Abstract
To elucidate the role of amino acid residues adjacent to the catalytic site of pepsin-like enzymes, we analyzed and compared the crystal structures of these enzymes, their complexes with inhibitors, and zymogens in the active site area (a total of 82 structures). In addition to the water molecule (W1) located between the active carboxyls and playing a role of the nucleophile during catalytic reaction, another water molecule (W2) at the vicinity of the active groups was found to be completely conserved. This water molecule plays an essential role in formation of a chain of hydrogen-bonded residues between the active site flap and the active carboxyls on ligand binding. These data suggest a new approach to understanding the role of residues around the catalytic site, which can assist the development of the catalytic reaction. The influence of groups adjacent to the active carboxyls is manifested by pepsin activity at pH 1.0. Some features of pepsin-like enzymes and their mutants are discussed in the framework of the approach.
Keywords: Aspartic proteases, pepsin-like enzymes, protein three-dimensional structures, comparison of protein structures, active site region of pepsin-like enzymes
Pepsin-like enzymes are aspartic proteases, which belong to the A1 family of peptidases (Rawlings and Barrett 1998). This family comprises proteins with a three-dimensional structure close to that of pepsin. The single molecular chain of these enzymes forms two domains with different amino acid sequences, but basically similar folds. In contrast, molecules of retroviral aspartic proteases, making up the A2 family, consist of two identical monomers resembling pepsin domains. The catalytic site of pepsin-like enzymes is formed at the junction of the domains and contains two aspartic acid residues, Asp 32 and Asp 215 (in pepsin numbering), one in each domain. An essential member of the active site is the water molecule bound between the active carboxyls, which becomes deprotonated on substrate binding to initiate the general base catalysis (Antonov et al. 1978, 1981). In accordance with the accepted mechanism of pepsin-like enzyme function (Suguna et al. 1987b; Davies 1990), Asp 215 has to be charged, whereas Asp 32 has to be protonated. A remarkable property of this catalytic center is adaptation for the action in a wide range of pH from pH 1.0 up to pH 7.0.
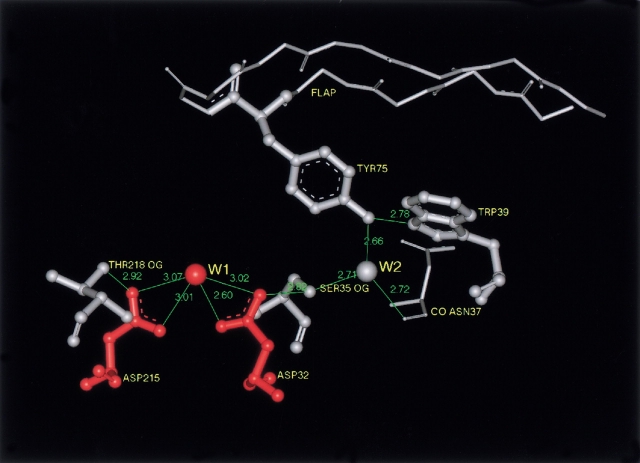
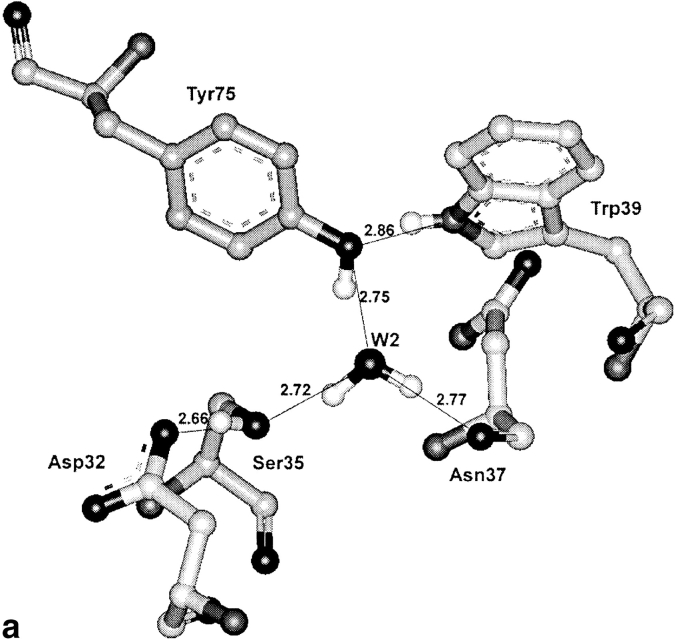
The three-dimensional structure of the porcine pepsin active site area is presented in Figure 1 ▶ (pdb code: 4pep; Sielecki et al. 1990). The active water molecule is labeled W1. As can be seen, the hydroxyl groups of Ser 35 and Thr 218 are near the active carboxyls of Asp 32 and Asp 215, respectively. Unlike Thr 218, whose hydroxyl can make only one hydrogen bond with the outer oxygen (Oδ2) of the Asp 215 carboxyl, the hydroxyl of Ser 35 is at a hydrogen-bonding distance from the outer oxygen of the Asp 32 carboxyl (Oδ1) and another water molecule, W2. The W2 is hydrogen-bonded itself with the carbonyl oxygen of Asn 37 and the hydroxyl of the active site flap residue Tyr 75, while the hydroxyl of Tyr 75 is fixed by a hydrogen bond with the Trp 39 Ne1 atom. The active carboxyls have additional hydrogen bonds (not shown in Fig. 1 ▶), which connect their inner oxygens with the NH groups of glycine residues located in the Asp-Thr-Gly segments of both active loops. The conserved nature of these bonds and their role were discussed previously (Davies 1990).
Fig. 1.
The catalytic site and the surrounding region of unbound porcine pepsin in monoclinic crystals (pdb code: 4pep; resolution 1.8 Å, Sielecki et al. 1990). In these crystals, the flap is involved in intermolecular contacts, and it is close to the molecular surface. The hydroxyl of Tyr 75 forms hydrogen bonds with the interior of the molecule, resulting in formation of a continuous chain of hydrogen-bonded residues Trp 39–Tyr 75–W2–Ser 35–Asp 32 extending from the flap to the active site. The same chain of hydrogen bonds is a characteristic property of all complexes of pepsin-like enzymes with inhibitors. The Thr 218 hydroxyl group, locating at a hydrogen-bond distance from the Asp 215 carboxyl, protects this carboxyl from protonation in acid media.
The residues outlined in Figure 1 ▶ are conserved in pepsin-like enzymes, with a few exceptions. Protein engineering experiments revealed that all of them have a marked influence on enzymatic activity. A prominent drop in kcat and small changes in KM were detected for two mutants, Ser 35 Ala and Thr 218 Ala of porcine pepsin and rhizopuspepsin (pepsin numbering of residues is used for all enzymes discussed in this article), whereas the pH optimum and pKa values of the active carboxyls changed only slightly (Lin et al. 1992). The authors of this work hypothesized that the hydrogen bonds formed by the active carboxyls with Ser 35 and Thr 218 did not control their ionization properties but provided the structural rigidity in the catalytic apparatus. However, the presence of Ala 218 instead of Thr 218 in some renins suggests that this explanation cannot be applied to all pepsin-like enzymes. The opposite point of view was used to explain the decrease in activity of the chymosin mutant Thr 218 Ala in acidic media (Mantafounis and Pitts 1990). The decrease in activity was interpreted as a consequence of a partial sacrifice of the negative charge of Asp 215 at low pH, which supposedly was stabilized by a hydrogen bond with the Thr 218 hydroxyl. This conclusion was discussed also in studies on mechanism of endothiapepsin, and the same role to stabilize the negative charge of Asp 32 was prescribed to the Ser 35 hydroxyl after formation of the tetrahedral intermediate (Veerapandian et al. 1992). The role of Ser 35 in the unbound enzyme was not considered. Attempts to change the pH optimum of the pepsin-like enzymes by point mutation experiments resulted in a small shift of this parameter (Mantafounis and Pitts 1990; Lin et al. 1992). Perutz (1992) proposed a convincing explanation of this result, suggesting that the pH optimum of pepsin-like enzymes is determined by a combined contribution of many polar residues in molecules. Special calculations in more recent studies (Yang et al. 1997) support this point of view.
Several Tyr 75 mutants were studied during protein engineering experiments with chymosin (Suzuki et al. 1989) and Rhizomucor pusillus protease (Park et al. 1996). The replacement of Tyr 75 with various amino acid residues (Thr, Ile, Val) in chymosin resulted in a complete loss of activity, whereas the mutant Tyr 75 Phe caused marked changes in the kinetic parameters, depending on the substrates used. The replacements of Tyr 75 by 17 residues in R. pusillus protease resulted in negligible activity for 15 mutants, weakened activity for the Tyr 75 Phe mutant, and increased catalytic efficiency for the Tyr 75 Asn mutant. A marked drop in kcat was observed for rat renin mutants Tyr 75 His, Tyr 75 Phe, and especially for the mutant Tyr 75 Ala (Suzuki et al. 1996). The results of all these experiments were explained by the special role of Tyr 75 in stabilizing the transition state of the substrate during the catalytic reaction, as proposed in studies of endothiapepsin (Blundell et al. 1987). However, the exceptional case of the increased catalytic efficiency of the Tyr 75 Asn mutant of R. pusillus protease was not interpreted.
