Abstract
IMP-1 β-lactamase is a zinc metallo-enzyme encoded by the transferable blaIMP-1 gene, which confers resistance to virtually all β-lactam antibiotics including carbapenems. To understand how IMP-1 recognizes and hydrolyzes β-lactam antibiotics it is important to determine which amino acid residues are critical for catalysis and which residues control substrate specificity. We randomized 27 individual codons in the blaIMP-1 gene to create libraries that contain all possible amino acid substitutions at residue positions in and near the active site of IMP-1. Mutants from the random libraries were selected for the ability to confer ampicillin resistance to Escherichia coli. Of the positions randomized, >50% do not tolerate amino acid substitutions, suggesting they are essential for IMP-1 function. The remaining positions tolerate amino acid substitutions and may influence the substrate specificity of the enzyme. Interestingly, kinetic studies for one of the functional mutants, Asn233Ala, indicate that an alanine substitution at this position significantly increases catalytic efficiency as compared with the wild-type enzyme.
Keywords: β-Lactam antibiotics; metallo-β-lactamases; structure, function; antibiotic resistance; carbapenems; randomization mutagenesis
The synthesis of β-lactamases is the most common bacterial defense mechanism against β-lactam antibiotics. These enzymes catalyze the irreversible hydrolysis of the amide bond of the β-lactam ring to create ineffective antimicrobial agents (Bush et al. 1995). Genes encoding β-lactamases can be found on the bacterial chromosome or on plasmids (Jacoby 1994). β-Lactamases are divided on the basis of their amino acid sequences and catalytic mechanisms into four classes: A, B, C, and D. Classes A, C, and D contain an active-site serine, whereas class B metallo-enzymes require one or two zinc ions in the active site (Ambler 1980). Metallo-β-lactamases show a broad-spectrum substrate profile and are resistant to the action of conventional β-lactamase inhibitors. Unlike other β-lactamases, most metallo-β-lactamases can efficiently hydrolyze carbapenems such as imipenem and meropenem (Matagne et al. 1999). The carbapenems are broad-spectrum β-lactam antibiotics often used as a last resort to treat serious Gram-negative bacterial infections. In 1991, the blaIMP-1 gene was the first transferable metallo-β-lactamase identified in clinical isolates (Watanabe et al. 1991). It is commonly found in Enterobacteriaceae (Serratia marcescens, Klebsiella pneumoniae, Citrobacter freundii), Pseudomonas aeruginosa, and other Gram-negative bacteria that cause nosocomial outbreaks (Osano et al. 1994; Arakawa et al. 1995; Ito et al. 1995; Marumo et al. 1995; Hirakata et al. 1998). The blaIMP-1 gene was found to reside on a mobile gene cassette within an integron (Arakawa et al. 1995; Laraki et al. 1999a). Integrons are mobile genetic units that use a site-specific recombination system to facilitate the transfer of gene cassettes among plasmids, transposons, and bacterial genomes (Hall and Collis 1995). The selective pressure of antibiotic therapy can result in the rapid dissemination of antibiotic-resistance genes via transfer of these highly mobile gene cassettes. Therefore, the blaIMP-1 gene is poised to spread quickly among clinically relevant bacterial pathogens, which emphasizes the need for novel and effective inhibitors.
Recently, variants of IMP-1 have been found in various geographical settings. For example, the blaIMP-2 gene, which is similar to blaIMP-1, was found on an integronborne gene cassette in a bacterial isolate from Italy (Riccio et al. 2000). Although the cassettes appear to have originated from separate sources, IMP-2 nevertheless shares 85% amino acid identity with IMP-1 and also shows similar kinetic parameters for several β-lactam substrates (Riccio et al. 2000). In Japan, the blaIMP-3 gene was identified on a transferable plasmid from the Gram-negative pathogen Shigella flexneri (O'Hara et al. 1998). The IMP-1 and IMP-3 β-lactamases are 99% identical at the amino acid level; however, the IMP-3 enzyme shows a restricted substrate profile with low catalytic efficiency for penicillins and carbapenems (O'Hara et al. 1998). It has been suggested that IMP-3 has evolved into a more specific, cephalosporinase-type enzyme (Iyobe et al. 2000). Most recently, a new transmissible metallo-β-lactamase, IMP-4, that shares 96% amino acid identity with IMP-1 was found in Enterobacteriaceae and Acinetobacter isolates in China (Chu et al. 2001; Hawkey et al. 2001). The IMP-4 enzyme possesses a broad-spectrum of activity that includes penicillins, cephalosporins, and carbapenems (Chu et al. 2001). Finally, Acinetobacter baumannii clinical isolates producing a metallo-enzyme with characteristics similar to IMP-1 have been reported in Portugal (Da Silva et al. 1999).
Structures have been determined by X-ray crystallography for Bacteroides fragilis (Concha et al. 1996; Carfi et al. 1998a; Fitzgerald et al. 1998), Bacillus cereus (Carfi et al. 1995, 1998b; Fabiane et al. 1998), Stenotrophomonas maltophilia (Ullah et al. 1998), and, most recently, IMP-1 metallo-β-lactamase (Concha et al. 2000). Based on these structures, the location of active-site zinc atoms has been described. One zinc ion (Zn1) is coordinated by His116, His118, and His196, and a second zinc (Zn2) is coordinated by residues Asp120, Cys221, and His263 (standard numbering of class B β-lactamases; Galleni et al. 2001). The cleavage of the amide bond in the β-lactam ring proceeds via nucleophilic attack of a Zn1-activated hydroxide on the carbonyl carbon atom of the β-lactam ring to yield a tetrahedral intermediate. The oxyanion formed at the β-lactam carbonyl oxygen is thought to be stabilized by Asn233 and Zn1. Zn2 potentially interacts with the lone pair electrons of the β-lactam nitrogen atom. Residue Asp120 may accept the proton of the activated water molecule to protonate the nitrogen of the cleaved β-lactam ring to release the hydrolyzed substrate and regenerate the free enzyme (Fabiane et al. 1998). The exact mechanism of this enzyme, however, has yet to be clearly defined.
