Abstract
Amide hydrogen exchange and mass spectrometry have been used to study the pH-induced structural changes in the capsid of brome mosaic virus (BMV). Capsid protein was labeled in a structurally sensitive way by incubating intact viral particles in D2O at pH 5.4 and 7.3. Deuterium levels in the intact coat protein and its proteolytic fragments were determined by mass spectrometry. The largest deuterium increases induced by structural alteration occurred in the regions around the quasi-threefold axes, which are located at the center of the asymmetric unit. The increased levels of deuterium indicate loosening of structure in these regions. This observation confirms the previously proposed swelling model for BMV and cowpea chlorotic mottle virus (CCMV) and is consistent with the structure of swollen CCMV recently determined by cryo-electron microscopy and image reconstruction. Structural changes in the extended N- and C-terminal arms were also detected and compared with the results obtained with other swollen plant viruses. This study demonstrates that protein fragmentation/amide hydrogen exchange is a useful tool for probing structural changes in viral capsids.
Keywords: Brome mosaic virus, amide hydrogen exchange, mass spectrometry, viral capsid
Most viruses consist of a genome enclosed in a protein shell or capsid. The life cycle of a virus involves several essential stages, including binding to a cellular surface, internalization into the cytoplasm, disassembly to release the viral genome for replication, reassembly of the capsid, and externalization of progeny virions to initiate a new round of replication. Most stages of the life cycle are accompanied by structural changes in the viral capsid. For example, assembly and disassembly inside the cell are processes that involve major structural changes in the capsid. Identification and characterization of structural changes in viral capsids are essential to understand and control the viral life cycle.
X-Ray diffraction and electron microscopy (EM) combined with image reconstruction have been the principal tools used to detect structural changes in viral capsids (Giranda et al. 1992; Mancini et al. 1997). Although NMR may be used to determine the structures of some capsid proteins as isolated subunits, it has not been suitable for high-resolution structural studies of assembled virus particles. The high-resolution structures derived from X-ray diffraction provide an important starting point for our understanding of the relation between structure and function. However, these structures provide only a static picture of the virion under non-physiological conditions. Other methods, such as ultracentrifugation (Incardona and Kaesberg 1964), limited proteolysis (Chidlow and Tremaine 1971; Lewis et al. 1998), and various spectroscopic methods (Vriend et al. 1982; Tuma and Thomas 1997; Leimkuhler et al. 2000; Perez et al. 2000), have been used to detect structural changes in viral capsids. Although the spatial resolution of these methods is low, they are essential tools for studies aimed at relating viral structure and function and are compatible with a wide range of experimental conditions.
Isotope exchange at peptide amide linkages is another method for detecting protein structural changes under diverse conditions (Englander et al. 1997; Li and Woodward 1999). Although hydrogen exchange (HX) has most often been detected by NMR, detection by mass spectrometry (MS) has become increasingly important (Smith et al. 1997; Coyle et al. 1999; Engen et al. 1999; Halgand et al. 1999). The rates at which hydrogens located at peptide amide linkages in proteins undergo isotopic exchange are highly sensitive to protein structure. Principal factors affecting hydrogen exchange rates include intramolecular hydrogen bonding and solvent accessibility. Changes in these structural features have been detected by analyzing the intact protein. Although this approach gives important information on the extent and cooperativity of structural changes, it provides no information on the locations of structural changes. However, the spatial resolution of HX MS can be substantially improved when the labeled protein is fragmented prior to mass analysis (Zhang and Smith 1993).
Brome mosaic virus (BMV) and cowpea chlorotic mottle virus (CCMV) are simple plant viruses with similar capsid structures (Krol et al. 1999).Both viruses have 180 chemically identical coat protein subunits arranged to form an icosahedral surface. The capsid volume of both BMV and CCMV increases as the pH is increased from 5 to 7 (Lane 1981). Understanding the molecular basis of capsid swelling, which is a common feature of many plant viruses, is required for our understanding of capsid assembly and disassembly processes. Combining the X-ray structure of the native virus with the cryo-electron microscopy (cryo-EM) image of the swollen CCMV has provided detailed structural information on the pH-induced swelling of CCMV (Speir et al. 1995). Such information has not been reported for BMV until now.
The present study explores the limits of HX MS for detecting structural changes in viral capsids. Results of this study also establish the similarity of mechanisms responsible for pH-induced swelling in BMV and CCMV. This report includes a description of a method for separating the effects of pH on capsid structure and the intrinsic rate of amide hydrogen exchange. This approach was used to follow hydrogen exchange in intact BMV equilibrated at pH 5 and 7. The labeled virus capsid was rapidly disassembled under conditions that minimized isotope exchange. Changes in deuterium levels in the labeled capsid protein and its peptic fragments were used to identify regions undergoing structural change with expansion of the capsid.
Results
BMV capsid protein sequence and peptic fragmentation
Prior to hydrogen exchange experiments, the BMV capsid protein was isolated from the intact virus of a greenhouse culture of BMV and analyzed by mass spectrometry to verify the amino acid sequence. Analysis of intact virus particles by reversed phase HPLC indicated one major component eluting at 68% acetonitrile. The fraction corresponding to the major peak was collected, concentrated, and analyzed by electrospray ionization mass spectrometry (ESIMS). The molecular mass was found to be 20,264 ± 2 D, which is 12 D greater than the mass calculated from the reported BMV sequence (Ahlquist et al. 1981; Dasgupta and Kaesberg 1982). This difference showed that the sequence of BMV used in this study did not match the published sequence. Enzymatic digestion and peptide mapping were performed to identify the sequence differences or modifications. Trypsin and Lys C digestions produced fragments that covered 100% of the sequence. Analyses of peptides found in these digests by MS and collision-induced dissociation (CID) MS/MS indicated the following modifications: (1) elimination of the N-terminal methionine; (2) acetylation of the N-terminal serine; and (3) substitution of arginine for tryptophan 23 (data not shown).
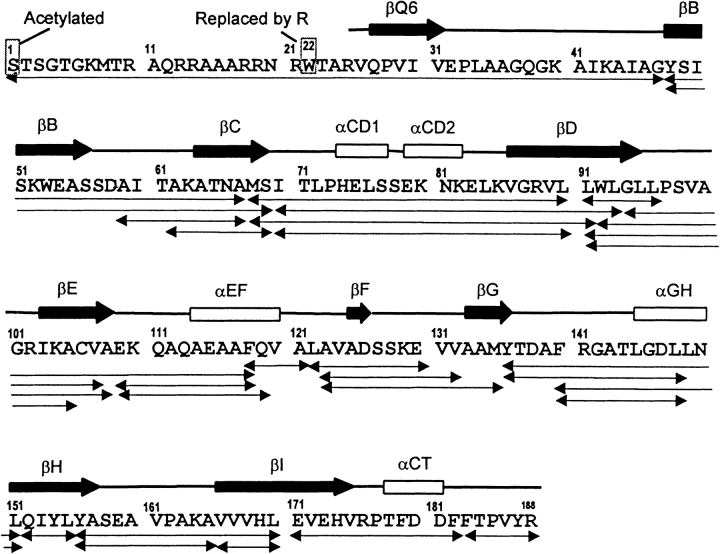
A similar peptide-mapping approach was used to identify peptic fragments formed when the BMV capsid protein was digested under conditions used to quench isotope exchange (0°C, pH 2.5). Digestion with pepsin produced approximately 30 peptic fragments, which covered 99% of the BMV capsid protein sequence. Identification of these fragments was verified by CID MS/MS and C-terminal sequencing. The correct amino acid sequence of this BMV isolate and the peptic fragments used in this study are presented in Figure 1 ▶.
