Abstract
The major serine proteinase inhibitor from bell pepper (Capsicum annuum, paprika) seeds was isolated, characterized, and sequenced, and its disulfide bond topology was determined. PSI-1.2 is a 52-amino-acid-long, cysteine-rich polypeptide that inhibits both trypsin (Ki = 4.6 × 10−9 M) and chymotrypsin (Ki = 1.1 × 10−8 M) and is a circularly permuted member of the potato type II inhibitor family. Mature proteins of this family are produced from precursor proteins containing two to eight repeat units that are proteolytically cleaved within, rather than between, the repeats. In contrast, PSI-1.2 corresponds to a complete repeat that was predicted as the putative ancestral protein of the potato type II family. To our knowledge, this is the first case in which two proteins related to each other by circular permutation are shown to exist in the same organism and are expressed within the same organ. PSI-1.2 is not derived from any of the known precursors, and it contains a unique amphiphilic segment in one of its loops. A systematic comparison of the related precursor repeat-sequences reveals common evolutionary patterns that are in agreement with the ancestral gene-duplication hypothesis.
Keywords: Protein chemistry, protein structure, proteinase inhibitors, evolution
Most plants produce proteinase inhibitors as a defense against insects and pathogenic organisms (Ryan 1980Ryan 1989). One of the well-studied groups is that of the serine proteinase inhibitors belonging to the so-called potato type II (PT-II) family (Richardson 1977; Casaretto and Corcuera 1995; Bowles 1998). The members of this group are ∼50-residue-long, cysteine-rich polypeptides that inhibit both trypsin and α-chymotrypsin. Their three-dimensional structure is characterized by four disulfide bonds, a triple-stranded β-sheet, and a long loop that interacts with the active site of the proteinase (Greenblatt et al. 1989; Bode and Huber 1992; Nielsen et al. 1994 Nielsen et al. 1995). These proteins are expressed in various tissues of the members of the Solanaceae family, sometimes at very high levels (Graham et al. 1985a; Keil et al. 1986; Johnson and Ryan 1990; Balandin et al. 1995; Miller et al. 2000). Several genes encoding PT-II proteins have been ideied (for a recent overview, see Miller et al. 2000). These genes usually encode a larger precursor protein, which is subsequently cleaved into several slightly different PT-II inhibitor molecules (Table 1). The precursors contain a variable number of cysteine-rich repeats that we had termed inhibitor precursor (IP)-repeats after the Swiss-Prot name of the parent proteins (Murvai et al. 1999). Proteolytic cleavage of the precursor occurs within, rather than between, repeats (Atkinson et al. 1993). It has been hypothesized that the gene encoding the precursor protein arose from the duplication of a gene originally encoding a single IP-repeat, which was then followed by a frame shift in the proteolytic processing (Scanlon et al. 1999). To support this hypothesis, Scanlon and associates designed and expressed in Escherichia coli an hypothetical ancestral protein corresponding to a single repeat of the Nicotiana alata precursor protein and determined its three-dimensional structure by nuclear magnetic resonance (NMR). This product actually inhibits trypsin and chymotrypsin, and its fold is very similar to that of the naturally occurring inhibitors (Scanlon et al. 1999). In other terms, the N. alata repeat has the ability to fold both as a structural repeat (similar to mature PT-II inhibitors) and as a sequence repeat (similar to aPI1; Scanlon et al. 1999).
Table 1.
Members of the potato proteinase inhibitor type II family used in the phylogenetic analysis.