The class B metallo-β-lactamases can be grouped into three functional subgroups based on metal requirements. In the active site of the B. cereus β-lactamase, Zn1 is tightly coordinated and Zn2 is loosely coordinated; interestingly, this enzyme appears to be an intermediate between a mononuclear and binuclear β-lactamase, because it can function with either one or two zinc ions (Fabiane et al. 1998). The second functional group is defined by the B. fragilis metallo-β-lactamase, which possesses two zinc sites, each with similar binding affinity (Kd < 10 μM) and each required for function (Crowder et al. 1996). Likewise, the L1 β-lactamase from S. maltophilia requires both zinc ions for activity (Ullah et al. 1998). In contrast, for the Aeromonas hydrophila AE036 β-lactamase, zinc binding at the Zn1 site (Kd < 20 nM) is required for activity, but zinc binding at the Zn2 site drastically reduces the activity of the enzyme (Hernandez Valladares et al. 1997). Like the B. fragilis enzyme, IMP-1 is a binuclear enzyme and appears to require both zinc ions (Concha et al. 2000); however, it is not clear if IMP-1 can function catalytically as a monozinc enzyme.
Although X-ray crystallography provides an accurate view of protein structure, it does not supply a complete understanding of structure–function relationships. Site-directed mutagenesis can be used to determine whether a protein will tolerate certain amino acid substitutions and thereby to infer the importance of the position for the structure and function of the enzyme. We have randomized the codons for 27 individual amino acid residues that lie in or near the active site of IMP-1 metallo-β-lactamase. The libraries were sorted based on the ability of mutants to confer ampicillin resistance to Escherichia coli. DNA sequencing of functional random mutants was employed to identify the residues necessary for β-lactam hydrolysis.
Results
Codon randomization and selection
The randomization procedure generates libraries containing all possible amino acid substitutions for the amino acid residue position of interest. Individual codons in the blaIMP-1 gene were randomized using overlap extension polymerase chain reaction (PCR; Ho et al. 1989). Functional random mutants were then selected from the libraries by growth in the presence of antibiotic and sequenced to determine the spectrum of amino acid substitutions at the randomized position that are consistent with β-lactam hydrolysis. The rationale for the approach is that a wide range of different amino acids will be identified at a position among the selected mutants if the position is not important for the structure and function of the enzyme. In contrast, if a position is critical for the structure and function of the enzyme, only one amino acid type will be identified among the selected mutants because all other substitutions will lead to nonfunctional enzymes that do not provide for resistance and therefore do not pass the antibiotic selection.
Previous studies based on a sequence alignment of five metallo-β-lactamases indicate that only nine residues: Leu95, His118, Asp120, Gly123, Leu145, His196, Gly232, Asn233, and His263, are strictly conserved among this family of metallo-β-lactamases (Carfi et al. 1995). In addition, residues His116, Lys224, and Cys221 are predominantly conserved in the alignment. All of these residues were targeted for randomization. Other residues were selected for their potential involvement in substrate binding because of their location near the active-site cleft.
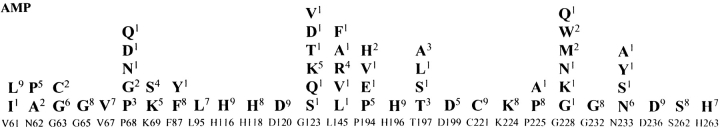
Each of the 27 random libraries was introduced into E. coli XL1-Blue cells (Bullock et al. 1987). Initially, an average of nine naïve mutants from each library were sequenced to verify that the codon of interest had been mutagenized and to ensure the library was not obviously biased toward a certain sequence. The amino acid residues of the naïve mutants from each library are shown in Figure 1 ▶. Functional random mutants were then selected for the ability to hydrolyze ampicillin. These mutants were selected by spreading E. coli containing a random library on Luria-Bertani (LB) agar plates and placing a paper disk saturated with 3 mg/mL of ampicillin in the center of each plate. The antibiotic diffused from the disk, killing susceptible bacteria and creating a zone of clearing. Mutants resistant to ampicillin grew as colonies within the zone of clearing. The susceptibility of each selected mutant was confirmed by determining the ampicillin minimum inhibitory concentration (MIC) using twofold dilutions (Table 1). The MIC values of the selected mutants were comparable to that of E. coli containing the wild-type enzyme. Therefore, the selection method is an effective means of identifying mutants with wild-type levels of function. An average of nine mutants from each library were sequenced to determine the range of amino acid substitutions at each randomized position that are consistent with ampicillin hydrolysis by the enzyme. The amino acid residues of the functional mutants from each library are shown in Figure 2 ▶.
Fig. 1.
Summary of sequencing results for the naïve IMP-1 random libraries. An average of 9 random mutants were sequenced from each naïve library. The amino acids found among the random mutants are shown below the labeled amino acid position. Each labeled amino acid position represents a distinct library. Stop codons and frameshift mutations are represented by an asterisk. The numbers in superscript represent the number of occurrences of the mutant among sequenced clones.
Table 1.
Ampicillin susceptibility of a set of E. coli XL-1 Blue IMP-1 random mutants, including wild-type and the vector control using two-fold LB-Cm broth dilutions
| Mutants | MIC (μg/ml) |
| pGR32 | 1 |
| Wild-type | 16 |
| Leul45Ala | 32 |
| Leu145Phe | 32 |
| Leu145Arg | 16 |
| Asn233Tyr | 16 |
| Asn233Ala | 32 |
| Asn233Ser | 64 |
| Gly232Asn | 4 |
| Val61His | 1 |
| Val61Leu | 8 |
Fig. 2.
Summary of sequencing results of mutants selected from the IMP-1 random libraries based on their ability to confer ampicillin resistance to Escherichia coli. The amino acids found among the functional mutants are shown above the labeled amino acid position. Each labeled amino acid position represents a distinct library. The numbers in superscript represent the number of times the mutant appeared in the selection.