Fig. 1.
Amino acid sequence of BMV capsid protein indicating changes found in the sequence and the peptic fragments used in this study. The α-helices and β-strands are illustrated schematically above the sequence.
Designing experiments to detect pH-induced structural changes by hydrogen exchange
The pH-induced swelling of the BMV capsid was detected from the changes in HX rates found when the intact virus particles were incubated in D2O at pD 5.4 and 7.3. The experimental procedure used in this study was designed to account for the fact that amide hydrogen exchange rates in unfolded polypeptides depend on pH. The general background on amide hydrogen exchange in proteins has been described (Englander and Kallenbach 1984; Miller and Dill 1995; Li and Woodward 1999). It is important to note that the present labeling experiments were performed under conditions strongly favoring the folded forms of the capsid protein (i.e., EX2 conditions). Under these conditions, the deuterium level at any peptide amide linkage is given by equation 1:
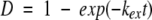 |
1 |
where kex is the phenomenological rate constant for isotope exchange and t is the time the protein was exposed to D2O. The phenomenological rate constant may be expressed as follows:
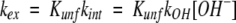 |
2 |
where Kunf is an equilibrium constant describing structural changes that facilitate isotope exchange, kint is the intrinsic isotope exchange rate constant, and kOH is the rate constant for base-catalyzed isotope exchange. The intrinsic exchange rate constant, kint, is given by the product of kOH and [OH−] (Bai et al. 1993). Insertion of Kunf, which may be viewed as the probability for various structural changes that enable isotope exchange in a folded polypeptide, provides the link between hydrogen exchange rates and protein structure. Structural changes enabling isotope exchange may be global unfolding or minor fluctuations that allow exchange at individual amide linkages. The goal of this research was to assess changes in Kunf as the pH was increased from 5 to 7. However, equation 2 indicates that kint at pH 7 is 100-fold greater than it is at pH 5. Substituting equation 2 into equation 1 gives equation 3:
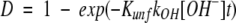 |
3 |
which indicates that pH-induced structural changes in folded proteins may be detected by hydrogen exchange if the exposure times were chosen so that the products of [OH−] and t were constant. This general approach has been used previously to compare hydrogen exchange results obtained at different temperatures (Zhang and Smith 1993,Zhang and Smith 1996).
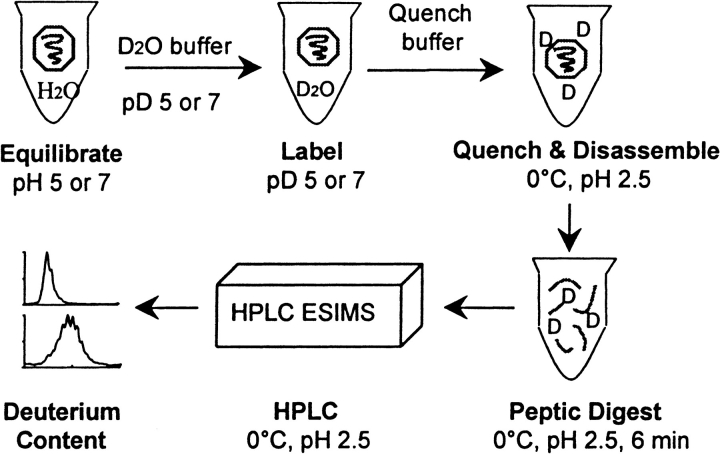
The experimental procedure used to detect pH-induced swelling of the BMV capsid is illustrated in Figure 2 ▶. To initiate isotope exchange, solutions of equilibrated intact BMV were diluted 20-fold into two exchange buffers. The pD of these labeling solutions was either 5.43 or 7.30. Following various labeling times, the solution was acidified both to quench the isotope exchange reaction and to disassemble the virus particles. The labeled capsid protein was analyzed either as the intact protein or as its peptic fragments by HPLC ESIMS, always maintaining conditions required to quench isotope exchange. Changes in deuterium levels found following labeling at pD 5.43 or 7.30 were used to detect structural changes in the capsid. To null the dependence of kint on pD, the labeling times were chosen to keep the product of [OH−] and t constant. That is, the labeling time for pH 5.43 was 74.1 times greater than the labeling time used for pH 7.30. Incubation times used in this study are given in Table 1.
Fig. 2.
General procedure used to label the BMV capsid protein in the intact viral particles and to determine deuterium levels at peptide amide linkages by HPLC ESI MS.
Table 1.
Exchange times used to label intact BMV in D2O at pD 5.43 and 7.30 a
| pD 7.30 (min) | PD 5.43 (min) |
| 0.33 | 24.5 |
| 1.00 | 74.1 |
| 5.50 | 407.6 |
| 30.00 | 2223.0 |
| 90.00 | 7113.6 |
a The labeling time at pD 5.43 was 74.1 times longer than the labeling time at pD 7.30.
Analysis of intact capsid protein labeled in assembled virus particles
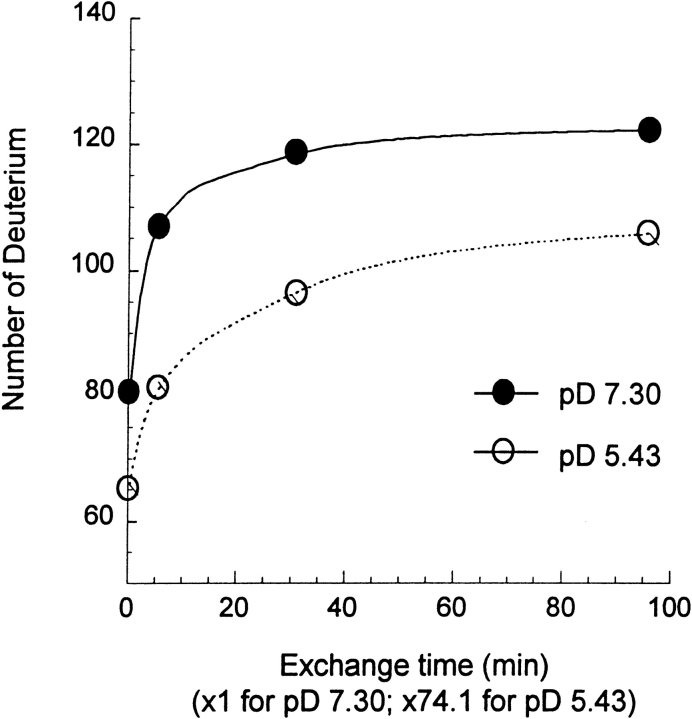
Global changes in the BMV capsid were detected from changes in the deuterium levels found by analyzing the intact protein by HPLC ESIMS. Closed data points in Figure 3 ▶ indicate the deuterium levels when the intact virus particles were labeled at pD 7.30 for 0.33–96 min. These results, which have been adjusted for deuterium loss (11%) during the HPLC step, reflect the number of deuteriums located at peptide amide linkages. Deuterium located in the side chains, as well as at the N and C termini, was lost during HPLC. The results in Figure 3 ▶ show that the deuterium level rose sharply during the shortest labeling times, but reached a level of only 123 after labeling for 96 min at pD 7.30. Finding only 123 deuteriums in this protein that has 180 amide hydrogens (7 of 188 residues are proline) indicates that isotope exchange was incomplete on this time scale. These results show that exchange in the virus particle occurs over a time scale of several orders of magnitude, which is typical for folded proteins.