| Entry name | Accession No. | Protein name (gene name) | Species, tissue | No. of repeats | Reference |
| S72492 (PIR) | S72492 | probable proteinase inhibitor precursor–tomato, AT2 protein (PIN2) | L. esculentum (tomato), shoot | 2 | Brandstadter et al. 1996 |
| O82735 | O82735 | potato (S. tuberosum) mRNA 2 for proteinase inhibitor II [fragment] | S. tubersum (potato). | 2 | Sanchez-Serrano et al. 1986 |
| IP25_SOLTU | Q41488 | proteinase inhibitor type II P303.51 [precursor] | S. tuberosum (potato), tuber | 2 | Jongsma et al. 1995 |
| IP27_SOLTU | Q43652 | proteinase inhibitor type II CM7 [precursor] (PIN2–CM7) | S. tuberosum (potato), leaf | 2 | Murray and Christeller 1994 |
| IP2Y_SOLTU | Q41489 | proteinase inhibitor type II [precursor] | S. tuberosum (potato) | 2 | ds |
| IP2K_SOLTU 4SGB_I(PDB) | P01080 | proteinase inhibitor type II K [precursor], IIK, (PIN2K) | S. tuberosum (potato) | 2 | Keil et al. 1986 |
| IP2X_SOLTU | Q00782 | proteinase inhibitor type II [precursor] | S. tuberosum (potato) | 2+ | Choi et al. 1990 |
| IP2T_SOLTU | Q41435 | proteinase inhibitor type II T [precursor], (PIN2T) | S. tuberosum (potato) | 2 | Park and Thornburg et al. 1996 |
| IP21_LYCES | P05119 | wound-induced proteinase inhibitor type II [precursor] | L. esculentum (tomato), leaf | Graham et al. 1985b | |
| IP21_TOBAC | Q40561 | proteinase inhibitor type II [precursor] | N. tabacum (common tobacco), leaf | 3 | Balandin et al. 1995 |
| IP22_LYCES | Q43710 | proteinase inhibitor type II TR8 [precursor] (ARPI) | L. esculentum (tomato), seedling, root | 3 | Taylor et al. 1993 |
| IP23_LYCES | Q43502 | proteinase inhibitor type II CEVI57 [precursor] (CEVI57) | L. esculentum (tomato), leaf | 3 | Gadea et al. 1996 |
| X99095 | X99095 | proteinase inhibitor type II (pin2) gene | S. tuberosum (potato), leaf | 3 | Dammann et al. 1997 |
| Q9SDL4, IP21_CAPAN | Q9SDL4 | Hypothetical 22.5 KDA protein (CAPIN II) | C. annuum (paprika, bell pepper), seed | 3` | ds |
| IP22_CAPAN | Q49146 | wound-induced proteinase inhibitor II [precursor] | C. annuum (paprika, bell pepper), pericarp | 3` | ds |
| Q9SQ77 | Q9SQ77 | proteinase inhibitor [precursor] | N. alata (winged tobacco) (Persian tobacco), stigma | 4 | Miller et al. 2000 |
| Q40378 | Q40378 | proteinase inhibitor [precursor] | N. alata (winged tobacco) (Persian tobacco), stigma, styles | 6 | Atkinson et al. 1993 |
| Q9SDW7, 1CE3 (PDB) | Q9SDW7 | proteinase inhibitor type II precursor NGPI-2 [fragment] | N. glutinosa (tobacco) | 6 | Choi et al. 1993 |
| Q9SDW8 | Q9SDW8 | proteinase inhibitor type II precursor NGPI-1 [fragment] | N. glutinosa (tobacco) | 8 | Choi et al. 1993 |
| PSI-1.2 | n.a. | Paprika seed proteinase inhibitor | C. annuum (paprika, bell pepper) seed | ? | this work. |
Capsicum annuum (bell pepper, paprika) seeds express several regular PT-II inhibitors, including PSI-1.1 (Antcheva et al. 1996). Here we report the isolation, amino acid sequencing, disulfide bond topology, and characterization of PSI-1.2, a proteinase inhibitor that corresponds to a single IP sequence repeat and thus has a fold similar to the putative ancestral inhibitor protein aPI1. To our knowledge, this is the first case in which two proteins related to each other by circular permutation are shown to exist in the same organism. The structure of the PSI-1.2 protein is discussed with the help of a structural model as well as in the light of a systematic comparison of IP sequence repeats.
Results
Isolation and characterization
Isolation of PSI-1.2 was based on a procedure previously developed for bell pepper seed inhibitors (Antcheva et al. 1996). This process is based on affinity chromatography on α-chymotrypsin-Sepharose, and yields two main fractions shown in Figure 1 ▶. Mass spectrometry analysis indicated that fraction I contains PSI-1.1, a member of the PT-II family of inhibitors that had been ideied previously (Antcheva et al. 1996). Fraction II contained two products designated as PSI-1.2A and PSI-1.2B. These components were repurified by narrow-bore reverse phase–high-performance liquid chromatography (RP-HPLC) before sequencing (data not shown).
Fig. 1.