Amino acid residues critical for β-lactamase structure and function
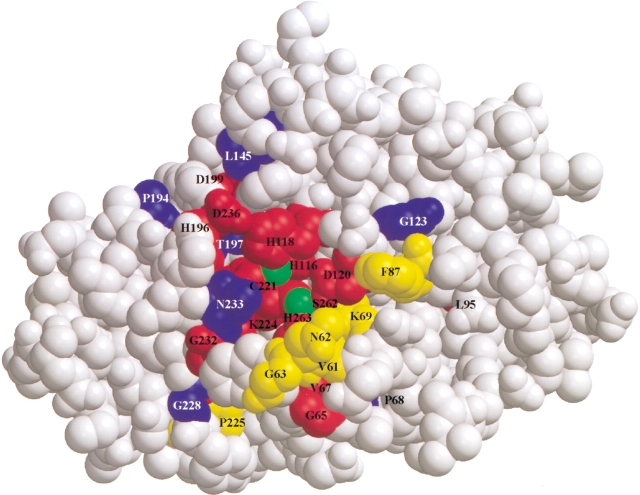
Based on the results of the 27 mutagenesis experiments, the amino acid positions were divided into three groups: critical, that is, only the wild-type amino acid was identified among selected mutants; restrictive, 1–2 substitutions allowed; and tolerant, 3–6 substitutions allowed (Fig. 3 ▶). In total, 14 of the 27 mutated residue positions are believed to be critical for ampicillin hydrolysis by the IMP-1 metallo-β-lactamase because only the wild-type amino acid residue is found at these positions among the sequenced mutants. The set of essential residues includes the putative catalytic residues His116, His118, Asp120, His196, Cys221, Lys224, and His263, which confirms the critical role of these residues for the function of the enzyme. Residues His116, His118, His196, His263, and Cys221 are positioned to coordinate Zn1 and Zn2 within the active site (Concha et al. 2000). The requirement for all of the chelating residues indicates that the coordination of both zincs is required for wild-type levels of ampicillin hydrolysis by IMP-1 enzyme. Residue Asp120 is believed to link Zn1 and Zn2 by hydrogen-bonding to the hydroxide nucleophile (Fabiane et al. 1998), whereas the interaction of the Lys224 side chain with the carboxyl group of the β-lactam is thought to be important for the binding and positioning of substrate (Concha et al. 2000). The fact that these positions do not tolerate substitutions is consistent with their critical role in β-lactam hydrolysis.
Fig. 3.
Spacefill diagram of the IMP-1 metallo-β-lactamase structure showing the mutagenized residues of the 27 random libraries. The residues are divided into three groups: critical, represents positions that do not allow any substitutions (red); restrictive, represents positions that allow 1–2 substitutions (yellow); and tolerant, represents positions that allow 3–6 substitutions (blue). The two green spheres represent zinc molecules. The figure was rendered using the Molscript and Raster3d programs (Kraulis 1991; Merritt and Murphy 1994) using coordinates from the Protein Data Bank accession code 1DD6 (Concha et al. 2000).
Several other residue positions including Gly65, Val67, Leu95, Asp199, Gly232, Asp236, and Ser262 do not tolerate residue substitutions and therefore make critical contributions to enzyme structure and function. The side chains of residues Asp236 and His118 are 2.7 Å apart and most likely form a hydrogen bond. This hydrogen bond may fix His118 in position to ligand Zn1. A substitution at this position would interfere with the orientation of His118 and consequently contribute to a decrease in catalytic efficiency. Residues Gly65, Val67, Leu95, Gly232, and Ser262 are not directly in the active-site pocket but most likely influence substrate binding. MIC values for mutants containing substitutions at these positions support a critical role for the wild-type residue. For example, the MIC value for Gly232Asn is 4 μg/mL compared with 16 μg/mL for E. coli containing the wild-type IMP-1 enzyme (Table 1). The flap, a flexible mobile loop consisting of two antiparallel β-strands connected by a β-turn, is made up of residues 58–67 (Concha et al. 2000). The flap is believed to be involved in substrate binding, and residues Gly65 and Val67 are part of the flap. Hence, substitutions at these positions may affect substrate binding by disrupting the flap structure. Residue Ser262 is located near Cys221 and Asp84, which are known to play an important structural role and may influence zinc binding in the B. cereus, B. fragilis, and S. maltophilia Zn-β-lactamases (Chantalat et al. 2000; Concha et al. 2000). However, it is unclear what role Ser262 plays in this region; it may be structural or involved in enzymatic activity. Nevertheless, it is clear that Ser262 is an essential residue for ampicillin hydrolysis. Leu95 is an essential residue that is not in the active site, but is buried in a hydrophobic region on the opposite end of the active site. Leu95 most likely is not involved in catalysis, but, rather, may be required for structural stability.
In total, more than half of the 27 residues randomized were found to be critical for function. This result indicates that the active site of IMP-1 β-lactamase is highly optimized for β-lactam hydrolysis and, therefore, is generally intolerant of amino acid substitutions.
At several residue positions only a few amino acid types are consistent with wild-type levels of ampicillin hydrolysis. This restrictive substitution group includes positions Val61, Asn62, Gly63, Lys69, Phe87, and Pro225 (Figs. 2, 3 ▶ ▶). Note that Val61, Asn62, and Gly63 reside within the flap region of the enzyme (Concha et al. 2000). The fact that a large percentage of residues in the flap either do not tolerate substitutions or only permit a restricted range of substitutions indicates that the flap plays a key role in the structure and function of the enzyme. It is also of interest that, at residues Val61 and Asn62, a non-wild-type amino acid was found to predominate among the functional mutants (Fig. 2 ▶). At position 61, leucine was found in 90% of the mutants sequenced, whereas the wild-type valine was not observed. Similarly, proline and alanine substitutions occurred multiple times at position 62, but the wild-type asparagine was not observed. The fact that non-wild-type residues predominate among mutants selected for ampicillin hydrolysis indicates that E. coli containing these mutants outcompete E. coli containing wild-type IMP-1. Therefore, substitutions at positions 61 and 62 may result in enzymes with increased catalytic efficiency for ampicillin hydrolysis. Hence, positions 61 and 62 may influence the substrate specificity of the enzyme. Taken together, the results support an important role for the flap region for substrate binding and catalysis.
Finally, at several residue positions a relatively broad range of amino acids was found to be consistent with ampicillin hydrolysis by the enzyme. This substitution-tolerant group includes positions Pro68, Gly123, Leu145, Pro194, Thr197, Gly228, and Asn233 (Figs. 2, 3 ▶ ▶). Residue Pro68 is located between Val67 and Lys69 on a β-strand that forms part of the hydrophobic pocket of the mobile flap. Hence, not all positions in the flap are critical for enzyme function. The most interesting of the positions that tolerate amino acid substitutions is Asn233. This residue is strongly conserved among class B β-lactamases and is believed to be involved in substrate binding (Carfi et al. 1995; Concha et al. 1996, 2000). Surprisingly, substitution of an alanine, tyrosine, or serine was observed at this position. In addition, MIC measurements indicated that E. coli strains containing these mutant enzymes possess ampicillin resistance levels that are equal to or greater that the strain containing wild-type IMP-1 (Table 1). To confirm that substitutions at this position function at a level similar to wild type, the Asn233Ala mutant was purified for further characterization by kinetic analysis.