The activity decreased also after the replacement of Trp 39 with a set of residues in R. pusillus protease (Park et al. 1997). Trp 39 was suggested to stabilize the position of the Tyr 75 phenolic ring by the hydrogen bond between the Trp 39 Ne1atom and the Tyr 75 hydroxyl. However, the natural replacement of Trp 39 by alanine in β-secretases (Rawlings and Barrett 1998) does not prevent these enzymes from being active.
The controversial interpretation of the role of residues surrounding the catalytic site of pepsin-like enzymes prompted us to perform a detailed structural analysis of groups in the active site area. The initial purpose of the present work was to understand the combined functional property of pepsin residues, which form a continuous chain of hydrogen bonds, with the active carboxyls being members of this chain (Fig. 1 ▶). However, the problem was more complicated than we originally had thought, and data known only for pepsin were not enough to solve it. The experimental approach involving a protein engineering introduction of the chain of hydrogen-bonded residues observed in pepsin into HIV-1 protease molecules (Dergousova et al. 1997) and a subsequent structural analysis of the mutant met many difficulties. Therefore, at this stage, all known three-dimensional structures of pepsin-like proteases, their complexes with inhibitors, and zymogens have been inspected and compared in the active site area (a total of 82 structures; Bernstein et al. 1977; Berman et al. 2000). Some observations and hypotheses ensuing from this study are described.
Results
First, we determined the degree of porcine pepsin conservation for the interactions shown in Figure 1 ▶. The threonine residue at the position 218 in pepsin numbering is present in all analyzed structures except renins, where it is replaced by alanine (human) or serine (mouse). With few exceptions, the location of the Thr 218 hydroxyl at a hydrogen bond distance from the Asp 215 carboxyl is a common feature of all active enzymes and their complexes with inhibitors. The exceptions among active enzymes include Atlantic cod pepsin (pdb code: 1am5; Karlsen et al. 1998) and cathepsin D crystallized at alkaline pH (pdb code: 1lyw; Lee et al. 1998), where this distance is larger than that of a hydrogen bond. In Rhizomucor miehei unbound protease (pdb code: 2asi; Yang et al. 1997), Thr 218 is present as an unusual rotamer, and its hydroxyl group does not form any hydrogen bond. The exceptions among complexes with inhibitors comprise Saccharomyces cerevisiae protease with the helical IA3 inhibitor (pdb codes: 1dpj and 1dp5; Li et al. 2000) and mouse submaxillary renin complexed with CH-66 inhibitor (pdb code: 1smr; Dealwis et al. 1994), where Thr 218 is replaced by Ser 218.
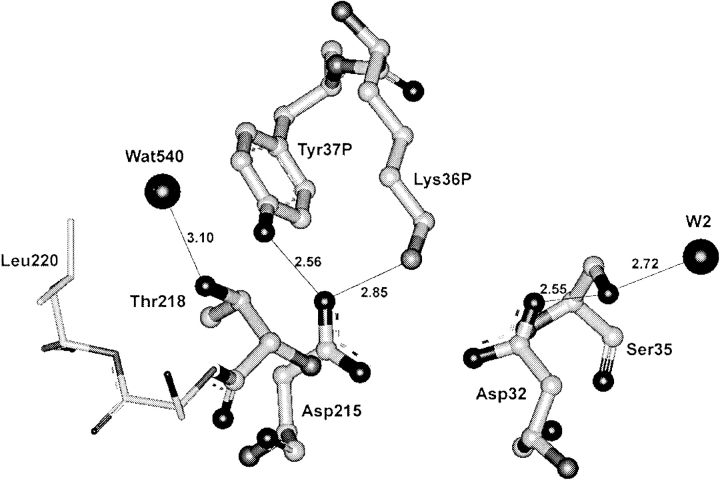
This last exception deserves special attention. The replacement of threonine by serine at position 218 in mouse submaxillary renin cannot affect the ability of the hydroxyl to form a hydrogen bond with the Asp 215 carboxyl. However, our analysis has shown the Ser 218 hydroxyl to turn around the dihedral angle χ1 to form a hydrogen bond with the main chain carbonyl oxygen of Phe 220 at the opposite side of Asp 215 in the complex of this renin with the inhibitor (pdb code: 1smr; Dealwis et al. 1994). The distance between the inhibitor and Ser 218 is too large to induce such reorientation. The nonstandard for these enzymes' rotamer of Thr 218 is present also in the complexes of S. cerevisiae protease with helical inhibitor IA3 crystallized at pH 6.6 (pdb codes: 1dpj and 1dp5; Li et al. 2000). The presence of the same nonstandard rotamer of Thr 218 is the property of zymogens (the exception is proplasmepsin that has an arrangement of residues in the region of plasmepsin active site unusual for other zymogens (pdb code 1pfz; Bernstein et al. 1999)), as shown in Figure 2 ▶ (pdb code: 2psg, Sielecki et al. 1991; pdb code: 3psg, Hartsuck et al. 1992; pdb code, 1htr; Moore et al. 1995; pdb code: 1qdm, Kervinen et al. 1999), and the intermediate of progastricsin activation (pdb code: 1avf, Khan et al. 1997).
Fig. 2.
The conformation of Thr 218 residue in porcine pepsinogen (pdb code: 3psg; resolution 1.65 Å, Hartsuck et al. 1992). In pepsinogen, a hydrogen bond between Thr 218 and Asp 215 is not possible because Asp 215 is involved in hydrogen bonds with Lys 36 and Tyr 37 of the propart. The hydroxyl group of Thr 218 can form only a weak hydrogen bond with water molecule W540, although it is close to Ser 219 NH group. In this structure, Thr 218 residue is present as a rotamer unusual for active enzymes.
Ser 35 is completely conserved in all pepsin-like enzymes. The position of this residue in the N-terminal domain is symmetrical to that of Thr 218 in the C-terminal domain; however, the interaction patterns formed by Ser 35 and Thr 218 are not quite similar. The proximity of the Ser 35 hydroxyl to the Asp 32 carboxyl found in porcine pepsin was observed in all complexes with inhibitors, but not in all active enzyme structures. The most conserved interaction of the Ser 35 hydroxyl is a hydrogen bond with the water molecule W2 (Fig. 1 ▶) found in all analyzed structures. Here, we show that this internal water molecule is a characteristic feature of all pepsin-like enzymes, their complexes with inhibitors, and zymogens. This characteristic feature first was revealed in our previous studies based on limited data (Andreeva et al. 1995; Kashparov et al. 1997); however, it has not been discussed in any other publication to our knowledge. In some structures, Ser 35 is present as a nonstandard rotamer, but in all of them the Ser 35 hydroxyl is located within a hydrogen-bond distance from W2.
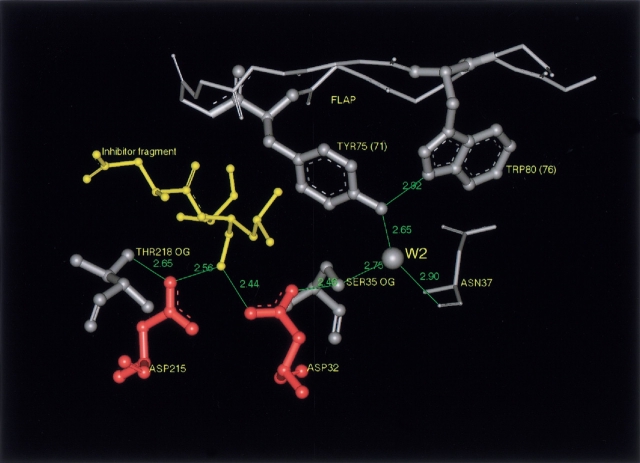
Besides Ser 35, water molecule W2 interacts with the carbonyl oxygen of a residue at the 37th position in pepsin numbering, and this bond is conserved. In complexes of enzymes with inhibitors, W2 also forms the third hydrogen bond with the hydroxyl of the conserved residue Tyr 75, as in porcine pepsin (Fig. 1 ▶), and the Tyr 75 hydroxyl itself accepts a hydrogen bond from the Trp 39 Nɛ1atom. These bonds provide the formation of a continuous chain of hydrogen-bonded residues, Trp 39–Tyr 75–W2–Ser 35–Asp 32, connecting the flap with the catalytic site. Summarizing our observations on the structure of the active site area in complexes of pepsin-like enzymes with inhibitors, we can advocate that the formation of this chain is their essential feature (an unique exception is the enlarged distance between the Ser 35 hydroxyl and Asp 32 carboxyl in the complex of saccharopepsin with 081282 inhibitor [pdb code: 2jxr, Aguilar et al. 1997]). We have revealed that in memapsin 2 (β-secretase), where tryptophane 39 is replaced by alanine, the Trp residue at the 80th position in pepsin numbering (Trp 76 in memapsin numbering) forms a hydrogen bond with the Tyr 75 hydroxyl as is shown in Figure 3 ▶ for the complex of this enzyme with the inhibitor OM99-2 (pdb code: 1fkn, Hong et al. 2000). The temperature factors for all members of this chain, including water molecule W2, decrease markedly on ligand binding in all enzymes. The greatest changes are observed for Ser 35, W2, and the Tyr 75 hydroxyl (Table 1).