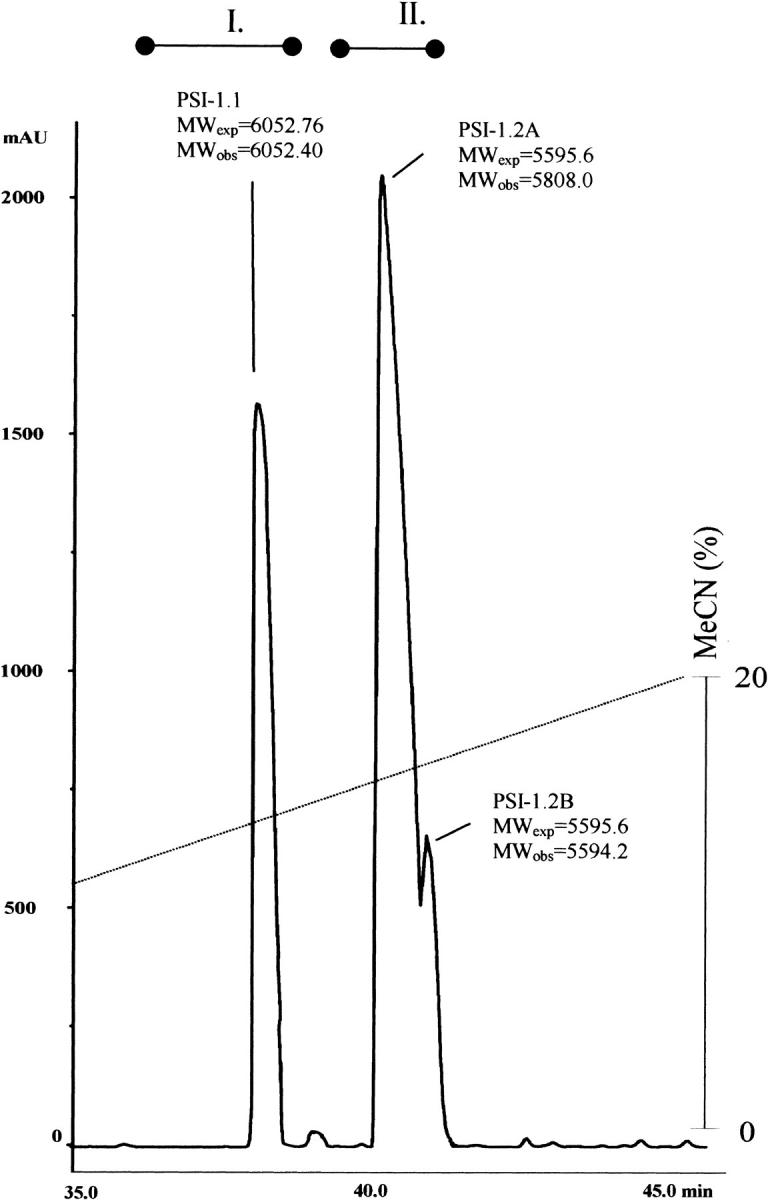
Separation of various proteinase inhibitors from Capsicum annuum seeds by reverse phase–high-performance liquid chromatography. The observed molecular weights (MWobs) were determined by mass spectrometry. The expected molecular weights (MWexp) are based on the sequences shown in Figs. 2, 3 ▶ ▶. The bars over the elution profile correspond to fractions I and II.
Protein sequencing
The major inhibitor fraction (II in Fig. 1 ▶) consisted of two overlapping peaks. Initial sequencing attempts of the larger peak (A) failed to detect a sequenceable N terminus. A sample was thus reduced, pyridylethylated, and digested separately with either CNBr in 70% HCOOH or trypsin. The resulting peptides (Fig. 2 ▶, PSI-1.2A-F1 and PSI-1.2A-F2, respectively) were isolated by narrow-bore RP-HPLC and sequenced. The smaller peak (B), on the other hand, gave a full sequence of 52 amino acids, identical with that of peak A. A comparison of the sequence (Fig. 2 ▶) and the observed molecular mass (Fig. 1 ▶) indicates that the difference between peak A and peak B results from an unideied N-terminal modification of peak A. The PSI-1.2 sequence (Fig. 2 ▶) has eight cysteines, same as in the previously isolated PSI-1.1 (Antcheva et al. 1996).