Fig. 3.
The number of deuteriums found at peptide amide linkages in intact BMV capsid protein following labeling of the intact virus in D2O for various times. The X axis indicates the labeling times used at pD 7.30 (×1) and pD 5.43 (×74.1).
Deuterium levels found in the intact capsid protein following labeling of the virus particles at pH 5.43 for 25 min to 118 h are also presented in Figure 3 ▶ (open data points). The time scale used to plot these results has been divided by 74.1 to adjust for the different values of kint at pD 5.43 and 7.30. Although the general behavior found for hydrogen exchange at pD 5.43 and 7.30 is similar, the deuterium levels at pD 7.30 are much greater than the deuterium levels found at pD 5.43. This difference (15%–25%) shows that the BMV capsid structures at pD 5.43 and 7.30 are substantially different. The higher level of deuterium found at pD 7.30 indicates that the structure at pD 7.30 is more open or flexible than the structure at pD 5.43. The estimated uncertainty in these measurements is represented by the size of the data points in Figure 3 ▶.
Analysis of peptic fragments of labeled capsid protein
To detect localized structural changes in the BMV coat protein, the labeled virus was disassembled into the monomer subunits, which were digested with pepsin and analyzed by directly coupled HPLC ESIMS. The digestion and HPLC were performed under conditions that minimized isotope exchange at peptide amide linkages (Bai et al. 1993). Deuterium levels in the peptic fragments were deduced from their molecular masses. The extent of artificial exchange occurring during digestion and HPLC was determined by analyzing under the same conditions BMV coat protein that either had no deuterium or was completely exchanged in D2O. Although the extent of artificial exchange depended on the amino acid sequences of the peptides, the deuterium recovery averaged over all peptic fragments was 67%.
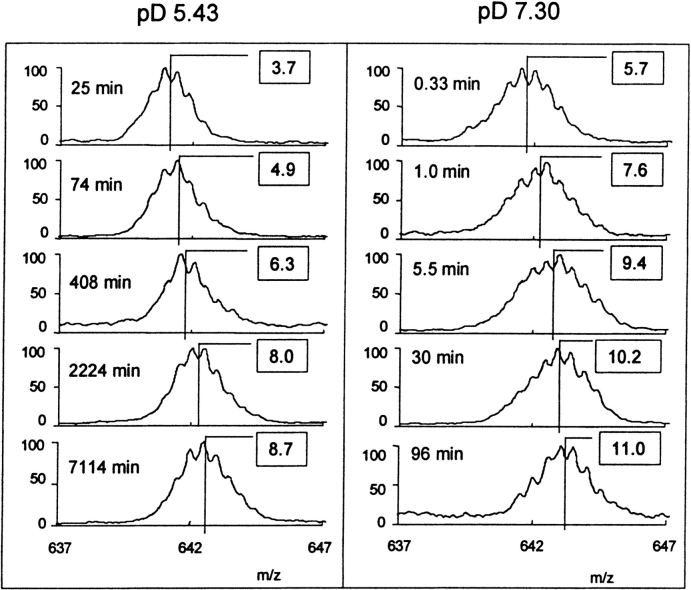
The deuterium levels found in these peptic fragments, following labeling of the intact BMV particles in D2O at pD 5 and 7, were used to detect pH-dependent changes in the structure of the capsid protein. Mass spectra of one representative peptic fragment, including residues 123–135, are presented in Figure 4 ▶. These spectra, as well as those of all peptic fragments used in this study, exhibited a single envelope of isotopic peaks, consistent with a high level of structural homogeneity (Miranker et al. 1993; Zhang et al. 1996). That is, isotope exchange in backbone regions represented by any peptic fragment was the same for all molecules, indicating a high level of structural homogeneity in the 180 monomers comprising the BMV capsid. The deuterium levels in this peptic fragment (Fig. 4 ▶, boxes) were determined from the centroids of the envelopes of isotope peaks. Results presented in Figure 4 ▶ show how the deuterium level in the segment including residues 123–135 increased with labeling time. The labeling times, which are given next to each spectrum, were chosen so that the labeling time at pD 5.43 was a factor of 74.1 greater than the labeling time at pD 7.30 (see Table 1). To detect structural changes in the region including residues 123–135, the deuterium level found in this segment from BMV incubated for 74 min at pD 5.43 is compared with the deuterium level found in the same segment from BMV incubated for 1 min at pD 7.30. The data in Figure 4 ▶ show that the deuterium level increased from 4.9 to 7.6 when the pD was increased from 5.43 to 7.30. This increase in deuterium level indicates a general loosening of the BMV capsid protein in the region including residues 123–135.
Fig. 4.
Mass spectra for the doubly charged peptic fragment including residues 123–135 of the BMV capsid protein labeled in the assembled virus at pD 5.43 and pD 7.30 for various times. The labeling times were selected to facilitate direct comparison of deuterium levels found following labeling at the different pDs. The deuterium levels in this segment (presented in boxes) were determined from the centroids (solid vertical lines) of the envelopes of isotope peaks.
The width of the envelopes of isotope peaks presented in Figure 4 ▶ indicates the intermolecular distribution of deuterium in this region of the protein. Peak widths presented in Figure 4 ▶ show that the distribution is broader for labeling at pD 7.30 than it is at pD 5.43. The broader peak at the higher pD is consistent with the higher levels of deuterium found under these conditions. As discussed previously, the peak width is attributable to the number of amide linkages that are only partially deuterated (Zhang et al. 1996).