Overexpression and purification of the IMP-1 metallo-β-lactamase
To facilitate purification of the IMP-1 enzyme, the intact wild-type blaIMP-1 gene was cloned into an isopropyl-1-thio-β-D-galactopyranoside (IPTG)-inducible expression vector (Petrosino et al. 1999), and the enzyme was expressed in E. coli RB791 (Brent and Ptashne 1981; Amann et al. 1983). The metallo-β-lactamases were expressed by inducing mid-log-phase cultures with IPTG and then incubating at 25°C to avoid cell lysis (Wang and Benkovic 1998). Most of the protein remained insoluble in the periplasm and cytoplasm; however, enough soluble protein was detectable in the supernatant to permit efficient purification by ion exchange and gel filtration chromatography. Activity of the protein was qualitatively monitored using nitrocefin, a chromogenic substrate, and enzyme inhibition by EDTA, a zinc chelator, was confirmed.
Kinetic studies for purified Asn233Ala mutant and wild-type IMP-1 metallo-enzymes
The values of the steady-state parameters, kcat, Km, and kcat/Km, for purified wild-type IMP-1 for four β-lactam antibiotics are presented in Table 2. The wild-type IMP-1 enzyme showed kinetic parameters similar to previously published values (Marumo et al. 1995; Laraki et al. 1999b; Haruta et al. 2000). The kinetic parameters for the Asn233Ala enzyme are presented in Table 3. The Asn233Ala mutant was selected for the ability to hydrolyze ampicillin, and therefore, it was expected that the enzyme would hydrolyze this substrate at near wild-type levels. Consistent with this expectation, E. coli containing the Asn233Ala mutant showed an approximately twofold increase in ampicillin resistance compared with wild-type (Table 1). Surprisingly, the catalytic efficiency (kcat/Km) for ampicillin hydrolysis by the Asn233Ala enzyme was found to be sixfold higher than that of the wild-type enzyme. The improved efficiency is owing to both increased turnover and improved binding of substrate. Therefore, rather than being critical for ampicillin hydrolysis, an asparagine at position 233 is, in fact, suboptimal compared with the alanine substitution.
Table 2.
Kinetic parameters of the purified wild-type IMP-1 metallo-β-lactamase at pH 7.5 and 30°C
| Antibiotics | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
| Ampicillin | 66 ± 22 | 231 ± 33 | 0.29 |
| Nitrocefin | 228 ± 11 | 9 ± 3 | 26.3 |
| Cefotaxime | 20 ± 3 | 14 ± 2 | 1.4 |
| Cephaloridine | 71 ± 14 | 39 ± 8 | 1.8 |
Table 3.
Kinetic parameters of the purified mutant Asn233Ala IMP-1 metallo-β-lactamase at pH 7.5 and 30°C
| Antibiotics | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
| Ampicillin | 230 ± 20 | 110 ± 18 | 2.1 |
| Nitrocefin | 1488 ± 488 | 13 ± 5 | 118 |
| Cefotaxime | 495 ± 40 | 30 ± 4 | 16.3 |
| Cephaloridine | 654 ± 141 | 148 ± 32 | 4.4 |
It is also of interest that the alanine substitution resulted in increased catalytic efficiency for all of the substrates tested (Table 3). Therefore, an enzyme that efficiently hydrolyzes a range of substrates was obtained despite the fact that the selection was only for ampicillin hydrolysis. The increased catalytic efficiency is predominately attributable to a large increase in turnover (kcat) for each of the substrates (Table 3). In contrast, the Km values are slightly higher than wild type for all substrates except ampicillin. Taken together, these data clearly indicate that asparagine at position 233 is not critical for the binding and turnover of any of the tested substrates.
Discussion
Based on X-ray crystallographic structures, several models for substrate binding and catalysis for metallo-β-lactamases have been proposed (Carfi et al. 1995; Concha et al. 1996; Fabiane et al. 1998; Ullah et al. 1998). Structural studies indicate that the carbonyl oxygen of the β-lactam ring interacts with the side chain of Asn233 in the IMP-1-mercaptocarboxylate inhibitor crystal complex (Concha et al. 1996). Based on this observation, Asn233 and Zn1 are thought to form an oxyanion hole to polarize the carbonyl group of the β-lactam ring of the substrate, making the carbonyl carbon susceptible to nucleophilic attack (Concha et al. 1996). Mutagenesis studies performed for B. fragilis metallo-β-lactamase CcrA3 indicate that substitution of Asn233 with aspartate increases kcat for penicillin G and cephaloridine but decreases kcat for imipenem, whereas Km is increased for all three substrates (Yang et al. 1999). Yanchak et al. (2000) replaced Asn233 of the CcrA enzyme with alanine, leucine, or aspartate and observed increases in both kcat and Km for penicillin G and cephaloridine. Taken together, these studies indicate that substitutions at Asn233 of CcrA β-lactamase are detrimental for substrate binding but may increase turnover of penicillins and cephalosporins. The Asn233Ala substitution of IMP-1 also resulted in an increase in both kcat and Km values for cephaloridine hydrolysis. However, for ampicillin, cefotaxime, and nitrocefin, the Asn233Ala substitution resulted in large increases in kcat with only minor effects on Km. Hence, the Asn233Ala enzyme showed significant increases in catalytic efficiency for these substrates. These data indicate that, if Asn233 is acting to polarize the carbonyl oxygen of the substrate, other residues can compensate for the loss of this interaction, and, therefore, challenge the proposed essential role of wild-type asparagine at position 233. Perhaps the removal of the Asn233 side chain facilitates another interaction that increases catalytic efficiency.
The ampicillin selection also indicated that tyrosine and serine are functional substitutions for position 233. The enzymatic activity of these mutants has yet to be tested. Based on MIC data, however, the mutants show wild-type or greater levels of activity (Table 1). Serine and tyrosine have a hydroxyl group in common that can potentially hydrogen-bond to the oxygen of the carbonyl bond. The phenol group of tyrosine is bulky, and it is unclear how it contributes to IMP-1 activity. Nevertheless, tyrosine replaces asparagine in the equivalent positions for the BlaB metallo-β-lactamase from Chryseobacterium meningosepticum and the IND-1 metallo-β-lactamase from Chryseobacterium indologenes (Wang et al. 1999).
