ± 2 |
| SpeAl–Glu33Ala | 5.8 ± 3 |
| SpeAl–Asp77Ala | 60 ± 30 |
| SpeAl–His106Ala | 120 ± 50 |
| SpeAl–His110Ala | 80 ± 30 |
| SpeAl–Asp77Ala, His106Ala | >150 |
Each value represents the mean ± SD of results from at least three distinct experiments.
A possible role for the zinc ion in MHC class II recognition?
SEA has two MHC class II binding sites, one at the C-terminus and the other at the N-terminus. The C-terminal SEA binding site contains a zinc ion, and has an approximate 100-fold higher affinity for MHC class II than the N-terminal MHC class II binding site (Abrahamsén et al. 1995). The N-terminal MHC class II binding site (generic site) does not contain zinc, and is similar to the MHC class II binding sites found in other superantigens, such as SEB and SEC.
There is evidence that SpeA1 contains a generic class II binding site. A previous study examining the affinity of various SpeA1 mutants for MHC class II demonstrated that mutations at SpeA1 residues 42 to 48 decrease binding to the MHC class II DQ molecule (Kline and Collins 1996). SpeA1 residue Leu 42 is conserved in the SEB generic MHC class II binding site. Therefore, it is probable that SpeA1 is similar to SEA, and has two distinct MHC class II binding sites: a generic site, and a zinc-mediated site discussed here. The position of the two predicted binding sites on the structure of SpeA1 is similar to the positions of the predicted MHC class II binding sites seen in the structure of SEC (Papageorgiou et al. 1995).
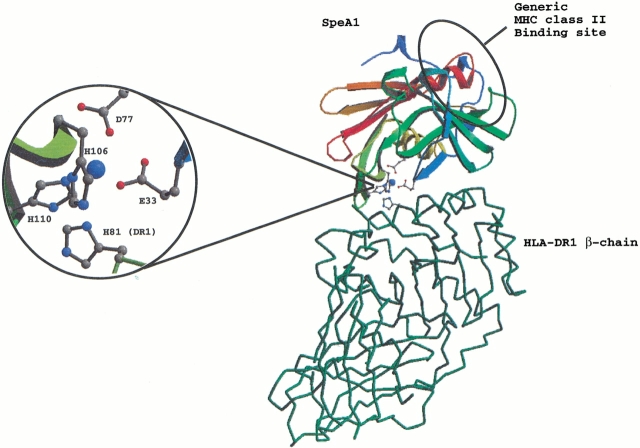
Mutagenesis studies have identified His 81 of the MHC class II β-chain as an important residue for the zinc-mediated binding of SEA to MHC class II molecule (Karp and Long 1992; Fraser 1993). To examine the possible interactions of SpeA1 with MHC class II molecules via its zinc site SpeA1 was docked onto mouse HLA-DR1 (Fig. 2 ▶). The theoretical model gives a good overall fit for the interaction of the zinc site of SpeA1 and His 81 of the DR1 β-chain. There are only minor clashes in the model involving residues Leu 111, Ala 112, and Ile 113 (from strands β5 and β6) of SpeA1 with part of the antigenic peptide bound in the peptide-binding groove of HLA-DR1. However, peptide antigen varies greatly throughout the MHC class II population (Chicz et al. 1992), and several peptides have been shown to either enhance or reduce the affinity of superantigens for MHC class II molecules, depending on their interactions with the superantigen (Wen et al. 1996). The modeled complex gives clues to the proposed zinc-mediated binding of SpeA1 to HLA-DR1 (Fig. 2 ▶). When His 81 of the DR1 β-chain binds to the toxin involving the zinc ion, there would be some rearrangement that would affect the overall geometry of the site, and therefore, the position of neighboring residues allowing the optimization of favorable contacts between the superantigen and the MHC class II molecule. It should be noted that in the present modeling exercise we used the coordinates of the SEB–DR1 complex (Jardetzky et al. 1994), and that SpeA1 has preferential binding for HLA-DQ. However, His 81 and its neighboring residues are conserved in HLA-DQ (Schiffenbauer et al. 1987).
Fig. 2.
Proposed model for DR1–SpeA1–zinc-mediated recognition. In this mode of binding, His 81 from the β-chain of MHC class II molecule would replace one of the zinc binding ligand from SpeA1 (inset).