Fig. 3.
The catalytic site and the surrounding region of inhibited memapsin 2 by OM 99-2 inhibitor (pdb code: 1fkn; resolution 1.9 Å, Hong et al. 2000). The figure shows the conserved nature of the chain of hydrogen bonds connecting the flap with the active site, which forms on inhibitor binding. Trp 80(76) plays the role of Trp 39, replaced in memapsin by Ala.
Table 1.
Temperature B factors of atoms in Å2 involved in continuous chain of hydrogen-bonded residues
| Enzyme | PDB code | Ser35 0γ | W2 | Tyr75 OH | Trp35 Nɛl |
| PP | 3app | 21.25 | 31.68 | 30.56 | 10.46 |
| PP-complex | 1apt | 6.76 | 5.86 | 12.28 | 6.60 |
| PP-complex | 1apu | 9.56 | 10.12 | 19.40 | 6.89 |
| PP-complex | 1apv | 3.86 | 8.40 | 9.67 | 7.10 |
| PP-complex | 1apw | 5.29 | 10.24 | 9.70 | 9.27 |
| PP-complex | 1ppk | 7.08 | 10.71 | 10.53 | 8.35 |
| PP-complex | 1ppl | 5.03 | 10.37 | 7.22 | 4.87 |
| PP-complex | 1ppm | 9.76 | 14.39 | 11.00 | 9.54 |
| PR | 2apr | 11.32 | 15.38 | 33.16 | 13.71 |
| PR-complex | 3apr | 9.20 | 9.68 | 14.11 | 10.19 |
| PR-complex | 4apr | 6.67 | 2.00 | 19.00 | 5.84 |
| PR-complex | 5apr | 9.52 | 8.68 | 22.07 | 9.66 |
| PR-complex | 6apr | 6.24 | 6.46 | 13.68 | 7.29 |
PP, penicillopepsin; PR, rhizopuspepsin.
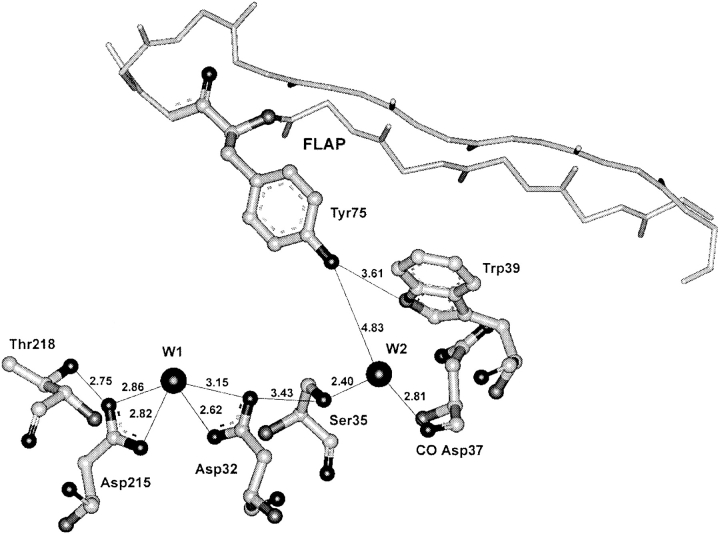
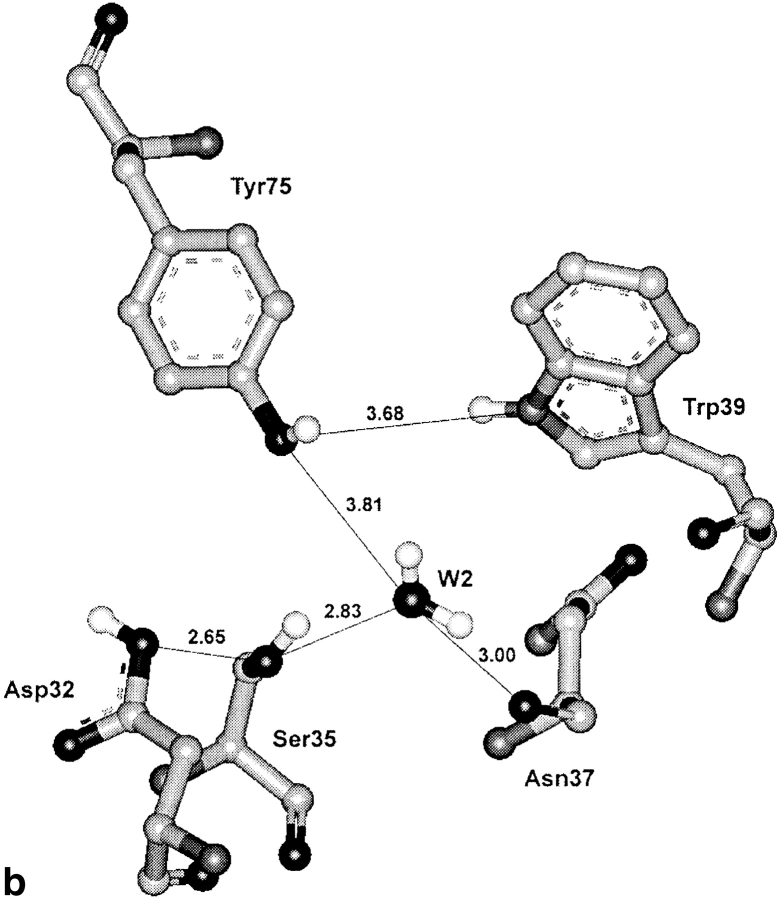
The continuous chain of hydrogen-bonded residues also was observed in several crystal structures of active enzymes, and porcine pepsin is the example. At the same time, in penicillopepsin (pdb code: 3app, James and Sielecki 1983) and in some other active enzyme structures, the flap is not fixed to the interior of the molecule by the intramolecular interactions, and the continuous chain of hydrogen-bonded residues is not formed (Fig. 4 ▶). In penicillopepsin, the distances from the Tyr 75 hydroxyl to W2 and Trp 39 are 4.83 and 3.61 Å, respectively. The position of the flexible flap in crystals of active enzymes is often dependent on molecular packing. However, in any case, the flap in free enzymes has at times to adopt in solution a conformation allowing a polypeptide substrate to occupy the cleft. At these moments, it cannot be fixed to the interior of molecules by the chain of intramolecular hydrogen bonds.
Fig. 4.
The catalytic site and the surrounding region of active penicillopepsin (pdb code: 3app; resolution 1.8 Å, James and Sielecki 1983) representing the arrangement of residues in the active site area, when the flap is close to the open position. The flap does not form contacts with the Trp 39 Oɛ1 atom or water molecule W2. At the same time, the distance between the Ser 35 Oγ atom and Asp 32 carboxyl is too large for the formation of a hydrogen bond.
One more structural property of active enzymes should be mentioned. In these structures, the absence of the hydrogen bond of Tyr 75 with W2 and Trp 39 correlates every time with the enlarged distance between the Ser 35 hydroxyl and the Asp 32 carboxyl, preventing the formation of a hydrogen bond.
Discussion
The accepted mechanism of pepsin-like enzyme function assumes that one of the active aspartic acid residues, acting as a general base (Asp 215), has to be charged, whereas the other one, acting as a general acid (Asp 32), has to be protonated (Suguna et al. 1987b; Davies 1990; James et al. 1992; Parris et al. 1992; Veerapandian et al. 1992). The same structure of the catalytic site and the surrounding region in all pepsin-like enzymes implies a universal mechanism of their action. It means that the situation when the Asp 215 carboxyl is charged and the Asp 32 carboxyl is protonated should hold in any conditions. Therefore, the first important role of residues adjacent to the catalytic centre of pepsin-like enzymes is to preserve the charged state of Asp 215 and the protonated state of Asp 32.
Are structural data consistent with this role? In all known structures of enzymes functioning at acidic pH, the Thr 218 hydroxyl takes up a guard position near the Asp 215 carboxyl at a distance of a hydrogen bond, as in porcine pepsin (Fig. 1 ▶). This hydrogen bond utilizes the anti lone pair electrons of the outer Oδ2 oxygen, while the syn lone pair of this oxygen is engaged in the hydrogen bond with the water molecule W1. As a result, the ability of the outer oxygen of Asp 215 to bind protons from bulky solvent becomes rather low. The analogous role of Thr 218 was proposed first in studies of chymosin mutant (Mantafounis and Pitts 1990) and discussed in the work on endothiapepsin mechanism (Veerapandian et al. 1992). The efficiency of such a mechanism of charge protection is shown by the activity of pepsin at pH 1.0. The preservation of the charged state of several carboxyl groups at low pH is the feature of pepsin (Sielecki et al. 1990; Andreeva and James 1991), but in the case of the active Asp 215 carboxyl it should be the property of all pepsin-like enzymes acting in acidic media. Therefore, the rearrangement of the Thr 218 hydroxyl group by changing the χ1 torsion angle and the formation of the hydrogen bond with the Asp 215 carboxyl is an important step in the activation of pepsinogen and other zymogens (cf. Figs. 1 and 2 ▶ ▶).