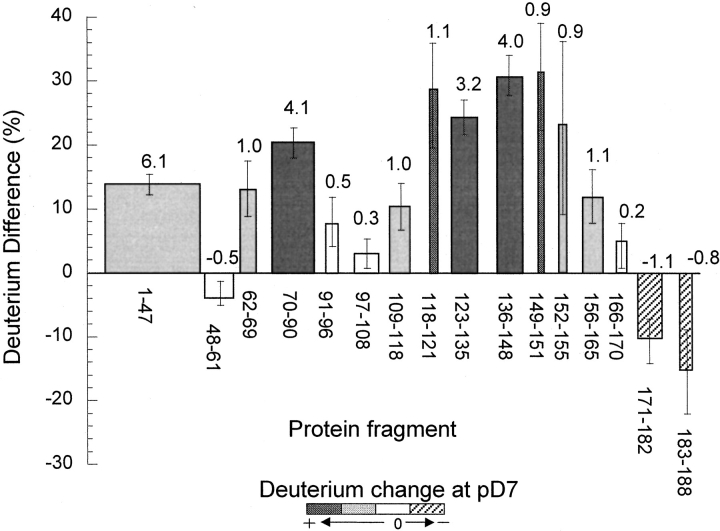
Differences in deuterium incorporation at pD 5.43 and 7.30 are presented in Figure 5 ▶ for a set of 16 segments that cover nearly the entire backbone of the BMV capsid protein. Many additional peptides with amino acid sequences overlapping with these peptides gave similar results. The spatial resolution, which is generally limited by the lengths of the peptic fragments, was enhanced in three regions by using the differences in deuterium levels found in overlapping peptides. The change in deuterium level, indicated by the Y axis in Figure 5 ▶, is expressed as the percent of the total number of amide hydrogens in each peptic fragment. As indicated by the results presented in Figure 4 ▶, the pH-induced change in deuterium levels changed with the labeling time. To simplify the discussion, only the results for labeling times where the change was greatest are presented in Figure 5 ▶. The change in the number of deuteriums found in each segment at the different pDs is given above each bar.
Fig. 5.
Changes in deuterium levels found in segments of the BMV capsid protein following labeling of the intact virus particles at pD 5.43 or 7.30. The changes are plotted as rectangular boxes, where the height indicates the change expressed as a percentage of the total number of amide hydrogens in the peptide and the width indicates the length of the backbone segment. The changes in deuterium levels expressed as the number of moles of deuterium per mole of peptide are given above each bar. Uncertainties estimated for each segment are indicated by the error bars, and shading indicates regions where the largest or smallest changes were found.
It is useful to present changes in deuterium levels in both relative and absolute units because each method has specific merits. Expressing the deuterium difference as the average difference per amide linkage facilitates the comparison of long and short fragments. However, showing the difference in the absolute number of deuteriums facilitates comparing the deuterium difference with the uncertainty of the measurement, which is typically <0.2 D for singly charged ions. This uncertainty was one of the criteria used to identify detectable, pH-induced structural changes. The 0.2-D uncertainty is attributed to uncertainties in sample preparation, instrument calibration, and data processing. The uncertainty in the determination of the change in deuterium level in each segment, expressed as percent, is indicated by the error bars in Figure 5 ▶.
Results presented in Figure 5 ▶ show that most segments (residues 1–170, 90% of the protein backbone) exhibited increased deuterium levels at pD 7, suggesting a looser or more flexible structure of the protein subunits in the swollen capsid. However, peptides corresponding to residues 171–188 in the C terminus appear to incorporate less deuterium at pH 7 than at pH 5, indicating a more compact or ordered structure in this region. Among the segments exhibiting increases in deuterium, three categories were defined based on the magnitude of the change. Moderate deuterium changes (less than 10%) were found for segments including residues 48–61, 91–108, and 166–170, indicating minimal structural changes. Intermediate increases (10%–20%) were found for segments including residues 1–47, 62–69, 109–118, and 152–165, indicating more substantial structural changes. Large increases in deuterium level were found in segments including residues 70–90 and 118–151, indicating extensive structural changes. These three categories of exchange are indicated by shading in Figure 5 ▶.
Discussion
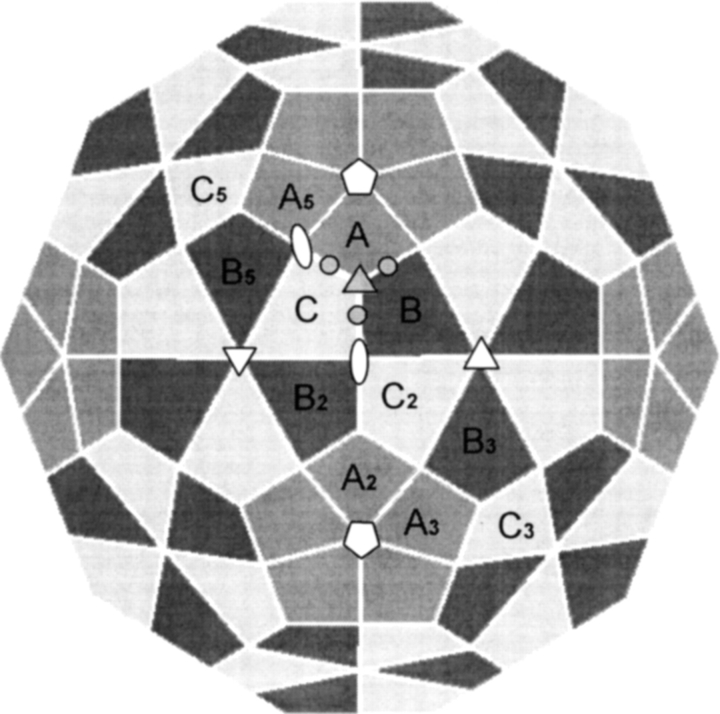
Bromoviruses are a group of small spherical viruses that have the same general hexamer–pentamer, T = 3 surface lattice class (Bancroft 1970). That is, the 180 coat protein subunits that make up the capsids are organized into 12 pentamers and 20 hexamers forming a shell with T = 3 icosahedral symmetry. A truncated model illustrating some of the pentamers and hexamers is presented in Figure 6 ▶. Although all 180 subunits have nominally identical structures, symmetry considerations lead to designation of three groups, A, B, and C. The A and B groups make up the hexamers, and the C groups make up the pentamers. The icosahedral threefold (quasi-sixfold) and fivefold axes of the hexamers and pentamers are illustrated in Figure 6 ▶ as triangles and pentagons, respectively. Points of twofold symmetry arising from dimers are indicated as ovals. The threefold axis at the center of the icosahedral asymmetric unit (central triangle in Fig. 6 ▶) is not exact (quasi-threefold axis) because of the different rotation axes outside of its local environment.
Fig. 6.
A truncated icosahedral model for BMV capsid. Polygons in three different gray scales (A, B, and C) represent chemically identical protein subunits occupying slightly different geometrical environments. Positions of fivefold, threefold, and twofold rotation axes are marked as pentagons, triangles, and ovals, respectively. Subscripts are used to relate subunits by icosahedral symmetry (A to A5 by fivefold rotation). The asymmetric unit is defined as a quasiequivalent trimer of A, B, and C. The quasi-threefold axis is indicated by the shaded triangle at the center of the trimer. The small circles indicate putative calcium-binding sites in the CCMV model (Speir et al. 1995).