There are at least four naturally occurring SpeA alleles, and three of these viz., speA1, speA2, and speA3 encode for toxins that differ from each other by a single amino acid (Musser et al. 1991). SpeA3 differs from SpeA1 by a single Val to Ile substitution at position 76. SpeA2 differs from SpeA1 by a single Gly to Ser substitution at position 80. In the SpeA1 structure, both Val 76 and Gly 80 are buried. The allelic variants of SpeA show different binding affinities for HLA-DQ with Kis of 104, 55, and 13 μM for SpeA 1, 2, and 3, respectively. SpeA3 is the most mitogenic of these alleles, and appears to exhibit higher affinity for the HLA-DQ than SpeA1 (Kline and Collins 1996). It is tempting to suggest that the proximity of this allele specific amino acid residue to the zinc binding site of SpeA3 (adjacent to Asp 77) may play a role in its increased affinity for HLA-DQ. Furthermore, it has been shown that a variant of SpeA1, Asp 77-Ala, has a much lower ability to bind to HLA-DQ (Hartwig and Fleisher 1993). In addition, it is interesting to note that Ser 80 in SpeA2 is also located in close proximity to the zinc binding site of SpeA1 and to residues implicated in TCR binding. Clearly, the role of these two residues in zinc-mediated MHC class II binding needs further investigation. However, the presence of a zinc site does offer an alternate mode of MHC class II recognition by SpeA1, and perhaps the possibility of additional interactions with MHC class II molecules to accommodate multiple TCR molecules.
Materials and methods
Protein crystallization
Recombinant SpeA1 with an N-terminal His tag was expressed in E. coli BL21 (DE3) (Novagen), and purified by immobilized metal affinity chromatography as described previously by Papageorgiou et al. (1999). Crystals of SpeA1 were grown at 16°C using the hanging drop vapor diffusion method (Papageorgiou et al. 1999). The crystallization conditions with a reservoir solution containing 16%–19% PEG 3350, 0.1 M sodium cacodylate buffer (pH 6.5), and 16% isopropanol gave small, irregular crystals. These crystals were used for microseeding of hanging drops equilibrated against a reservoir solution containing 17% PEG 8000, 0.2 M ammonium sulphate and 0.1 M sodium cacodylate buffer (pH 6.5). Crystals grown under these conditions were soaked in a mother liquor solution containing 2.5 mM zinc chloride (final concentration) for 12 h prior to data collection.
Data collection and refinement
X-ray diffraction data to 2.8 Å were collected at 100 K using the crystallization buffer containing 15% glycerol as a cryoprotectant at EMBL (c/o DESY), on beamline X11 equipped with a MAR345 image plate. Forty-four images were collected using dose mode (480–560 sec/image), with an oscillation range of 2° per image. A second data set was collected at Max II, Max Lab to 2.9 Å resolution using a MAR345 image plate. The exposure time was 480 sec/image, and the oscillation range was 1.0°. Data processing, scaling, and merging of the two data sets to 2.8 Å resolution was performed using the HKL package (Otwinowski and Minor 1997). The final Rmerge was 10.8%, with an overall completeness of 95.8% (Table 1). Phases were determined using the structure of native SpeA1 at 2.6 Å (Papageorgiou et al. 1999) as a starting model. The initial model was subjected to rigid body refinement. Calculation of a (|Fo| − |Fc|) electron density map at this stage revealed extra density at the predicted zinc binding site of the toxin for each of the four molecules in the asymmetric unit. The structure was refined by simulated annealing using tight noncrystallographic symmetry (NCS) restraints and the maximum likelihood target as implemented in the program CNS (Brünger et al. 1998). Rfree and Rcryst were used to monitor the progress of refinement (Brünger et al. 1987). The refinement was pursued with simulated annealing and at the final stages with B-factor refinement and release of the noncrystallographic restraints. SigmaA-weighted electron density maps (|Fo| − |Fc| and 2|Fo| − |Fc|) were calculated after each cycle of refinement and visualized using the program "O" (Jones et al. 1991). Water molecules were added to the model towards the end of refinement with the aid of difference electron density maps. The final model has a crystallographic R-factor (Rcryst) of 21.4% for all data from 40 to 2.8 Å resolution, and an Rfree of 28.0% for 5% of the data omitted (Table 1).