The absence of a hydrogen bond between the residue at the 218 position and the Asp 215 carboxyl in all renins because of the change of Thr 218 to Ala or to a nonstandard rotameric form of Ser 218 excludes the proposed mechanism of the charge protection. However, these enzymes act at neutral pH, and additional stabilization of the charged state of the Asp 215 carboxyl is not significant for them. In contrast, for pepsin, chymosin, and rhizopuspepsin, the ability of Thr 218 Ala mutants to keep the charged state of the Asp 215 carboxyl decreases substantially on lowering the pH, which results in a decrease of activity, shown in a decrease of kcat. Some residual activity of the mutants can be explained by the occasional appearance of a charge at the Asp 215 carboxyl owing to fluctuation processes.
In previous studies, the role of Thr 218 and Ser 35 was considered the same, because of the symmetrical arrangement of these residues in relation to the active carboxyls. However, the absence of a hydrogen bond between the Ser 35 hydroxyl and the Asp 32 hydroxyl in free enzymes (Fig. 4 ▶) can make the role of Ser 35 opposite that of Thr 218, as it does not impede the protonation of Asp 32 in acidic media. At the same time, the functional properties of the Ser 35 residue, which, unlike Thr 218, is absolutely conserved in all pepsin-like enzymes regardless of their acting media, seem to be more complex than that of Thr 218.
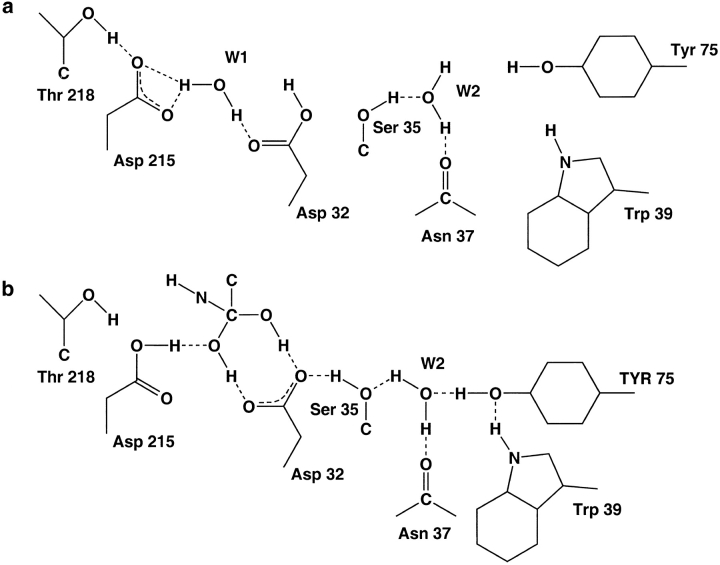
Structural data suggest the existence of a mechanism assisting the proton leaving from the Asp 32 carboxyl at the initial stage of the catalysis and proton acceptance after substrate cleavage. It follows from the ability of the Ser 35 hydroxyl and the water molecule W2 to exchange their donor and acceptor roles while being hydrogen-bonded (Fig. 5a,b ▶). If the relative orientation of Ser 35 and W2 is such as shown in Figure 5a ▶, where W2 donates a proton to the Ser 35 hydroxyl, then this hydroxyl donates a proton to the anti lone pair electrons of the Asp 32 outer Oδ1 carboxyl oxygen, thus enhancing its acidic properties. However, in the opposite situation (Fig. 5b ▶), W2 accepts a proton from the Ser 35 hydroxyl. As a result, Ser 35 does not prevent the protonation of Oδ1 atom. In the case of unbound enzymes acting at neutral pH, the captured proton can be trapped between Asp 32 and Ser 35, which stabilizes the protonated state of the active Asp 32 carboxyl. Although this property is not yet experimentally proven for unbound pepsin-like enzymes acting at elevated pH, we emphasize that such a possibility follows from the structure.
Fig. 5.
(a) Scheme showing the arrangement of hydrogen bonds in the active site area of pepsin-like enzymes during formation of enzyme–substrate complex when Tyr 75 approaches Trp 39 and water molecule W2. At the same time, Ser 35 becomes close to the Asp 32 carboxyl. Trp 39 donates proton to the Tyr 75 hydroxyl and forces it to donate proton to water molecule W2. As a result, Ser 35 becomes a proton donor to the Asp 32 carboxyl, enhancing its acidic properties (Asp 32 presumes to leave its proton participating in formation of the gem-diol structure of the tetrahedral intermediate). Hydrogen-bond distances shown in both a and b are not relevant to any structure. In the scheme, they show only the presence of hydrogen bonds. (b) Scheme showing a possible reorientation of water molecule W2 after moving of Tyr 75 out of the interior of a molecule. Reorientation of the proton of the Ser 35 hydroxyl results in some increase of basic properties of the Asp 32 carboxyl, which helps to return its initial protonated state after the catalytic reaction.
The high temperature factor of W2 in free enzymes suggests that this water molecule can have rotational freedom (Table 1). What happens when the substrate binds in the active site cleft? When the flap takes up the closed conformation, the hydroxyl of Tyr 75 approaches the Trp 39 Nɛ1 atom and W2. The hydrogen bond formed between the Tyr 75 hydroxyl and W2 fixes the orientation of this water molecule. As Trp 39 donates its proton to the Tyr 75 hydroxyl, it forces Tyr 75 to become a proton donor to W2, making this water donate protons to the Ser 35 hydroxyl and the carbonyl oxygen of the residue at the 37th position. The Ser 35 hydroxyl, approaching the Asp 32 carboxyl after substrate binding, turns to donate a proton to the Oδ1 carboxyl oxygen of Asp 32 (Figs. 5a and 6b ▶ ▶), enhancing its acidic properties and assisting the transfer of a proton from the Asp 32 carboxyl to the carbonyl oxygen of the scissile bond. This bond is concomitantly attacked by W1 polarized into a nucleophilic state by the charged Asp 215 carboxyl (Fig. 6 ▶). The same consideration is relevant to memapsin, in which Trp at the 80th position in pepsin numbering plays the role of Trp 39 (Fig. 3 ▶). The chain of the hydrogen bonds, Trp 39–Tyr 75–W2–Ser 35, acts as a bridge for transmitting a signal from the flap to the active site. In memapsins, this chain of hydrogen bonds is formed by Trp 80(76)–Tyr 75(71)–W2–Ser 35.
Fig. 6.
Formula representation of the two steps of the catalytic reaction, showing the interaction of the catalytic groups with the surrounding residues. (a) The initial step before substrate binding. (b) Tetrahedral intermediate (Davies 1990; Parris et al. 1992; James et al. 1992; Veerapandian et al. 1992) and its interaction with groups, adjacent to the catalytic site. We suppose the hydrogen bond of the Asp 215 carboxyl with the Thr 218 carboxyl is weakened after substrate binding, while the bond of the Asp 32 carboxyl with the Ser 35 hydroxyl becomes stronger.
After substrate cleavage, a proton should be accepted by the Asp 32 carboxyl to return the active site to its initial state. The Ser 35 hydroxyl can assist this process if it alters a proton orientation toward W2. It is possible that W2 also changes its orientation, which may be initiated by the movement of the flap (Tyr 75) after substrate cleavage and release of the C-terminal part of the product. One can suppose that these movements are concerted, and they force Ser 35 to become a proton donor to W2. Therefore, the W2 molecule, returning a rotational degree of freedom, may act as a switch, utilizing consecutively its donor and acceptor properties in a hydrogen bond with Ser 35 during the catalytic cycle.
Analogously, Thr 218 can assist proton acceptance by the Asp 215 carboxyl from the gem-diol unit of the tetrahedral intermediate if the hydrogen bond Thr 218–Asp 215 becomes weaker after substrate binding. In the unique work on the experimental localization of hydrogen positions in transition state analogs of penicillopepsin (pdb code: 1bxo, Khan et al. 1998), the hydrogen of the Thr 218 hydroxyl is turned away from the Asp 215 carboxyl. The Thr 218 hydroxyl also can form a hydrogen bond with a substrate in some complex structures (Fraser et al. 1992; Aguilar et al. 1997).
This consideration shows that the formation of a chain of hydrogen-bonded residues Trp 39–Tyr 75–W2–Ser 35–Asp 32 (or Trp 80(76)–Tyr 75(71)–W2–Ser 35–Asp 32 in memapsins) can be an important step for the reaction catalyzed by pepsin-like enzymes. The breakage of this chain by a mutation of any of these residues results in a decrease of enzymatic activity (kcat). An increase in the catalytic efficiency of R. pusillus protease after the replacement Tyr 75 Asn suggests that the chain of hydrogen bonds remains on this mutation.