Analysis of the CCMV crystal structure has revealed the general structure of bromovirus coat protein subunits, as well as many details of their interactions between subunits (Speir et al. 1995). The structure of the capsid protein subunit is an eight-stranded, antiparallel β-barrel with the N- and C-terminal arms extending outward (Fig. 7a ▶). The N- and C-terminal arms of the capsid subunits interweave into a network to stabilize the capsid. For example, six N-terminal short β-strands cluster into a parallel β-tube structure, which stabilizes the hexameric units. The extended C-terminal arm (residues 180–190) appears to interact with the adjacent twofold related subunit by penetrating into a gap formed by the N-terminal arm and the β-barrel core.
Fig. 7.
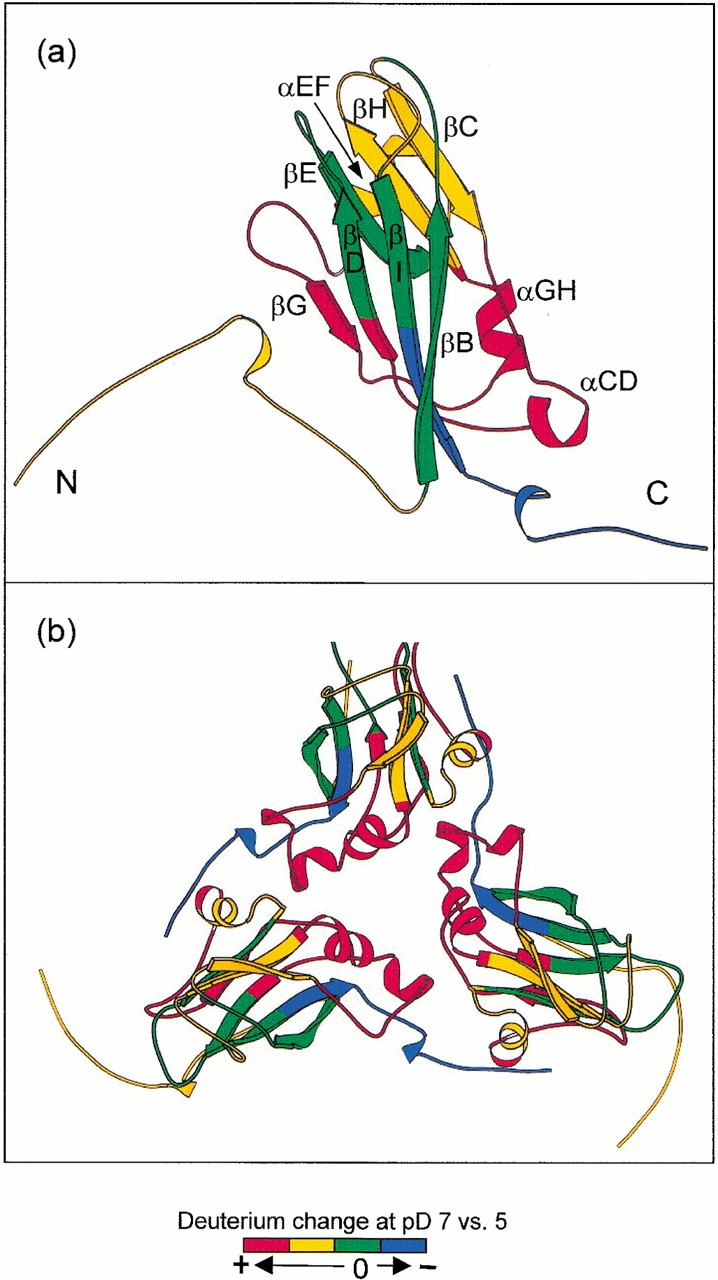
Ribbon plot of the BMV capsid protein indicating changes in deuterium levels found following labeling of intact BMV particles in D2O at pD 5.43 or 7.30. (a) Capsid protein monomer; (b) asymmetric unit trimer. These structures are based on the X-ray crystal structure of the closely related virus CCMV (PDB 1cwp). Orientation: (a) Virus exterior is at the top of the diagram. (b) Viewed from the exterior down the quasi-threefold axis. The different colors indicate the extent of pH-induced structural change sensed by hydrogen exchange.
Capsid expansion with increasing pH in small RNA plant viruses such as BMV and CCMV has been studied extensively (Incardona and Kaesberg 1964; Bancroft et al. 1967; Adolph 1975; Hsu et al. 1976; Hull 1977). In an early study, it was noted that BMV underwent a reversible decrease in its sedimentation coefficient when the pH was increased from 5 to 7 (Incardona and Kaesberg 1964). Measurements of diffusion and viscosity coefficients of swollen BMV together with X-ray scattering experiments established that the hydrodynamic radius of the virus particles increased by about 10% while the molecular mass was unchanged. Virus swelling has also been studied by various spectroscopic methods, such as circular dichroism, ultraviolet absorption, and proton NMR (Incardona et al. 1973; Vriend et al. 1982). Results of these studies provided strong evidence for a structural transition during swelling, but detailed structural information was not available. However, early titration measurements suggested that deprotonation of side-chain carboxylic acids may be involved with pH-induced capsid expansion (Jacrot 1975).
Our most detailed structural information on the swelling of bromoviruses comes from fitting the 28-Å resolution cryo-EM density map of the swollen capsid with the high-resolution atomic model of individual subunits of CCMV coat protein (Speir et al. 1995). This model indicates the presence of large holes at the center of the quasiequivalent trimer (Fig. 6 ▶, triangle at the trimer interface). Putative calcium-binding sites (small circles in Fig. 6 ▶) consisting of clusters of aspartic and glutamic acid residues were identified in the protein trimer interface. These acidic residues were assumed to be protonated at pH 5 but deprotonated at pH 7, creating electrostatic repulsion that triggered expansion of the capsid. However, pentamers and hexamers retain their shapes, and the dimeric protein–protein interactions remain largely intact in the swollen capsid. One of the goals of the present study was to determine whether a similar model may be used to explain acid-induced expansion of the BMV capsid.
Structural changes in BMV capsid monomers
To locate structural changes within BMV capsid subunits, the amino acid sequences of the CCMV and BMV coat protein were aligned. The high sequence identity (70%) of the proteins permits alignment with minimal ambiguity, indicating that the subunit structure of BMV may be represented by the atomic coordinates for CCMV. A similar scheme has been used to locate modified residues in the capsid of a BMV mutant (Flasinski et al. 1997).
Specific regions of the BMV backbone are colored in Figure 7a ▶ to indicate four categories of change in the deuterium levels found when the pH was increased from 5 to 7 (see Fig. 5 ▶). Segments exhibiting the largest increases (>20%) are colored red, and segments with intermediate (10%–20%) or minimal increases (<10%) in deuterium are indicated by yellow and green, respectively. Regions labeled blue in Figure 7a ▶ had decreased levels of deuterium at pH 7. The color scheme in Figure 7a ▶ offers a qualitative, low-resolution view of structural changes in various regions of BMV coat protein as the pH was increased from 5 to 7. The strong dependence of amide hydrogen exchange rates on intramolecular hydrogen bonding and access to the solvent suggests that the increased hydrogen exchange rates correlate with expansion of the viral capsid. The widespread distribution of regions exhibiting increased hydrogen exchange indicates that expansion of the capsid at the higher pH involves loosening of the structure along much of the capsid protein backbone.