The majority of the positions that are invariant in an alignment of metallo-β-lactamase amino acid sequences also do not tolerate substitutions in the randomization and selection experiments. These residues include the ligands of Zn1 and Zn2: His116, His118, His196, His263, and Cys221. Although comparative studies of monozinc and dizinc forms of IMP-1 β-lactamase have not been performed, these data indicate that the coordination of Zn1 and Zn2 is essential for function. These results are consistent with experiments showing that IMP-1 mutants with substitutions at His116, His118, and His196 show reduced hydrolysis for cephalothin in the absence of 1 mM zinc sulfate, but show wild-type activity in the presence of 1 mM zinc sulfate (Haruta et al. 2000). The randomization results are also consistent with the finding that substitution of the Zn2 ligands Asp120 and Cys221 generally leads to impaired cephalothin hydrolysis (Haruta et al. 2000). Taken together, the results strongly indicate that both Zn1 and Zn2 are required for maximal levels of β-lactam hydrolysis by IMP-1 β-lactamase.
Another residue that was found to be essential in the randomization experiments is Ser262. This residue is situated near residues Cys221, Asp84, and Lys69, all of which contribute to the structure and function of IMP-1 and may form an important network of hydrogen or electrostatic bonds (Concha et al. 2000). Interestingly, IMP-3 metallo-β-lactamase, an enzyme identical to IMP-1 except for two amino acid substitutions, Gly for Glu at position 126 and Gly for Ser at position 262, shows low hydrolytic activity for penicillins and carbapenems (Iyobe et al. 2000). It has also been shown that reversion of residue 262 to Ser restores catalytic efficiency toward ceftazidime, benzylpenicillin, ampicillin, and imipenem. However, the hydrolysis of cefotaxime and cephalothin is largely unaffected by the substitutions at either position 126 or 262 in IMP-3 (Iyobe et al. 2000). These data indicate that substitutions at Ser262 influence the substrate specificity of the enzyme. Therefore, selection of the Ser262 random library on cefotaxime or cephalothin may reveal functional substitutions that were not detected in the ampicillin selection.
The data obtained for residues in the flap (positions 58–67) of IMP-1 are of particular interest for further substrate-specificity experiments. It is clear that Gly65 and Val67 are essential for ampicillin hydrolysis because they do not tolerate substitutions. Residues Val61, Asn62, and Gly63 only tolerate one or two structurally conservative substitutions. This trend of either no substitutions or restrictive substitutions indicates that the flap is a sensitive structural component of the enzyme. Presently, there is no other collective information available on flap mutants from any metallo-β-lactamases; however, there is published data about the flap structure as a whole. It has been observed for the B. fragilis CcrA enzyme that there is a small but significant increase in the backbone heteronuclear NOE measurements for the flap upon inhibitor binding (Scrofani et al. 1999). This indicates that there is a decrease in flexibility of the flap, and therefore, that the flap likely participates in binding of substrate and in the shielding of the zinc sites from solvent (Scrofani et al. 1999). Scrofani et al. propose that the flap acts as a fishing rod to bind a wide range of different substrate molecules and accommodate them in an active site of variable width and depth.
Randomization of codons followed by a genetic selection for enzyme function is an efficient method to determine the spectrum of amino acids at a position that is consistent with enzyme function, and thereby, infer the importance of the position for the structure and function of the enzyme. Here we have randomized the codons for 27 positions in the IMP-1 enzyme and selected mutants based on ampicillin resistance. Previously, a similar comprehensive codon randomization and selection study was carried out for the class A TEM-1 β-lactamase (Cantu et al. 1996, 1997; Huang et al. 1996). Ampicillin selection data for 30 positions that encompass the active site of TEM-1 indicate that 33% of the positions do not tolerate any substitutions and are therefore critical for function, 30% of the positions tolerate one or two substitutions, and 37% of the positions tolerate 3 or more substitutions (Cantu et al. 1997). In comparison, the IMP-1 ampicillin selection data indicate that 52% of the residues do not tolerate substitutions, 22% of the positions tolerate one or two substitutions, and 26% tolerate more than two substitutions. Based on this comparison, it appears that the active-site region of the IMP-1 metallo-β-lactamase is more sensitive to substitution than the active site of the class A TEM-1 β-lactamase. These differences presumably reflect structural and mechanistic differences between the class A and class B enzymes.
It is interesting to note that 14 of the 27 randomized positions were found to be essential for ampicillin hydrolysis, but only nine positions are strictly conserved in the alignment of metallo-β-lactamases (Carfi et al. 1995). This observation may be owing to different selective pressures on metallo-β-lactamases in nature. The selection in the laboratory was for ampicillin resistance, but metallo-β-lactamases as a group are likely to be under pressure to hydrolyze other β-lactam antibiotics. The nine positions strictly conserved in the alignment may reflect the minimal core of residues necessary for hydrolysis of all β-lactams, whereas the additional positions found to be critical in the laboratory selections may reflect requirements unique to a subset of β-lactams that includes ampicillin. It is also possible that reducing the ampicillin concentration in the laboratory selections would result in a wider range of substitutions at any particular position among the selected mutants and thereby reduce the number of positions that are found to be critical.
The relative intolerance of the IMP-1 β-lactamase for amino acid substitutions may have important implications for the evolution of antibiotic resistance. The TEM-1 β-lactamase is an efficient penicillinase and cephalosporinase, yet it does not possess the broad substrate profile of IMP-1 β-lactamase. However, mutant derivatives of TEM-1 with expanded substrate profiles have evolved in response to antibiotic therapy (Jacoby 1994). TEM-1 has evolved because the active-site region can tolerate amino acid substitutions, which allows alternative amino acids to be sampled by the enzyme (Huang et al. 1996). In contrast, the evolution of the IMP-1 enzyme may be more constrained by the relatively stringent amino acid sequence requirements of the active-site region. Thus, antibiotics and inhibitors designed to combat IMP-1 mediated resistance may enjoy a longer period of efficacy before bacteria containing the IMP-1 enzyme evolve resistance.
Clearly, evolving β-lactamases such as TEM-1, and enzymes with broad substrate profiles such as IMP-1, pose a serious threat to antimicrobial therapy and challenge our current use of antibiotics. Therefore, the detailed characterization of such enzymes is vital for the design of new β-lactam inhibitors.