Of course, the possibility of directly verifying these hypotheses is limited by only one known structure of pepsin-like enzyme, containing experimental data on hydrogen positions, except for water molecules and carboxyl groups (pdb code: 1bxo, Khan et al. 1998). Theoretically, calculated hydrogen positions included in some structures are obviously not suitable for the purpose. However, the many self-consistent data support the chosen approach to explaining the role of residues adjacent to the catalytic site of pepsin-like enzymes. They also show how the structure of the active site can be adapted for the function in a wide range of pH from 1.0 up to 7.0. A large variety of conditions in living organisms, in which pepsin-like enzymes should act, made the development of interactions helping their function very important. The physiological adaptation of the same structural motif for the action in diverse conditions can be a gain of evolution. Simple homodimeric structures of retroviral aspartic proteases do not posses such a regulating system; their function in cells is limited to a narrow interval of pH common to all of them not far from the pKa of carboxyl groups, and the asymmetry of the active site arises after substrate binding.
Materials and methods
The three-dimensional structures of pepsin-like enzymes deposited with Protein Data Bank (Bernstein et al. 1977; Berman et al. 2000) and used in this work are listed in Table 2. Data corresponding to the highest resolution are analyzed for proteins studied by various groups of authors. Medium resolution data are not considered. The internal coordinate system (ICS) approach is applied to compare three-dimensional structures (Andreeva and Pechik 1995). (The exception is the coordinate file If34 corresponding to the complex of porcine pepsin with Ascaris inhibitor.).
Table 2.
Atomic coordinate PDB files of pepsin-like proteases
| Protein | PDB ID | Resolution (Å) | Reference |
| Zymogens | |||
| Porcine pepsinogen | 2psg | 1.8 | Sielecki et al. 1991 |
| Porcine pepsinogen | 3psg | 1.65 | Hartsuck et al. 1992 |
| Human progastricsin (hpgc) | 1htr | 1.62 | Moore et al. 1995 |
| Intermediated of hpgc activ. | 1avf | 2.4 | Khan et al. 1997 |
| Prophytepsin | 1qdm | 2.3 | Kervinen et al. 1999 |
| Proplasmepsin | 1pfz | 1.85 | Bernstein et al. 1999 |
| Pepsins | |||
| Human pepsin (hp) | 1psn | 2.2 | Fujinaga et al. 1995 |
| Complexes of hp with: | |||
| Pepstatin | 1pso | 2.0 | Fujinaga et al. 1995 |
| Phosphonate inhibitor | 1qrp | 1.96 | Fujinaga et al. 2000 |
| Porcine pepsin (pp) | |||
| Porcine pepsin monoclinic | 3pep | 2.3 | Abad-Zapatero et al. 1990 |
| Porcine pepsin monoclinic | 4pep | 1.8 | Sielecki et al. 1990 |
| Porcine pepsin hexagonal | 5pep | 2.3 | Cooper et al. 1990 |
| Complexes of pp with: | |||
| Inhibitor of renin | 1psa | 2.9 | Chen et al. 1992 |
| Ascaris inhibitor-3 | 1f34 | 2.45 | Ng et al. 2000 |
| Atlantic cod pepsin | 1am5 | 2.15 | Karlsen et al. 1998 |
| Chymosin | |||
| Bovine chymosin | 1cms | 2.3 | Gilliland et al. 1990 |
| Bovine chymosin | 4cms | 2.2 | Newman et al. 1991 |
| Bovine chymosin mutant | 3cms | 2.0 | Ŝtrop et al. 1990 |
| Complex with reduced peptide | 1czi | 2.3 | Groves et al. 1998 |
| Cathepsin D | |||
| Human cathepsin D | 1lya | 2.5 | Baldwin et al. 1993 |
| Complex with pepstatin | 1lyb | 2.5 | Baldwin et al. 1993 |
| Cathepsin D at alkaline pH | 1lyw | 2.5 | Lee et al. 1998 |
| Renin | |||
| Human renin tetragonal | 2ren | 2.5 | Sielecki et al. 1989 |
| Human renin cubic | 1bbs | 2.8 | Dhanaraj et al. 1992 |
| Complexes with renin | |||
| 1908 inhibitor | 1bil | 2.4 | Tong et al. 1995a |
| 2151 inhibitor | 1bim | 2.8 | Tong et al. 1995a |
| GP 38,560 inhibitor | 1rne | 2.4 | Rahuel et al. 1991 |
| Polyhydroxymonoamide 980 | 1hrn | 1.8 | Tong et al. 1995b |
| Mouse renin with CH-66 inh. | 1smr | 2.0 | Dealwis et al. 1994 |
| Memapsin (β-secretase) | |||
| Complex with OM99-2 inh | 1fkn | 1.9 | Hong et al. 2000 |
| Rhizopuspepsin | |||
| Rhizopuspepsin | 2apr | 1.8 | Suguna et al. 1987a |
| Complexes with: | |||
| Reduced peptide inhibitor | 3apr | 1.8 | Suguna et al. 1987b |
| Renin inhibitor | 4apr | 2.5 | Suguna et al. 1992 |
| Renin inhibitor | 5apr | 2.1 | Suguna et al. 1992 |
| Pepstatin | 6apr | 2.5 | Suguna et al. 1992 |
| Penicillopepsin | |||
| Penicillopepsin | 3app | 1.8 | James and Sielecki 1983 |
| Complexes with: | |||
| Iva-Val-Val-Lyssta-OEt | 1apt | 1.8 | James et al. 1985 |
| Iva-Val-Val-Sta-Oet | 1apu | 1.8 | James et al. 1982 |
| Difluorostatine inhibitor | 1apv | 1.8 | James et al. 1992 |
| Difluorostatone inhibitor | 1apw | 1.8 | James et al. 1992 |
| Iva-Val-Val-Sta(P)-Oet | 1ppk | 1.8 | Fraser et al. 1992 |
| Iva-Val-Val-Leu(P)-O(Phe)-OMe | 1ppl | 1.7 | Fraser et al. 1992 |
| Cbz-Ala-Ala-Leu(P)-OPhe-OMe | 1ppm | 1.7 | Fraser et al. 1992 |
| Macrocyclic inhibitor PPi4 | 1bxo | 0.95 | Khan et al. 1998 |
| Macrocylic inh.1 at 100°K | 2wea | 1.25 | Ding et al. 1998 |
| Macrocyclic inh.1 at 293°K | 2wed | 1.5 | Ding et al. 1998 |
| Acyclic deriv. (3) of inh.1 | 2web | 1.5 | Ding et al. 1998 |
| Acyclic deriv. (4) of inh.1 | 2wec | 1.5 | Ding et al. 1998 |
| Macrocyclic inh.PPi3 | 1bxq | 1.41 | Khan et al. 1998 |
| Endothiapepsin | |||
| Endothiapepsin | 4ape | 2.1 | Blundell et al. 1990 |
| Complexes with: | |||
| Inhibitor PD-125754 | 1eed | 2.0 | Cooper et al. 1992 |
| Inhibitor Pd 130328 | 1ent | 1.9 | Bailey and Cooper 1994 |
| Inhibitor PS1 | 1epl | 2.0 | Bailey and Cooper 1994 |
| Inhibitor PS2 | 1epm | 1.6 | Bailey and Cooper 1994 |
| Inhibitor CP-80, 794 | 1epn | 1.6 | Bailey and Cooper 1994 |
| Inhibitor CP-81, 282 | 1epo | 2.0 | Veerapandian et al. 1992 |
| Inhibitor PD-130693 | 1epp | 1.91 | Lunney et al. 1993 |
| Inhibitor PD-133,450 | 1epq | 1.9 | Lunney et al. 1993 |
| Inhibitor PD-135,040 | 1epr | 2.3 | Bailey and Cooper 1994 |
| Inhibitor H-77 | 1er8 | 2.0 | Cooper et al. 1989 |
| Inhibitor L-364,099 | 2er0 | 3.0 | Cooper et al. 1989 |
| Inhibitor H-256 | 2er6 | 2.0 | Cooper et al. 1987 |
| Inhibitor H-261 | 2er7 | 1.6 | Veerapandian et al. 1990 |
| Inhibitor L-363,564 | 2er9 | 2.2 | Cooper et al. 1989 |
| Inhibitor CP-71,362 | 3er3 | 2.0 | Bailey and Cooper 1994 |
| Inhibitor H-189 | 3er5 | 1.8 | Bailey et al. 1993 |
| Inhibitor PD-125,967 | 4er1 | 2.0 | Cooper et al. 1992 |
| Inhibitor pepstatin A | 4er2 | 2.0 | Bailey et al. 1993 |
| Inhibitor H-142 | 4er4 | 2.1 | Foundling et al. 1987 |
| Inhibitor BW 624 | 5er1 | 2.0 | Cooper et al. 1988 |
| Inhibitor CP-69,799 | 5er2 | 1.8 | Ŝali et al. 1989 |
| Rhizomucor proteases | |||
| Rhizomucor miehei proteinase | 2ast | 2.15 | Yang et al. 1997 |
| Complex with pepstatin | 2rmp | 2.7 | Yang and Quail 1999 |
| Mucor pusillus proteinase | 1mpp | 2.0 | Newman et al. 1993 |
| Saccharopepsin | |||
| Saccharopepsin complexes with | |||
| Inhibitor 081282 | 2jxr | 2.4 | Aguilar et al. 1997 |
| Helical inhibitor IA3 | 1dp5 | 2.2 | Li et al. 2000 |
| Truncated inhibitor IA3 | 1dpj | 1.8 | Li et al. 2000 |
| Plant enzyme cardosin A | 1b5f | 1.72 | Frazao et al. 1999 |
| Plasmepsin complexes with pepstatin: | |||
| From Plasmodium vivax | 1qs8 | 2.5 | Khazanovich-Bernstein et al. (in prep.) |
| From Plasmodium falciparum | 1sme | 2.7 | Silva et al. 1996 |
| Candidapepsin complexes | |||
| With inhibitor A70450 | 1eag | 2.1 | Cutfield et al. 1995 |
| With inhibitor A70450 | 1zap | 2.5 | Abad-Zapatero et al. 1996 |
ICS is the right-handed orthogonal internal coordinate system based on PDB atomic coordinates of three reference points p1(x1,y1,z1), p2(x2,y2,z2), and p3(x3,y3,z3) corresponding to positions of certain atoms, identical in compared structures. The conversion of PDB atomic coordinates into ICS coordinates can be performed by the standard coordinate translation and rotation procedures. In the program used, the origin of ICS is placed to the position of the first point. The first coordinate axis coincides with the vector, connecting the first and the third reference points. The direction of the second coordinate axis corresponds to the vector perpendicular to the plane, containing all reference points. The third axis is defined as the vector product of the first two.