Regions exhibiting different levels of hydrogen exchange can be related to specific structural elements. For example, the largest increases in deuterium (shown in red) occurred in backbone segments including three α-helices (αCD, most of αEF and αGH), two β-strands (βF and βG), and loops that connect them. Intermediate changes in deuterium levels were found in the extended N-terminal arm including part of βC, βH, the HI loop, and part of αEF (shown in yellow). The yellow β-barrel segments were found adjacent to the red regions, indicating that the larger structural changes in the red areas caused weakening of the β-barrel fold. However, finding only small changes in several β-strands (green regions including βB, βD, βE, and βI) shows that the major part of the canonical antiparallel β-barrel core remained essentially intact in the expanded form of the capsid.
The increased deuterium level in the N terminus may reflect weakening of the β-hexameric structure that stabilizes the hexamer. Modeling of X-ray and electron microscopy results indicates that the parallel β-tube is partially disrupted in swollen CCMV (Speir et al. 1995). In addition, proteolysis experiments of swollen BMV produced primarily N-terminal peptides, suggesting increased exposure of the N terminus in the expanded virus (Chidlow and Tremaine 1971; Agrawal and Tremaine 1972). However, the expanded BMV capsid, though loosely structured, is still an assembled form, suggesting that the protein–protein hexameric interaction is retained in the swollen virus.
Two fragments in the C terminus including residues 171–188 (blue in Fig. 7 ▶) exhibited decreased levels of deuterium at pH 7. This decrease in hydrogen exchange suggests tightening of structure in the vicinity of the C terminus as the virion expands. The CCMV crystal structure indicates that the C terminus (residues 181–190) is clamped between the N-terminal arm (residues 36–54) and the β-strand wall (βB-I-D-G) of the adjacent twofold related protein subunit. This inter-subunit interaction may be the principal factor responsible for the high stability of dimers. A closer look at the exchange data from the clamping site also supports strong dimer interactions. The clamping site consists of a β-strand wall (βB-I-D-G) and the N-terminal arm. The βB, βI, and βD strands (shown in green in Fig. 7 ▶) showed minimal deuterium change, indicating the stability of the β-wall structure. The deuterium level in the peptide segment containing the βB strand decreased slightly (see residues 48–61 in Fig. 5 ▶). Although the deuterium level in the long N-terminal segment increased with swelling of the virus, it is possible that exchange in some regions within this segment decreased with capsid expansion.
The present hydrogen exchange results suggest that this inter-subunit interaction remains strong in the expanded virus. This view is consistent with results obtained by other methods. Cryo-EM of swollen CCMV indicates a strong protein–protein dimeric interaction (Speir et al. 1995). Increased structure within the N-terminal segment with virion expansion also agrees with an X-ray study of the swollen state of tomato bushy stunt virus (TBSV), which suggests formation of new secondary structural elements in the N terminus in a similar pH-induced swelling process (Robinson and Harrison 1982). This persistent interaction of the C terminus of one subunit with a pocket in an adjacent subunit is consistent with the high stability of the dimer in solution after disassembly of the swollen virus (Bancroft 1970).
Structural changes in the BMV capsid trimer interface
Both cryo-EM studies of CCMV (Speir et al. 1995) and crystallographic studies of swollen TBSV (Robinson and Harrison 1982) indicate that large structural changes occur in the trimer interface with swelling. This region is indicated by the triangle joining two hexamers and one pentamer in Figure 6 ▶. To examine the relation between the present hydrogen exchange results and structural changes in the trimer interface of BMV, a ribbon plot of the three subunits (A, B, and C) comprising this interface is presented in Figure 7b ▶. The quasi-threefold axis is located at the center of Figure 7b ▶. The color scheme, which is the same as used in Figure 7a ▶, shows how the deuterium levels changed with swelling of the BMV capsid. This presentation shows that regions with the largest increases of deuterium surround the trimer interface, indicating that weakening of protein–protein interactions in the trimer interface is an important part of the swelling and possibly disassembly processes in BMV.
Insight on the general mechanism for pH-induced swelling of BMV and related plant viruses may be traced to early titration experiments, which suggested that a group of acidic residues (Asp and Glue) had anomalously high pKas (Bancroft 1970). The abnormally high pKa of these carboxyl groups probably reflects the clustering effect of acidic groups and the presence of phosphate residues from RNA in the vicinity. The X-ray crystal structure of CCMV indicates clusters of carboxyl groups between neighboring subunits in the capsid quasiequivalent trimer interface (Speir et al. 1995). These carboxyl clusters, each containing three acidic residues (two Glu and one Asp) and two Glns, were modeled as calcium-binding sites. Aligning the sequence of BMV and CCMV coat proteins also led us to similar carboxyl clusters (Glu 81, Glu 85, Asp 149, and Gln 155) in the trimer interface. It is noteworthy that the helical structures containing these carboxyl groups are also the regions with the largest deuterium increases (Fig. 7b ▶). This observation with the titration results points to a swelling model in which deprotonation increases the number of carboxylate anions that repel each other, displacing near-by structural units. Finding the largest increases in hydrogen exchange in the trimer interface indicates that a similar mechanism may be responsible for swelling of BMV.
Conclusion
The pH-induced structural changes in BMV capsids have been studied by incubating the intact virus in D2O for various times followed by determining the extent of H/D exchange by mass spectrometry. Analysis of the intact capsid protein revealed substantially increased levels of deuterium in the expanded virus, indicating a general destabilization of the expanded capsid. Peptic fragmentation of the labeled capsid protein led to estimates of destabilization along the entire backbone of the protein. The largest deuterium increases were found in the region surrounding the quasi-threefold axis, which is located at the center of the icosahedral asymmetric trimer. The increased hydrogen exchange is attributed to substantially reduced stability in this region. This observation agrees with a proposed swelling model for small spherical plant virus such as BMV and CCMV, suggesting that deprotonation of Asp and Glu residues leads to expansion of the virus. This model is also consistent with recent cryo-EM and image reconstruction studies of CCMV. Although expansion of the virion was accompanied by increased deuterium levels along most of the polypeptide backbone, reduced deuterium levels were found in the C terminus. These results suggest that the C terminus and its surrounding pocket in an adjacent subunit retain a high level of stability in the expanded virion, consistent with the strong protein–protein dimer interaction between twofold related neighboring subunits.
This study points to a potentially important role for HX MS in studies of viral capsids. Although X-ray crystallography and cryo-EM with image reconstruction continue to be the principal tools for high-resolution structural studies of viruses, HX MS will be particularly useful for studying virions under a wide range of physiological and nonphysiological conditions. Potential applications of hydrogen exchange and mass spectrometry are numerous. For example, some antiviral agents bind to specific regions of viral capsids, blocking certain steps in the virus life cycle such as uncoating or cell attachment (Badger et al. 1988). Binding of antiviral agents normally induces conformational changes in the binding pocket as well as other areas of the coat protein. The present results suggest that HX MS may be ideal for determining the fingerprint of a particular type of drug binding and may prove useful for drug screening.