Materials and methods
Bacterial strains and plasmids
E. coli XL1-Blue [F`:: Tn10 proA+B+ lacIq Δ(lacZ) M15/recA1, endA1, gyrA96 (Nalr), thi-1c, hsdR17, supE44, relA1, lac] (Bullock et al. 1987; Stratagene) was used for transformation of the ligation reaction of the pCM01 plasmid. E. coli RB791 (strain W3110 lacIqL8) was used for expression and purification of IMP-1 β-lactamase (Brent and Ptashne 1981; Amann et al. 1983). The vector used for expression of IMP-1 β-lactamase, pGR32, was previously described (Petrosino et al. 1999).
Construction and cloning of pGR32 and blaIMP-1 gene
Two oligonucleotide primers—Imp-XbaI, 5`-GGCGGGTCTAGAAATTTAGTTGCTTGGTTTTGATGG-3` (containing a XbaI site), and Imp-Sac10, 5`-GCGGGAGCTCGTGGAAACGGATGAAGGCAC-3` (containing a SacI site)—were used to amplify the blaIMP-1 gene, including the native promoter of the gene, from the pBCAM-52E plasmid by PCR (Laraki et al. 1999a). The PCR product was digested with XbaI and SacI and ligated into the cloning site of the pGR32 vector that contains an IPTG-inducible Ptrc promoter for large-scale expression of the enzyme. The Ptrc promoter is tightly regulated by the presence of the lacIq-gene-encoded product present in pGR32. Therefore, in the absence of IPTG, transcription of the gene is mediated by the native promoter from the integron. The ligation product was then transformed into E. coli RB791 cells by electroporation, and plasmid minipreps were performed to isolate plasmid DNA from transformants. The blaIMP-1 gene was PCR-amplified from the plasmid DNA of transformants and sequenced by the Sanger method to confirm the correct sequence.
Random mutagenesis procedures
Codons were randomized by overlap-extension PCR as previously described (Ho et al. 1989) using AdvanTaq DNA polymerase as specified by the manufacturer (CLONTECH Laboratories). Briefly, primers complementary to internal and external sequences in wild-type blaIMP-1 were used to PCR-amplify two DNA fragments with overlapping ends. Specific mutations in the nucleotide sequence of the internal complementary primers incorporated nucleotide changes NNN, where N represented any of the four base pairs at the codon position to be randomized (Huang et al. 2000). The reaction products were analyzed on a 1% agarose gel containing 0.2 μg/mL ethidium bromide run in 1× TAE (40 mM Tris-acetate, 1 mM EDTA). The two PCR reactions containing the overlapping fragments were then combined in a single PCR reaction and amplified using the external primers Imp-XbaI and Imp-Sac10 to create randomized fusion products. The fusion products were then digested with XbaI and SacI and ligated into the multiple cloning site of the pGR32 vector. The ligation products (library DNA) were transformed into E. coli XL1Blue cells by electroporation. The transformants were grown at 37°C in LB-Cm, then plated and pooled for storage at −80°C. Plasmid minipreps were performed to isolate plasmid DNA from the mutant transformants. The blaIMP-1 gene containing the randomized codon was PCR-amplified from the plasmid DNA of transformants and sequenced to determine the sequence of naïve mutants and, after antibiotic selection, functional mutants.
The probability that a given sequence occurs is calculated using the Poisson distribution P = λxe−λ/x!. For these calculations, λ = np, where n is the pool size, p is the probability of a codon occurring, and x is the number of times the codon sequence occurs in pool size n. For these calculations, x = 0. The probability that the given sequence occurs is then P = 1 − e−np, which is the probability that the sequence occurs one or more times in the pool (Palzkill and Botstein 1992). Using these calculations, a library randomized for one codon has a 1/64 probability of occurring; therefore, for one codon sequence to be represented with a 99% probability, 294 transformants are needed. The average size of the random libraries is 104–105 cfu/mL.
Selection of active mutants
Selection for ampicillin-resistant random mutants was carried out by the disk diffusion method. Mutants were grown at 37°C overnight in LB broth containing 12.5 μg/mL chlorampenicol (Cm) from single colonies. A 1:9 dilution in LB-Cm was made from the overnight mutant cultures, and 100 μL of the dilution was spread on LB-Cm agar plates. IPTG was not added to the liquid cultures or the agar plates, and therefore the Ptrc promoter was not induced. The presence of the native, constitutive integron promoter within the construct ensures transcription of the IMP-1 gene. Disks saturated with 100 μL of 3 mg/mL ampicillin were then applied to the center of LB-Cm agar plates, and the plates were incubated at 37°C overnight. The antibiotic in the disk diffused outward forming a concentration gradient on the agar plate that created a zone of clearing in which susceptible bacteria were inhibited from growing. The mutant transformants that grew closest to the disk within the zone of clearing were considered to have wild-type or greater than wild-type IMP-1 hydrolytic activity. MIC measurements confirmed that the selected mutants possessed ampicillin resistance levels similar to wild type (Table 1). The resistant mutants were isolated from the zone of clearing and sequenced to determine the identity of their randomized target region.
Minimum inhibitory concentration determinations
The MIC values, in micrograms per milliliter (μg/mL), for each mutant were determined by using twofold dilutions in LB-Cm for ampicillin concentrations ranging from 0.5 to 256 μg/mL. An inoculum of 104 cfu/mL of E. coli XL1 Blue cells containing plasmid-carried mutant blaIMP-1 genes, the wild-type blaIMP-1 gene, or the pGR32 vector were added to the serial dilutions. The MICs were read after ∼16 h of incubation at 37°C. Data for a set of mutants are presented in Table 1.
Overexpression and purification of soluble wild-type metallo-β -lactamase IMP-1
The recombinant plasmid pCM01 was transformed into E. coli RB791 cells by electroporation. A single colony was then used to inoculate 25 mL of LB broth containing 12.5 μg/mL Cm and allowed to grow at 37°C overnight with shaking. The overnight culture was diluted 1:500 into 1 L of fresh LB broth containing 12.5 μg/mL Cm. The cell culture was grown at 37°C to an OD600 of ∼0.6. The culture was then induced with IPTG for protein expression to a final concentration of 0.5 mM and further incubated at 25°C for 4 h. The culture was centrifuged at 5000g for 20 min. The cultured supernatant was concentrated to ∼100 mL using an Amicon concentrator (Millipore Corp.) with a YM-10 membrane at 55 psi. The resulting solution was dialyzed overnight against 4 L of Buffer H (50 mM HEPES at pH 7.5, 50 μM ZnSO4). The dialyzed supernatant was filtered and then loaded onto a HiTrap SP sepharose cation-exchange column (Amersham Pharmacia Biotech) that had been washed with Buffer H containing 1 M NaCl and equilibrated with Buffer H. The column was washed with Buffer H, and the enzyme was eluted with a linear gradient of 0–0.5 M NaCl. The active fractions were pooled, concentrated to 2 mL using the Amicon concentrator with a YM-10 membrane, and dialyzed against 4 L of Buffer H. The solution was then loaded onto a G-75 Sephadex gel permeation column (Amersham Pharmacia Biotech) that had been equilibrated with Buffer H. After loading, the column was eluted with Buffer H at a flow rate of 0.3 mL/min. The purified active enzyme was pooled, concentrated with a YM-10 Amicon membrane to a final volume of 1 mL, and stored in aliquots at −80°C.