The relations of ICS (Xics,Yics,Zics) and PDB (x,y,z) coordinates are determined by the following formula:
 |
 |
 |
where cosα1, ..., cosγ3 are direction cosines of the internal system coordinate axes in relation to the PDB axes. A good use of the ICS depends on the strategy of reference points selection and the quality of analyzed structures.
One of the advantages of the ICS approach is the convenience of the work with a large family of homologous proteins. A set of atomic coordinate files for their structures, converted to common ICS system, forms a mini ICS bank. Atomic positions in any new structure described in terms of the ICS coordinates become automatically superimposed, including water molecules, with all structures of the family presented in the ICS bank.
The conception of the internal coordinate system, its advantages and pitfalls for comparison of protein structures, and the strategy for the appropriate selection of reference points are described in the previous publication (Andreeva and Pechik 1995). The internal coordinate system used in the current work is based on the position of origin close to the active site.
Visual analysis of compared structures was performed with WebLabViewer (WebLabViewer Pro 3.0; Molecular Simulations Inc.) and Swiss-PdbViewer (Swiss-PdbViewer 3.5, Glaxo Welcome Research and Development S.A.) programs.
Acknowledgments
We are grateful to Dr. A.Yu. Borisov, L.M. Ginodman, I.V. Kashparov, and I.V. Pechik for helpful discussions. We thank Dr. I.V. Pechik for the help with the figures. The work was supported by the Grants of Russian Foundation of Fundamental Researches N 99-04-49241 and the Grant of Ministry of Science of Russian Federation N 00-15-97835.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.25801.
Note added in proof
Extraordinary structural properties of free chymosin molecule, where Tyr75 was found to occlude S1/S3 substrate binding pockets, are not discussed in this paper, being a subject of special studies.
References
- Abad-Zapatero, C., Rydel, T.J., and Erickson, J.W. 1990. Revised 2.3 Å structure of porcine pepsin. Evidence for a flexible subdomain. Proteins Struct. Funct. Genet. 8 62–81. [DOI] [PubMed] [Google Scholar]
- Abad-Zapatero, C., Goldman, R., Muchmore, S.W., Hutchins, C., Stewart, K., Navaza, J., Payne, C.D., and Ray, T.E. 1996. Structure of a secreted aspartic proteinase from Candida albicans complexed with a potent inhibitor: Implications for the design of antifungal agents. Protein Sci. 5 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar, C.F., Cronin, N.B., Badasso, M., Dreyer, T., Newman, M.P., Cooper, J.B., Hoover, D.J., Wood, S.P., Johnson, M.S., and Blundell, T.L. 1997. The three dimensional structure at 2.4 Å resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae. J. Mol. Biol. 257 899–915. [DOI] [PubMed] [Google Scholar]
- Andreeva, N.S. and James, M.N.G. 1991. Why does pepsin have a negative charge at very low pH? An analysis of conserved charged residues in aspartic proteinases. Adv. Exp. Med. Biol. 306 39–45. [DOI] [PubMed] [Google Scholar]
- Andreeva, N.S. and Pechik, I.V. 1995. Comparison of three-dimensional structures of flexible protein molecules. Mol. Biol. 22 650–657. [PubMed] [Google Scholar]
- Andreeva, N.S., Bochkarev, A.V., and Pechik, I.V. 1995. A new way of looking at aspartic proteinase structures: A comparison of pepsin structure to other aspartic proteinases in the near active site region. Adv. Exp. Med. Biol. 362 19–32. [DOI] [PubMed] [Google Scholar]
- Antonov, V.K., Ginodman, L.M., Kapitannikov, Y.V., Barshevskaya, T.N., Gurova, A.G., and Rumsh L.D. 1978. Mechanism of pepsin catalysis: General-base catalysis by the active site carboxylate ion. FEBS Lett. 88 87–90. [DOI] [PubMed] [Google Scholar]
- Antonov, V.K., Ginodman, L.M., Rumsh, L.D., Kapitannikov, Y.V., Barshevskaja, T.N., Yavashev, L.P., Gurova, A.G., and Volkova, L.I. 1981. Studies on mechanism of action of proteolytic enzymes using heavy oxygen exchange. Eur. J. Biochem. 117 195–200. [DOI] [PubMed] [Google Scholar]
- Bailey, D. and Cooper, J.B. 1994. A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica. Protein Sci. 3 2129–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, D., Cooper, J.B., Veerapandian, B., Blundell, T.L., Atrash, B., and Jones, D.M. 1993. X-ray crystallographic studies of complexes of pepstatine A and a statine-containing human renin inhibitor with endothiapepsin. Biochem. J. 289 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, E.T., Bhat, T.N., Gulnik, S., Hosur, M.V., Sowder, R.C., Cachau, R.E., Collins, J., Silva, A.M., and Erickson, J.W. 1993. Crystal structure of native and inhibited forms of human cathepsin D: Implications for lysosomal targeting and drug design. Proc. Natl. Acad. Sci. 90 6796–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shendyalov, I.N., and Bourne, P.E. 2000. Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, C., Koetzle, T., Williams, C.J.B., Meyer, E.F., Brice, M.D., Rogers, J.R., Kennard, O., Shimanouchi, T., and Tasumi, M. 1977. The Protein Data Bank, a computer-based archival file for macromolecular structures. J. Mol. Biol. 12 535–542. [DOI] [PubMed] [Google Scholar]
- Bernstein, N.K., Cherney, M., Loetsher, H., Ridley, R.G., and James, M.N.G. 1999. Crystal structure of the novel aspartic proteinase zymogen proplasmepsin from Plasmodium falciparum. Nat. Struct. Biol. 6 32–37. [DOI] [PubMed] [Google Scholar]
- Blundell, T.L., Cooper, J., Foundling, S.I., Jones, D.M., Atrash, B., and Szelke, M. 1987. On the rational design of renin inhibitors: X-ray studies of aspartic proteinases complexed with transition state analogues. Biochemistry 26 5585–5590. [DOI] [PubMed] [Google Scholar]
- Blundell, T.L., Jenkins, J.A., Sewell, B.T., Pearl, L.H., Cooper, J.B., Tickle, I.J., Veerapandian, B., and Wood, S.P. 1990. X-ray analysis of aspartic proteinases. I. The three-dimensional structure at 2.1 Å resolution of endothiapepsin. J. Mol. Biol. 211 919–941. [DOI] [PubMed] [Google Scholar]
- Chen, L., Erickson, J.W., Rydel, T.J., Park, C.H., Neidhart, D., Luly, J., and Abad-Zapatero, C. 1992. Structure of pepsin/renin inhibitor complex reveals a novel crystal packing induced by minor alterations in the inhibitor. Acta Crystallogr. B 48 476–488. [DOI] [PubMed] [Google Scholar]
- Cooper, J., Foundling, S., Hemmings, A., Blundell, T., Jones, D.M., Hallett, A., and Szelke, M. 1987. The structure of a synthetic pepsin inhibitor complexed with endothiapepsin. Eur. J. Biochem. 169 215–221. [DOI] [PubMed] [Google Scholar]
- Cooper, J.B., Foundling, S.I., Blundell, T.L., Arrowsmith, R.J., Harris, C.J., and Champness, J.N. 1988. A rational approach to the design of antihypertensives: X-ray studies of complexes between aspartic proteinases and aminoalcohol renin inhibitors. In Topics of medical chemistry. R. Soc. Chem., Special Publication, (ed. P.R. Leeming), pp. 308–313.