Materials and methods
Propagation and purification of BMV
Procedures used for BMV propagation and isolation have been described previously (Lane 1986). The concentration of the purified virus was 30 mg/mL in a solution that was 50 mM sodium acetate (pH 5) and 1 mM EDTA. For initial studies, the BMV coat protein was isolated from the virus by reversed phase HPLC (C4 column, 4.6 × 100 mm, 30%–70% in 20 min, 0.1% TFA). Fractions containing the protein were collected and concentrated in a vacuum concentrator.
Peptide mapping using trypsin, Lys C, and pepsin
Trypsin digestion of BMV coat protein was performed at 20°C (0.05 M phosphate buffer at pH 7.5) with an enzyme/substrate (E/S) weight-to-weight ratio of 1/50. Digestion time was only 10 min to limit the digestion in the N terminus, which would otherwise be cut into many tiny pieces because it is rich in arginine and lysine residues. Digestion with Lys C was performed at 37°C and pH 7.5 for 2.5 h (E/S = 1/50). Pepsin digestion was performed for 5 min (E/S = 1/1) using conditions required to minimize hydrogen exchange (0°C, 0.1 M phosphate buffer at pH 2.4). Peptides in the digests were fractionated by HPLC (C4 column, 4.6 × 250 mm, 0%–60% acetonitrile in 60 min, 0.1 % TFA) and collected for analysis by ESIMS (Finnigan LCQ ion trap). Peptides were identified by a combination of molecular mass analysis, collision-induced dissociation (CID) MS/MS, and C-terminal sequencing using carboxypeptidase B and Y. These measurements were made with a Finnigan LCQ ion trap mass spectrometer.
Hydrogen exchange/mass spectrometry
The general procedure used to detect structural changes in the BMV capsid by hydrogen exchange is illustrated in Figure 2 ▶. Intact virus particles (30 mg/mL) were equilibrated overnight in H2O buffer (100 mM phosphate at pH 5.1 or 7.5) prior to 20-fold dilution into D2O buffer (10 mM phosphate at pD 5.43 or 7.30) to initiate HD exchange. Reported pH and pD values were taken directly from the meter without correction. Exchange times used for labeling at pD 7.30 spanned the range from 20 sec to 1.5 h. To cancel the effect of pH on the intrinsic rate of exchange, the exchange time at pD 5.43 was 74.1 times greater than the exchange time at pD 7.30. Exchange times used in this study are given in Table 1. A typical error of measuring pH in this study was 0.02, which would cause deviations in exchange rate constants of less than 5%. Following incubation in D2O, an aliquot of the labeled BMV was mixed 1 : 1 with the D2O quench buffer (0°C, 0.2 M phosphate at pD 2.4, 6 M GdHCl) to quench isotope exchange. The GdHCl accelerated disassembly of the BMV capsid.
Intact labeled BMV coat protein was analyzed with a Finnigan LCQ mass spectrometer coupled to microbore reverse phase HPLC (C4, 1 × 50 mm; 0°C; 30%–70% acetonitrile in 4–5 min; 0.05% TFA; 40 μL/min). Approximately 30 μL (400 pmoles) of sample were injected. Because protic solvents were used for HPLC, labile deuterium in side chains and N- or C-terminal ends was replaced with protium (Englander et al. 1985; Zhang and Smith 1993). Deuterium back exchange from amide linkages was minimized by cooling the chromatographic system to 0°C, maintaining a pH of 2.4, and using a steep elution gradient. Two control samples were analyzed under the same conditions to assess the extent of exchange occurring during analysis, despite maintaining quench conditions. Results for these control samples, undeuterated protein (m0%), and totally exchanged protein (m100%) were used to adjust for the artificial exchange that occurred during analysis (Zhang and Smith 1993). The undeuterated control sample was prepared by diluting a solution of BMV into D2O buffer under quench conditions (0°C at pH 2.4). The completely exchanged control sample was prepared by diluting a solution of BMV into D2O phosphate buffer at pD 2.4 and incubating for 48 h at 37°C.
To investigate hydrogen exchange in different regions of the BMV capsid protein, the labeled virus was diluted twofold (0.1 M phosphate buffer at pH 2.4), digested with pepsin (E/S = 2/1 w/w, 5 min, 0°C), and analyzed by HPLC ESI MS. The peptides were separated by reversed phase chromatography (C18 1 × 50 mm; 0°C; 2%–60% acetonitrile in 6 min; 0.05% TFA; 40 μL/min). Adjustments for a small amount of deuterium loss during digestion and HPLC were made using results obtained for each of the control samples, as described above. This artificial loss of deuterium was 15%–20% in most peptides. The MS analyses were performed with a Micromass Autospec magnetic sector mass spectrometer equipped with a focal plane detector and standard ESI interface. Data were processed by centroiding isotopic distributions corresponding to the +1, +2, or +3 charge states of each deuterated peptide.
Acknowledgments
The authors are pleased to acknowledge assistance from J.A. Speir to generate Figure 6 ▶. The authors also thank Y.M. She and K. Standing for sharing unpublished results on the sequence of BMV. Part of the manuscript was prepared while D.L. Smith was on sabbatical leave at the Institute for Structural Biology in Grenoble, France. This work was supported by the National Institutes of Health (RO1 GM40384) and the Nebraska Center for Mass Spectrometry.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
BMV, brome mosaic virus
CCMV, cowpea chlorotic mottle virus
TBSV, tomato bushy stunt virus
CP, capsid protein
EM, electron microscopy
ESI, electrospray ionization
GdHCl, guanidine hydrochloride
HX, hydrogen exchange
HPLC, high performance liquid chromatography
MS, mass spectrometry
m/z, mass-to-charge ratio
TFA, trifluoroacetic acid
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps. 100101
References
- Adolph, K.W. 1975. Structural transitions of cowpea chlorotic mottle virus. J. Gen. Virol. 28 147–154. [Google Scholar]
- Agrawal, H.O. and Tremaine, J.H. 1972. Proteins of cowpea chlorotic mottle, broad bean mottle, and brome mosaic viruses. Virology 47 8–20. [DOI] [PubMed] [Google Scholar]
- Ahlquist, P., Luckow, V., and Kaesberg, P. 1981. Complete nucleotide sequence of brome mosaic virus RNA3. J. Mol. Biol. 153 23–38. [DOI] [PubMed] [Google Scholar]
- Badger, J., Minor, I., Kremer, M.J., Oliveira, M.A., Smith, T.J., Griffith, J.P., Guerin, D.M., Krishnaswamy, S., Luo, M., Rossmann, M.G., et al. 1988. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA 85 3304–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y., Milne, J.S., Mayne, L., and Englander, S.W. 1993. Primary structure effects on peptide group hydrogen exchange. Proteins: Struct. Funct. Genet. 17 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, J.B. 1970. The self-assembly of spherical plant viruses. Advan. Virus Res. 16 99–134. [DOI] [PubMed] [Google Scholar]