Kinetic parameters
The hydrolysis of ampicillin, cephaloridine, cefotaxime (Sigma), and nitrocefin (Becton Dickinson) antibiotics by wild-type and mutant IMP-1 metallo-β-lactamases was monitored at 235 nm, 260 nm, 260 nm, and 482 nm, respectively, using a DU 640 spectrophotometer (Beckman Coulter). All the reactions were performed in a total volume of 500 μL. The substrate and buffer (50 μM ZnSO4; 50 mM HEPES at pH 7.5; 20 μg/mL final concentration bovine serum albumin, BSA) were preincubated at 30°C for 5 min. All substrate stocks and dilutions were prepared in the assay buffer. Then 4% dimethylsulfoxide was added to the nitrocefin stock to facilitate dissolution. The steady-state kinetic parameters, Km and kcat, were derived from initial velocity measurements using the EnzymeKinetics v. 1.42 program. The absorbance/time data were converted into concentration per centimeter using extinction coefficients Δɛ = −820 M−1 cm−1, −10,000 M−1 cm−1, −7500 M−1 cm−1, and 15,000 M−1 cm−1 for ampicillin, cephaloridine, cefotaxime, and nitrocefin, respectively (Laraki et al. 1999b). The data obtained were from three independent experiments each consisting of duplicate assays.
Acknowledgments
We thank Gian Maria Rossolini, University of Siena, for providing the blaIMP-1 gene in the pBCAM-52E plasmid. We also thank Gary Rudgers and Matthew McKevitt for providing comments on the manuscript. This work was funded by NIH grant 1 F31 AI10535-01 to I.M. and NIH grant AI32956 to T.P.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.40884.
References
- Amann, E., Brosius, J., and Ptashne, M. 1983. Vectors bearing a hybrid trp–lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25 167–178. [DOI] [PubMed] [Google Scholar]
- Ambler, R.P. 1980. The structure of β-lactamases. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 289 321–331. [DOI] [PubMed] [Google Scholar]
- Arakawa, Y., Murakami, M., Suzuki, K., Ito, H., Wacharotayankun, R., Ohsuka, S., Kato, N., and Ohta, M. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMPAntimicrob. Agents Chemother. 39 1612–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent, R. and Ptashne, M. 1981. Mechanism of action of the lexA gene product. Proc. Natl. Acad. Sci. 78 4204–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock, W.O., Fernandez, J.M., and Short, J.M. 1987. XL1-Blue: A high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5 376–379. [Google Scholar]
- Bush, K., Jacoby, G.A., and Medeiros, A.A. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39 1211–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu, C., Huang, W., and Palzkill, T. 1996. Selection and characterization of amino acid substitutions at residues 237–240 of TEM-1 β-lactamase with altered substrate specificity for aztreonam and ceftazidime. J. Biol. Chem. 271 22538–22545. [DOI] [PubMed] [Google Scholar]
- Cantu, C., Huang, W., and Palzkill, T.1997. Cephalosporin substrate specificity determinants of TEM-1 β-lactamase. J. Biol. Chem. 272 29144–29150. [DOI] [PubMed] [Google Scholar]
- Carfi, A., Pares, S., Duee, E., Galleni, M., Duez, C., Frere, J.M., and Dideberg, O. 1995. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14 4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfi, A., Duee, E., Paul-Soto, R., Galleni, M., Frere, J.M., and Dideberg, O. 1998a. X-ray structure of the ZnII β-lactamase from Bacteroides fragilis in an orthorhombic crystal form. Acta Crystallogr. D Biol. Crystallogr. 54 45–57. [DOI] [PubMed] [Google Scholar]
- Carfi, A., Duee, E., Galleni, M., Frere, J.M., and Dideberg, O. 1998b. 1.85 Å resolution structure of the zinc (II) β-lactamase from Bacillus cereus. Acta Crystallogr. D Biol. Crystallogr. 54 313–323. [DOI] [PubMed] [Google Scholar]
- Chantalat, L., Duee, E., Galleni, M., Frere, J.M., and Dideberg, O. 2000. Structural effects of the active site mutation cysteine to serine in Bacillus cereus zinc-β-lactamase. Protein Sci. 9 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y.W., Afzal-Shah, M., Houang, E.T., Palepou, M.I., Lyon, D.J., Woodford, N., and Livermore, D.M. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45 710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha, N.O., Rasmussen, B.A., Bush, K., and Herzberg, O. 1996. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure 4 823–836. [DOI] [PubMed] [Google Scholar]
- Concha, N.O., Janson, C.A., Rowling, P., Pearson, S., Cheever, C.A., Clarke, B.P., Lewis, C., Galleni, M., Frere, J.M., Payne, D.J., Bateson, J.H., and Abdel-Meguid, S.S. 2000. Crystal structure of the IMP-1 metallo β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: Binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39 4288–4298. [DOI] [PubMed] [Google Scholar]
- Crowder, M.W., Wang, Z., Franklin, S.L., Zovinka, E.P., and Benkovic, S.J. 1996. Characterization of the metal-binding sites of the β-lactamase from Bacteroides fragilis. Biochemistry 35 12126–12132. [DOI] [PubMed] [Google Scholar]
- Da Silva, G.J., Leitao, G.J., and Peixe, L. 1999. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 37 2109–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiane, S.M., Sohi, M.K., Wan, T., Payne, D.J., Bateson, J.H., Mitchell, T., and Sutton, B.J. 1998. Crystal structure of the zinc-dependent β-lactamase from Bacillus cereus at 1.9 Å resolution: Binuclear active site with features of a mononuclear enzyme. Biochemistry 37 12404–12411. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, P.M., Wu, J.K., and Toney, J.H. 1998. Unanticipated inhibition of the metallo-β-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): A crystallographic study at 1.85-Å resolution. Biochemistry 37 6791–6800. [DOI] [PubMed] [Google Scholar]