- Cooper, J.B., Foundling, S.I., Blundell, T.L., Boger, J., Jupp, R.A., and Kay, J. 1989. X-ray analysis of aspartic proteinase-statine inhibitor complexes. Biochemistry 28 8596–8603. [DOI] [PubMed] [Google Scholar]
- Cooper, J.B., Khan, G., Taylor, G., Tickle, I.J., and Blundell, T.L. 1990. X-ray analysis of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution. J. Mol. Biol. 214 199–222. [DOI] [PubMed] [Google Scholar]
- Cooper, J., Quail, W., Frazao, C., Foundling, S.I., Blundell, T.L., Humblet, C., Lunney, E.A., Lowther, W.T., and Dunn, B.M. 1992. X-ray crystallographic analysis of inhibition of endothiapepsin by cyclohexyl renin inhibitors. Biochemistry 31 8142–8150. [DOI] [PubMed] [Google Scholar]
- Cutfield, S.M., Dodson, E.J., Anderson, B.F., Moody, P.C.E., Marshall, C.J., Sullivan, P.A., and Cutfield, J.F. 1995. The crystal structure of a major secreted aspartic proteinase from Candida albicans in complexes with two inhibitors. Structure 3 1261–1271. [DOI] [PubMed] [Google Scholar]
- Davies, D.R. 1990. The structure and function of the aspartic proteinases. Annu. Rev. Biophys. Chem. 19 189–215. [DOI] [PubMed] [Google Scholar]
- Dealwis, C.G., Frazao, C., Badasso, M., Cooper, J.B., Tickle, I.J., Driessen, H., and Blundell, T.L. 1994. X-ray analysis at 2 Å resolution of mouse submaxillary renin complexed with a decapeptide inhibitor CH-66 based on 4–16 fragment of rat angiotensinogen. J. Mol. Biol. 236 342–360. [DOI] [PubMed] [Google Scholar]
- Dergousova, N.I., Leonova, Y.F., Zinchenko, A.A., Rumsh, L.D., and Andreeva, N.S. 1997. Mutant of HIV-1 protease with new specific properties. Protein Pept. Lett. 4 321–328. [Google Scholar]
- Dhanaraj, V., Dealwis, C.G., Frazao, C., Badasso, M., Sibanda, B.L., Tickle, I.J., Cooper, J.B., Driessen, H.P.C., Newman, M., Aguilar, C., Wood, S.P., Blundell, T.L., Hobart, P.M., Geoghegan, K.F., Ammirati, M.J., Danley, D.E., O'Connor, B.A., and Hoover, D.J. 1992. X-ray analysis of peptide-inhibitor complexes define the structural basis of specificity for human and mouse renins. Nature 357 466–471 [DOI] [PubMed] [Google Scholar]
- Ding, J., Fraser, M.E., Meyer, J., Bartlett, P.A., and James, M.N.G. 1998. Macrocyclic inhibitors of penicillopepsin. 2. X-ray crystallographic analysis of penicillopepsin complexes with a P3–P1 macrocyclic peptidyl inhibitor and with its two acyclic analogues. J. Am. Chem. Soc. 120 4610–4621. [Google Scholar]
- Foundling, S.I., Cooper, J., Watson, F.E., Cleasby, A., Pearl, L.H., Sibanda, B.I., Hemmings, A., Wood, S.P., Blundell, T.L., Valler, M.J., Norey, C.J., Kay, J., Boger, J., Dunn, B.M., Leckie, B.J., Jones, D.M., Atrash, B., Hallett, A., and Szelke, M. 1987. High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes. Nature 327 349–352. [DOI] [PubMed] [Google Scholar]
- Fraser, M.E., Strynadka, N.C.J., Bartlett, P.A., Hanson, J.E., and James, M.N.G. 1992. Crystallographic analysis of transition state mimics bound to penicillopepsin: Phosphorous containing peptide analogues. Biochemistry 31 5201–5214. [DOI] [PubMed] [Google Scholar]
- Frazao, C., Bento, I., Costa, J., Soares, C.M., Verissimo, P., Faro, C., Pires, E., Cooper, J., and Carrondo, M.A. 1999. Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp containing aspartic proteinase from flowers of Cynara cardunculus L. J. Biol. Chem. 274 27694–27701. [DOI] [PubMed] [Google Scholar]
- Fujinaga, M., Chernaja, M., Tarasova, N.I., Mosimann, S.C., and James, M.N.G. 1995. Structural studies of human pepsin and its complex with pepstatin. Protein Sci. 4 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga, M., Cherney, M.M., Tarasova, N.I., Bartlett, P.A., Hanson, J.E., and James, M.N.G. 2000. Structural study of the complex between human pepsin and a phosphorous-containing peptidic transition-state analog. Acta Crystallogr. D Biol. Crystallogr. 56 272–279. [DOI] [PubMed] [Google Scholar]
- Gilliland, G.L., Winborn, E.L., Nachman, J., and Wlodawer, A. 1990. The three-dimensional structure of recombinant bovine chymosin at 2.3 Å resolution. Proteins Struct. Funct. Genet. 8 82–101. [DOI] [PubMed] [Google Scholar]
- Groves, M.R., Dhanaraj, V., Badasso, M., Nugent, P., Pitts, J.E., Hoover, D.J., and Blundell, T.L. 1998. A 2.3 Å resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active-site β-hairpin flap is rearranged when compared with the native crystal structure. Protein Eng. 11 833–840. [DOI] [PubMed] [Google Scholar]
- Hartsuck, J.A., Koelsch, G., and Remington, S.J. 1992. The high resolution crystal structure of porcine pepsinogen. Proteins Struct. Funct. Genet. 13 1–25. [DOI] [PubMed] [Google Scholar]
- Hong, L., Koelsch, G., Lin, X., Wu, S., Terzyan, S., Ghosh, A.K, Zhang, X.C., and Tang, J. 2000. Structure of the protease domain of memapsin 2 (β-secretase) complexed with inhibitor OM-99-2. Science 290 150–153. [DOI] [PubMed] [Google Scholar]
- James, M.N.G. and Sielecki, A.R. 1983. Structure and refinement of penicillopepsin at 1.8 Å resolution. J. Mol. Biol. 163 299–361. [DOI] [PubMed] [Google Scholar]
- James, M.N.G., Sielecki, A.R., Salituro, F., Rich, D.H., and Hofmann, T. 1982. Conformational flexibility in the active sites of aspartyl proteinases revealed by pepstatine fragment binding to penicillopepsin. Proc. Natl. Acad. Sci. 79 6137–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M.N.G., Sielecki, A.R., and Hofmann, T. 1985. X-ray diffraction studies on penicillopepsin and its complexes. The hydrolytic mechanism. In Aspartic proteinases and their inhibitors, (ed. V. Kostka), pp. 163–177. Walter de Gruyter, Berlin.