Equilibrium dialysis
The affinity of SpeA1 for 65Zn2+ (NEN; 171 mCi/mg) and SpeA1 mutants was determined by equilibrium dialysis using cells purchased from Amika. The native toxin and the mutants were purified as described above, and preparations were assayed for metal content using a Jarrell-Ash 965 ICP Plasma Emission Spectrophotometer (Chemical Analysis Laboratory, University of Georgia) prior to analysis.
Acknowledgments
This work is supported by the Medical Research Council (U.K.) Program grant (9540039) to K.R.A., a postgraduate studentship to M.B., and in part by a PHS grant (AI 42353) to C.M.C. from the National Institutes of Health (USA). The work at EMBL, Hamburg (HASYLAB, c/o DESY, Germany) and MAX Lab, Lund (Sweden) was supported by the European Union (EU TMR/LSF Grants). We are grateful to the staff at the Synchrotron Radiation Sources at Hamburg and Lund for their help during X-ray data collection, and members of Acharya group for the constructive criticisms of the manuscript.
The atomic coordinates of SpeA1–zinc complex have been deposited with the RCSB Protein Data Bank (accession code 1HA5).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.330101.
References
- Abrahamsén, L., Dohlsten, M., Segrén, S., Björk, P., Jonsson, E., and Kalland, T. 1995. Characterisation of two distinct MHC class II binding sites in the superantigen staphylococcal enterotoxin A. EMBO J. 14 2978–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, I.L., Nadassy, K., and Wodak, S.J. 1998. Analysis of zinc-binding sites in protein crystal structures. Protein Sci. 7 1700–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., GrosseKunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Kuriyan, J., and Karplus, M. 1987. Crystallographic R-factor refinement by molecular dynamics. Science 235 458–460. [DOI] [PubMed] [Google Scholar]
- Cavillin, A., Arozenius, H., Kristensson, K., Antonsson, P., Otzen, D.E., Bjork, P., and Forsburg, G. 2000. The spectral analysis and thermodynamic properties of Staphylococcal enterotoxin A, E and variants suggests that structural modifications are important. J. Biol. Chem. 275 1165–1172. [DOI] [PubMed] [Google Scholar]
- Chausee, M.S., Liu, J., Stevens, D.L., and Ferretti, J.J. 1996. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J. Infect. Dis. 173 901–908. [DOI] [PubMed] [Google Scholar]
- Chicz, R.M., Urban, R.G., Lane, W.S., Gorga, J.C., Stern, L.J., Vignall, D.A.A., and Strominger, J.L. 1992. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 358 764–768. [DOI] [PubMed] [Google Scholar]
- Earhart, C.A., Vath, G.M., Roggiani, M., Schlievert, P.M., and Ohlendorf, D.H. 2000. Structure of streptococcal pyrogenic exotoxin A reveals a novel metal cluster. Protein Sci. 9 1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf, R.M. 1997. An extensively modified version of Molscript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15 132–134. [DOI] [PubMed] [Google Scholar]
- Fraser, J.D. 1993. 49th forum in immunology—T-cell recognition of superantigens. Res. Immunol. 144 173–174. [DOI] [PubMed] [Google Scholar]
- Häkansson, M., Petersson, K., Nilsson, H., Forsberg, G., Björk, P., Antonsson, P., and Svensson, L.A. 2000. The crystal structure of staphylococcal enterotoxin H: Implications for binding properties to MHC class II and TCR molecules. J. Mol. Biol. 302 527–537. [DOI] [PubMed] [Google Scholar]
- Hartwig, U.F. and Fleisher, B. 1993. Mutations affecting MHC class II binding of the superantigen streptococcal erythrogenic toxin A. Eur. J. Immunol. 5 869–875. [DOI] [PubMed] [Google Scholar]
- Herman, A., Kappler, J.W., Marrack, P., and Pullen, A.M. 1991. Superantigens—Mechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol. 9 745–772. [DOI] [PubMed] [Google Scholar]