- Bancroft, J.B., Hills, G.J., and Markham, R. 1967. A study of the self-assembly process in a small spherical virus: Formation of organized structures from protein subunits in vitro. Virology 31 354. [DOI] [PubMed] [Google Scholar]
- Chidlow, J. and Tremaine, J.H. 1971. Limited hydrolysis of cowpea chlorotic mottle virus by trypsin and chymotrypsin. Virology 43 267–278. [DOI] [PubMed] [Google Scholar]
- Coyle, J.E., Texter, F.L., Ashcroft, A.E., Masselos, D., Robinson, C.V., and Radford, S.E. 1999. GroEL accelerates the refolding of hen lysozyme without changing its folding mechanism. Nat. Struct. Biol. 6 683–690. [DOI] [PubMed] [Google Scholar]
- Dasgupta, R. and Kaesberg, P. 1982. Complete nucleotide sequences of coat protein messenger RNAs of brome mosaic virus and cowpea chlorotic mottle virus. Nucleic Acids Res. 10 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen, J.R., Smithgall, T.E., Gmeiner, W.H., and Smith, D.L. 1999. Comparison of SH3 and SH2 domain dynamics when expressed alone or in an SH(3 + 2) construct: The role of protein dynamics in functional regulation. J. Mol. Biol. 287 645–656. [DOI] [PubMed] [Google Scholar]
- Englander, S.W. and Kallenbach, N.R. 1984. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16 521–655. [DOI] [PubMed] [Google Scholar]
- Englander, J.J., Rogero, J.R., and Englander, S.W. 1985. Protein hydrogen exchange studied by the fragment separation method. Anal. Biochem. 147 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander, S.W., Mayne, L., Bai, Y., and Sosnick, T.R. 1997. Hydrogen exchange: The modern legacy of Linderstrom–Lang. Protein Sci. 6 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flasinski, S., Dzianott, A., Speir, J.A., Johnson, J.E., and Bujarski, J.J. 1997. Structure-based rationale for the rescue of systemic movement of brome mosaic virus by spontaneous second-site mutations in the coat protein gene. J. Virol. 71 2500–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranda, V.L., Heinz, B.A., Oliveira, M.A., Minor, I., Kim, K.H., Kolatkar, P.R., Rossmann, M.G., and Rueckert, R.R. 1992. Acid-induced structural changes in human rhinovirus 14: Possible role in uncoating. Proc. Natl. Acad. Sci. USA 89 10213–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgand, F., Dumas, R., Biou, V., Andrieu, J.P., Thomazeau, K., Gagnon, J., Douce, R., and Forest, E. 1999. Characterization of the conformational changes of acetohydroxy acid isomeroreductase induced by the binding of Mg2+ ions, NADPH, and a competitive inhibitor. Biochemistry 38 6025–6034. [DOI] [PubMed] [Google Scholar]
- Hsu, C.H., Sehgal, O.P., and Pickett, E.E. 1976. Stabilizing effect of divalent metal ions on virions of southern bean mosaic virus. Virology 69 587–595. [DOI] [PubMed] [Google Scholar]
- Hull, R. 1977. The stabilization of the particles of turnip rosette virus and of other members of the southern bean mosaic virus group. Virology 79 58–66. [DOI] [PubMed] [Google Scholar]
- Incardona, N.L. and Kaesberg, P. 1964. A pH-induced structural change in bromegrass mosaic virus. Biophys. J. 4 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona, N.L., McKee, S., and Flanegan, J.B. 1973. Noncovalent interactions in viruses: Characterization of their role in the pH and thermally induced conformational changes in bromegrass mosaic virus. Virology 53 204–214. [DOI] [PubMed] [Google Scholar]
- Jacrot, B. 1975. Studies on the assembly of a spherical plant virus II. The mechanism of protein aggregation and virus swelling. J. Mol. Biol. 95 433–446. [DOI] [PubMed] [Google Scholar]
- Krol, M.A., Olson, N.H., Tate, J., Johnson, J.E., Baker, T.S., and Ahlquist, P. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96 13650–13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane L.C. 1981. Bromoviruses. In Handbook of plant virus infections and comparative diagnosis (ed. E. Kurstak), pp. 334–376. Elsevier/North-Holland, Amsterdam.
- ———. 1986. Propagation and purification of RNA plant viruses. Methods Enzymology 118 687–696. [Google Scholar]
- Leimkuhler, M., Goldbeck, A., Lechner, M.D., and Witz, J. 2000. Conformational changes preceding decapsidation of bromegrass mosaic virus under hydrostatic pressure: A small-angle neutron scattering study. J. Mol. Biol. 296 1295–1305. [DOI] [PubMed] [Google Scholar]
- Lewis, J.K., Bothner, B., Smith, T.J., and Siuzdak, G. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95 6774–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. and Woodward, C. 1999. The hydrogen exchange core and protein folding. Protein Sci. 8 1571–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini, E.J., Haas, F.D., and Fuller, S.D. 1997. High-resolution icosahedral reconstruction: Fulfilling the promise of cryo-electron microscopy. Structure 5 741–750. [DOI] [PubMed] [Google Scholar]
- Miller, D.W. and Dill, K.A. 1995. A statistical mechanical model for hydrogen exchange in globular proteins. Protein Sci. 4 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranker, A., Robinson, C.V., Radford, S.E., Aplin, R.T., and Dobson, C.M. 1993. Detection of transient protein folding populations by mass spectrometry. Science 262 896–900. [DOI] [PubMed] [Google Scholar]
- Perez, J., Defrenne, S., Witz, J., and Vachette, P. 2000. Detection and characterization of an intermediate conformation during the divalent ion-dependent swelling of tomato bushy stunt virus. Cell Mol. Biol. 46 937–948. [PubMed] [Google Scholar]
- Robinson, I.K. and Harrison, S.C. 1982. Structure of the expanded state of tomato bushy stunt virus. Nature 297 563–568. [Google Scholar]
- Smith, D.L., Deng, Y., and Zhang, Z. 1997. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J. Mass Spectrom. 32 135–146. [DOI] [PubMed] [Google Scholar]
- Speir, J.A., Munshi, S., Wang, G., Baker, T.S., and Johnson, J.E. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma, R. and Thomas, G.J., Jr. 1997. Mechanisms of virus assembly probed by Raman spectroscopy: The icosahedral bacteriophage P22. Biophys. Chem. 68 17–31. [DOI] [PubMed] [Google Scholar]
- Vriend, G., Hemminga, M.A., Verduin, B.J.M., and Schaafsma, T.J. 1982. Swelling of cowpea chlorotic mottle virus studied by proton nuclear magnetic resonance. FEBS Letters 146 319–321. [Google Scholar]
- Zhang, Z. and Smith, D.L. 1993. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 2 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1996. Thermal-induced unfolding domains in aldolase identified by amide hydrogen exchange and mass spectrometry. Protein Sci. 5 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Post, C.B., and Smith, D.L. 1996. Amide hydrogen exchange determined by mass spectrometry: Application to rabbit muscle aldolase. Biochemistry 35 779–791. [DOI] [PubMed] [Google Scholar]