- James, M.N.G., Sielecki, A.R., Hayakawa, K., and Gelb, M.H. 1992. Crystallographic analysis of transition state mimics bound to penicillopepsin: Difluorostatine- and difluorostatone-containing peptides. Biochemistry 31 3872–3886. [DOI] [PubMed] [Google Scholar]
- Karlsen, S., Hough, E., and Olsen, R.L. 1998. The crystal structure and proposed amino acid sequence of A pepsin from Atlantic cod. Acta Crystallogr. D Biol. Crystallogr. 54 32–46. [DOI] [PubMed] [Google Scholar]
- Kashparov, I.V., Popov, M.E., and Andreeva, N.S. 1997. On the role of water molecules at conserved positions near the active center of aspartic proteinases. Mol. Biol. 31 1030–1035. [PubMed] [Google Scholar]
- Kervinen, J., Tobin, J.J., Costa, J., Waugh, D.S., Wlodawer, A., and Zdanov, A.S. 1999. Crystal structure of plant aspartic protease prophytepsin: Inactivation and vacuolar targeting. EMBO J. 18 3947–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A.R., Cherney, M.M., Tarasova, N.I., and James, M.N.G. 1997. Structural characterization of activation `intermediate 2' on the pathway to human gastricsin. Nat. Struct. Biol. 4 1010–1015. [DOI] [PubMed] [Google Scholar]
- Khan, A.R., Parrish, J.C., Fraser, M.E., Smith, W.W., Bartlett, P.A., and James, M.N.G. 1998. Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes. Biochemistry 37 16839–16845. [DOI] [PubMed] [Google Scholar]
- Lee, A.Y., Gulnik, S.V., and Erickson, J.W. 1998. Conformational switching in an aspartic proteinase. Nat. Struct. Biol. 5 866–871. [DOI] [PubMed] [Google Scholar]
- Li, M., Phylip, L.H., Lees, W.E., Winther, J.R., Dunn, B.M., Wlodawer, A., and Gustchina, A.E. 2000. The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix. Nat. Struct. Biol. 7 113–116. [DOI] [PubMed] [Google Scholar]
- Lin, Y., Fusek, M., Lin, X., Hartsuck, J.A., Kezdy, F.J., and Tang, J. 1992. pH dependence of kinetic parameters of pepsin, rhizopuspepsin, and their active-site hydrogen bond mutants. J. Biol. Chem. 267 18413–18418. [PubMed] [Google Scholar]
- Lunney, E.A., Hamilton, H.W., Hodges, J.C., Kaltenbronn, J.S., Repine, J.T., Badasso, M., Cooper, J.B., Dealwis, C., Wallace, B.A., Lowther, W.T., Dunn, B.M., and Humblet, C. 1993. Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors. J. Med. Chem. 36 3809–3820. [DOI] [PubMed] [Google Scholar]
- Mantafounis, D. and Pitts, J. 1990. Protein engineering of chymosin: Modification of the pH optimum of enzyme catalysis. Protein Eng. 3 605–609. [DOI] [PubMed] [Google Scholar]
- Moore, S.A., Sielecki, A.R., Chernaia, M.M., Tarasova, N.I., and James, M.N.G. 1995. Crystal and molecular structure of human progastricsin at 1.62 Å resolution. J. Mol. Biol. 247 466–485. [DOI] [PubMed] [Google Scholar]
- Newman, M., Safro, M., Frazao, C., Khan, G., Zdanov, A., Tickle, I.J., Blundell, T.L., and Andreeva, N. 1991. X-ray analysis of aspartic proteinases IV. Structure and refinement at 2.2 Å resolution of bovine chymosin. J. Mol. Biol. 221 1295–1309. [PubMed] [Google Scholar]
- Newman, M., Watson, F., Roychowdhury, P., Jones, H., Badasso, M., Cleasby, A., Wood, S.P., Tickle, I.J., and Blundell, T.L. 1993. X-ray analyses of aspartic proteinases V. Structure and refinement at 2.0 Å resolution of the aspartic proteinase from Mucor pusillus. J. Mol. Biol. 230 260–283. [PubMed] [Google Scholar]
- Ng, K.K.S., Petersen, J.F.W., Cherney, M.M., Garen, C., Zalatoris, J.J., Rao-Naik, C., Dunn, B.M., Martzen, M.R., Peanasky, R.J., and James, M.N.G. 2000. Structural basis for inhibition of porcine pepsin by Ascaris pepsin inhibitor-3. Nat. Struct. Biol. 7 653–657. [DOI] [PubMed] [Google Scholar]
- Park, Y.N., Aikawa, J.I., Nishiyama, M., Horinouch, S., and Beppu, T. 1996. Involvement of a residue at position 75 in catalytic mechanism of a fungal aspartic proteinase, Rhizomucor pusillus pepsin. Replacement of tyrosine 75 on the flap by asparagine enhances catalytic efficiency. Protein Eng. 9 869–875. [DOI] [PubMed] [Google Scholar]
- Park, Y.N., Aikawa, J.I., Nishiyama, M., Horinouch, S., and Beppu, T. 1997. Site-directed mutagenesis of conserved Trp39 in Rhizomucor pusillus pepsin: Possible role of Trp39 in maintaining Tyr75 in the correct orientation for maximizing catalytic activity. J. Biochem. 121 118–121. [DOI] [PubMed] [Google Scholar]
- Parris, K.D., Hoover, D.J., Damon, D.B., and Davies, D.R. 1992. Synthesis and crystallographic analysis of two rhizopuspepsin inhibitor complexes. Biochemistry 31 8125–8141. [DOI] [PubMed] [Google Scholar]
- Perutz, M.F. 1992. Protein structures: New approaches to disease and therapy. W.H. Freeman & Co., New York.
- Rahuel, J., Priestle, J.P., and Grotter, M.G. 1991. The crystal structure of recombinant glycosylated human renin alone and in complex with a transition state analog inhibitor. J. Struct. Biol. 107 227–236. [DOI] [PubMed] [Google Scholar]
- Rawlings, N.D. and Barrett, A.J. 1998. MEROPS—The peptidase database. http://www.merops.co.uk/ [DOI] [PMC free article] [PubMed]
- Ŝali, A., Veerapandian, B., Cooper, J., Foundling, S.I., Hoover, D.J., and Blundell, T.L. 1989. High resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: The analysis of inhibitor binding and description of the rigid body shifts in the enzyme. EMBO J. 8 2179–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielecki, A.R., Hayakawa, K., Fujinaga, M., Murphy, M.E.P., Fraser, M., Muir, A.K., Carilli, C.T., Lewicki, J.A., Baxter, J.D., and James, M.N.G. 1989. Structure of recombinant human renin, a target for cardiovascular-active drugs, at 2.5 Å resolution. Science 243 1346–1351. [DOI] [PubMed] [Google Scholar]
- Sielecki, A.R., Fedorov, A.A., Bodhoo, A., Andreeva, N.S., and James, M.N.G. 1990. Molecular and crystal structures of monoclinic porcine pepsin crystals refined at 1.8 A resolution. J. Mol. Biol. 214 143–170. [DOI] [PubMed] [Google Scholar]
- Sielecki, A.R., Fujinaga, M., Read, R.J., and James, M.N.G. 1991. Refined structure of porcine pepsinogen at 1.8 Å resolution. J. Mol. Biol. 219 671–692. [DOI] [PubMed] [Google Scholar]
- Silva, A.M., Lee, A.Y., Gulnik, S.V., Majer, P., Collins, J., Bhat, T.N., Collins, P.J., Cachau, R.E., Luker, K.E., Gluzman, L.Y., Francis, S.E., Oksman, A., Goldberg, D.E., and Erickson, J.W. 1996. Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum. Proc. Natl. Acad. Sci. 93 10034–10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ŝtrop, P., Sedlacek, J., Stys, J., Kaderabkova, Z., Blaha, I., Pavlickova, L., Pohl, J., Fabry, M., Kostka, V., Newman, M., Frazao, C., Shearer, A., Tickle, I.J., and Blundell, T.L. 1990. Engineering enzyme subsite specificity: Preparation, kinetic characterization, and X-ray analysis at 2.0 A resolution of Val111Phe site-mutated calf chymosin. Biochemistry 29 9863–9871. [DOI] [PubMed] [Google Scholar]
- Suguna, K., Bott, R.B., Padlan, E.A., Subrananian, E., Sheriff, S., Cohen, G.H., and Davies, D.R. 1987a. Structure and refinement at 1.8 A resolution of the aspartic proteinase from Rhizopus chinensis. J. Mol. Biol. 196 877–900. [DOI] [PubMed] [Google Scholar]
- Suguna, K., Padlan, E.A., Smith, K.W., Carlson, W.D., and Davies, D.R. 1987b. Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: Implications for a mechanism of action. Proc. Natl. Acad. Sci. 84 7009–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suguna, K., Padlan, E.A., Bott, R., Rogers, J., Parris, K.D., and Davies, D.R. 1992. Structures of complexes of rhizopuspepsin with pepstatine and other statine-containing inhibitors. Proteins Struct. Funct. Genet. 13 195–205. [DOI] [PubMed] [Google Scholar]
- Suzuki, F., Goto, K., Shiratori, Y., Inagami, T., Murakami, K., and Nakamura, Y. 1996. Tyrrosine-83 of renin has an important role in renin-angiotensinogen reaction. Protein Pept. Lett. 3 45–49. [Google Scholar]
- Suzuki, J., Sasaki, K., Sasao, Y., Akio, H., Kawasaki, H., Nishiyama, M., Horinouchi, S., and Beppu, T. 1989. Alteration of catalytic properties of chymosin by site-directed mutagenesis. Protein Eng. 2 563–569. [DOI] [PubMed] [Google Scholar]
- Tong, L., Pav, S., Lamarre, D., Simoneau, B., Lavallee, P., and Jung, G. 1995a. Crystallographic studies on the binding modes of P2-P3 butanediamide renin inhibitors. J. Biol. Chem. 270 29520–29524. [DOI] [PubMed] [Google Scholar]
- Tong, L., Pav, S., Lamarre, D., Pilote, L., LaPlante, S., Anderson, P.C., and Jung, G. 1995b. High resolution crystal structure of recombinant human renin in complex with polyhydroxymonoamide inhibitors. J. Mol. Biol. 250 211–222. [DOI] [PubMed] [Google Scholar]
- Veerapandian, B., Cooper, J.B., Sali, A., and Blundell, T.L. 1990. X-ray analyses of aspartic proteinases. III. Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 Å resolution. J. Mol. Biol. 216 1017–1029. [DOI] [PubMed] [Google Scholar]
- Veerapandian, B., Cooper, J.B., Sali, A., Blundell, T.L., Rosati, R.L., Dominy, B.W., Damon, D.B., and Hoover, D.J. 1992. Direct observation by X-ray analysis of the tetrahedral `intermediate' of aspartic proteinases. Protein Sci. 1 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. and Quail, J.W. 1999. Crystal structure of the Rhizomucor miehei aspartic proteinase complexed with the inhibitor pepstatin A at 2.7 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55 625–630. [DOI] [PubMed] [Google Scholar]
- Yang, J., Teplyakov, A., and Quail, J.W. 1997. Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 Å resolution. J. Mol. Biol. 268 449–459. [DOI] [PubMed] [Google Scholar]