- Galleni, M., Lamotte-Brasseur, J., Rossolini, G.M., Spencer, J., Dideberg, O., and Frere, J. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, R.M. and Collis, C.M. 1995. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 15 593–600. [DOI] [PubMed] [Google Scholar]
- Haruta, S., Yamaguchi, H., Yamamoto, E.T., Eriguchi, Y., Nukaga, M., O'Hara, K., and Sawai, T. 2000. Functional analysis of the active site of a metallo-β-lactamase proliferating in Japan. Antimicrob. Agents Chemother. 44 2304–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey, P.M., Xiong, J., Ye, H., Li, H., and M'Zali, F.H. 2001. Occurrence of a new metallo-β-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China. FEMS Microbiol. Lett. 194 53–57. [DOI] [PubMed] [Google Scholar]
- Hernandez Valladares, M., Felici, A., Weber, G., Adolph, H.W., Zeppezauer, M., Rossolini, G.M., Amicosante, G., Frere, J.M., and Galleni, M. 1997. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β- lactamase activity and stability. Biochemistry 36 11534–11541. [DOI] [PubMed] [Google Scholar]
- Hirakata, Y., Izumikawa, K., Yamaguchi, T., Takemura, H., Tanaka, H., Yoshida, R., Matsuda, J., Nakano, M., Tomono, K., Maesaki, S., Kaku, M., Yamada, Y., Kamihira, S., and Kohno, S. 1998. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant Gram-negative rods carrying the metallo-β- lactamase gene blaIMP. Antimicrob. Agents Chemother. 42 2006–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 51–59. [DOI] [PubMed] [Google Scholar]
- Huang, W., Petrosino, J., Hirsch, M., Shenkin, P.S., and Palzkill, T. 1996. Amino acid sequence determinants of β-lactamase structure and activity. J. Mol. Biol. 258 688–703. [DOI] [PubMed] [Google Scholar]
- Huang, W., Zhang, Z., and Palzkill, T. 2000. Design of potent β-lactamase inhibitors by phage display of β-lactamase inhibitory protein. J. Biol. Chem. 275 14964–14968. [DOI] [PubMed] [Google Scholar]
- Ito, H., Arakawa, Y., Ohsuka, S., Wacharotayankun, R., Kato, N., and Ohta, M. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39 824–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyobe, S., Kusadokoro, H., Ozaki, J., Matsumura, N., Minami, S., Haruta, S., Sawai, T., and O'Hara, K. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44 2023–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, G.A. 1994. Extrachromosomal resistance in Gram-negative organisms: The evolution of β-lactamase. Trends Microbiol. 2 357–360. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallog. 24 946–950. [Google Scholar]
- Laraki, N., Galleni, M., Thamm, I., Riccio, M.L., Amicosante, G., Frere, J.M., and Rossolini, G.M. 1999a. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laraki, N., Franceschini, N., Rossolini, G.M., Santucci, P., Meunier, C., de Pauw, E., Amicosante, G., Frere, J.M., and Galleni, M. 1999b. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43 902–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumo, K., Takeda, A., Nakamura, Y., and Nakaya, K. 1995. Purification and characterization of metallo-β-lactamase from Serratia marcescens. Microbiol. Immunol. 39 27–33. [DOI] [PubMed] [Google Scholar]
- Matagne, A., Dubus, A., Galleni, M., and Frere, J. M. 1999. The β-lactamase cycle: A tale of selective pressure and bacterial ingenuity. Nat. Prod. Rep. 16 1–19. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster 3D Version 2.0: A program for photorealistic molecular graphics. Acta Crystall. D50 869–873. [DOI] [PubMed] [Google Scholar]
- O'Hara, K., Haruta, S., Sawai, T., Tsunoda, M., and Iyobe, S. 1998. Novel metallo β-lactamase mediated by a Shigella flexneri plasmid. FEMS Microbiol. Lett. 162 201–206. [DOI] [PubMed] [Google Scholar]
- Osano, E., Arakawa, Y., Wacharotayankun, R., Ohta, M., Horii, T., Ito, H., Yoshimura, F., and Kato, N. 1994. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill, T. and Botstein, D. 1992. Probing β-lactamase structure and function using random replacement mutagenesis. Proteins 14 29–44. [DOI] [PubMed] [Google Scholar]
- Petrosino, J., Rudgers, G., Gilbert, H., and Palzkill, T. 1999. Contributions of aspartate 49 and phenylalanine 142 residues of a tight binding inhibitory protein of β-lactamases. J. Biol. Chem. 274 2394–2400. [DOI] [PubMed] [Google Scholar]
- Riccio, M.L., Franceschini, N., Boschi, L., Caravelli, B., Cornaglia, G., Fontana, R., Amicosante, G., and Rossolini, G.M. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrofani, S.D., Chung, J., Huntley, J.J., Benkovic, S.J., Wright, P.E., and Dyson, H.J. 1999. NMR characterization of the metallo-β-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: Role of an active-site loop. Biochemistry 38 14507–14514. [DOI] [PubMed] [Google Scholar]
- Ullah, J.H., Walsh, T.R., Taylor, I.A., Emery, D.C., Verma, C.S., Gamblin, S.J., and Spencer, J. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J. Mol. Biol. 284 125–136. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Benkovic, S.J. 1998. Purification, characterization, and kinetic studies of a soluble Bacteroides fragilis metallo-β-lactamase that provides multiple antibiotic resistance. J. Biol. Chem. 273 22402–22408. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Fast, W., Valentine, A.M., and Benkovic, S.J. 1999. Metallo-β-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3 614–622. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., Iyobe, S., Inoue, M., and Mitsuhashi, S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanchak, M.P., Taylor, R.A., and Crowder, M.W. 2000. Mutational analysis of metallo-β-lactamase CcrA from Bacteroides fragilis. Biochemistry 39 11330–11339. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Keeney, D., Tang, X., Canfield, N., and Rasmussen, B.A. 1999. Kinetic properties and metal content of the metallo-β-lactamase CcrA harboring selective amino acid substitutions. J. Biol. Chem. 274 15706–15711. [DOI] [PubMed] [Google Scholar]