- Jardetzky, T.S., Brown, J.H., Gorga, J.C., Urban, R.G., Chi, Y.I., Stauffacher, C., Strominger, J.L., and Wiley, D.C. 1994. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature 368 711–718. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zon, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47 110–119. [DOI] [PubMed] [Google Scholar]
- Karp, D.R. and Long, E.O. 1992. Identification of HLA-DR1 β-chain residues critical for binding staphylococcal enterotoxins A and E. J. Exp. Med. 175 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, J.B. and Collins, C.M. 1996. Analysis of the superantigenic activity of mutant and allelic forms of Streptococcal pyrogenic exotoxin A. Infect. Immun. 64 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack, P. and Kappler, J. 1990. The staphylococcal enterotoxins and their relatives. Science 248 705–711. [DOI] [PubMed] [Google Scholar]
- Musser, J.M., Hauser, A.R., Kim, M.H., Schlievert, P.M., Nelson, K., and Selander, R.K. 1991. Streptococcus pyogenes causing toxic-shock like syndrome and their invasive diseases: Clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. 88 2668–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Papageorgiou, A.C. and Acharya, K.R. 2000. Microbial superantigens: From structure to function. Trends Microbiol. 8 369–375. [DOI] [PubMed] [Google Scholar]
- Papageorgiou, A.C., Acharya, K.R., Shapiro, R., Passalacqua, E.F., Brehm, R.D., and Tranter, H.S. 1995. Crystal structure of the superantigen enterotoxin C2 from Staphylococcus aureus reveals a zinc binding site. Structure 3 769–779. [DOI] [PubMed] [Google Scholar]
- Papageorgiou, A.C., Collins, C.M., Gutman, D.M., Kline, J.B., O'Brian, S.M., Tranter, H.S., and Acharya, K.R. 1999. Structural basis for the recognition of superantigen streptococcal pyrogenic exotoxin A1 (SpeA1) by MHC class II molecules and T-cell receptors. EMBO J. 18 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussell, A., Anderson, B.F., Baker, H.M., Fraser, J.D., and Baker, E.N. 1997. Crystal structure of the Streptococcal superantigen SpeC: Dimerisation and zinc binding suggest a novel mode of interaction with MHC class II molecules. Nat. Struct. Biol. 4 635–643. [DOI] [PubMed] [Google Scholar]
- Schad, E.M., Papageorgiou, A.C., Svensson, L.A., and Acharya, K.R. 1997. A structural and functional comparison of staphylococcal enterotoxins A and C2 reveals remarkable similarity and dissimilarity. J. Mol. Biol. 269 270–280. [DOI] [PubMed] [Google Scholar]
- Schad, E.M., Zaitseva, I., Zaitsev, V.N., Dohlsten, M., Kalland, T., Schlievert, P.M., Ohlendorf, D.H., and Svensson, L.A. 1995. Crystal structure of the superantigen Staphylococcal enterotoxin type A. EMBO J. 14 3292–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffenbauer, J., Didier, D.K., Klearman, M., Rice, K., Shuman, S., Tieber, V.L., Kittlesen, D.J., and Schwartz, B.D. 1987. Complete sequence of the HLA DQ alpha and beta cDNA from a DRS/DQW3 cell line. J. Immunol. 139 228–233. [PubMed] [Google Scholar]
- Sundström, M., Abrahamsen, L., Antonsson, P., Mehindate, K., Mourad, W., and Dohlsten, M. 1996a. The crystal structure of Staphylococcal enterotoxin type D reveals zinc-mediated homodimerisation. EMBO J. 15 6832–6840. [PMC free article] [PubMed] [Google Scholar]
- Sundström, M., Hallen, D., Svensson, A., Schad, E., and Dohlsten, M. 1996b. The co-crystal structure of Staphylococcal enterotoxin type A with zinc at 2.7 Å resolution. Implications for MHC class II binding. J. Biol. Chem. 271 32212–32216. [DOI] [PubMed] [Google Scholar]
- Stevens, D.L. 1995. Streptococcal toxic-shock syndrome—Spectrum of disease, pathogenesis and new concepts in treatment. Emerg. Infect. Dis. 1 109–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R., Cole, G.A., Surman, S., Blackman, M.A., and Woodland, D.L. 1996. MHC class II-associated peptides control the presentation of bacterial superantigens to T-cells. J. Exp. Med. 183 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]