Fig. 2.
The sequence of PSI-1.2 as determined by automated Edman sequencing after reduction and pyridylethylation. PSI-1.2B gave a complete sequence corresponding to its observed molecular weight determined by mass spectrometry. PSI-1.2A failed to produce an N-terminal signal, so its fragments obtained by CNBr cleavage (PSI-1.2A-F1) and trypsin (PSI-1.2A-F2) were subjected to sequencing.
The sequence of PSI-1.2 does not correspond to any published sequence found in the databases. On the other hand, the sequence search revealed that the previously determined PSI-1.1 is identical with one of the predicted proteolytic fragments of the recently published PT-II family precursor Q9SDL4 (Fig. 3A ▶). The Q9SDL4 precursor can, in principle, yield three mature PT-II proteins. Interestingly, PSI-1.1 is identical with the first putative cleavage product.
Fig. 3.
Multiple alignment of PSI-1.2 with selected PT-II sequences. (A multiple alignment comprising all inhibitor precursor IP-repeat sequences was deposited as supplemental material.) The dashed line indicates the region in which proteolytic cleavage of the IP-repeats takes place (PSI-1.2 is not cleaved in this region). The segments A, P, and B are explained in the text and also shown in Fig. 7 ▶. The conserved cysteines are highlighted in grey. Q9SQ77-R1 corresponds to the aPI1 (PDB code 1CE3), IP2K_SOLTU to PCI-1 (PDB code 4SGB), and PSI-1.1 to IP21_CAPAN.
Disulfide bridges of PSI-1.2
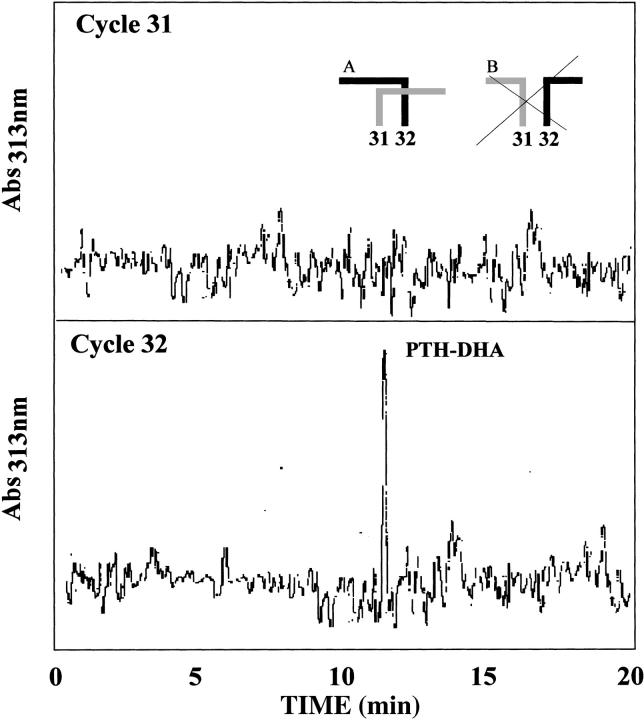
As PSI-1.2 is not identical with any natural mature proteinase inhibitor, its disulfide topology was experimentally determined from a set of enzymatic digests combined with mass spectrometry and N-terminal sequencing (Table 2). This analysis in itself did not yield the complete disulfide topology of the protein. In particular, the connectivity of two adjacent cysteines, Cys31 and Cys32, is not unambiguous (see inset in Fig. 4 ▶). Additional data were collected by Edman degradation combined with phenylhydantoin (PTH) analysis at 313 nm, which makes it possible to idey PTH-dehydroalanine (PTH-DHA), the β-elimination product of PTH-cystine. PTH-DHA forms when the process of N-terminal sequencing reaches a Cys residue that is disulfide bonded to a sequentially upstream Cys residue (Li and Liang 1999). This analysis showed PTH-DHA at positions 28, 32, 38, and 49; an example is shown in Figure 4 ▶. These results, combined with the data of Table 2, confirmed that PSI-1.2 has the disulfide topology Cys3-Cys32, Cys7-Cys28, Cys16-Cys38, Cys31-Cys49. The disulfide bridges of PSI-1.2 thus correspond to those of the aPI1 hypothetical ancestral protein (Scanlon et al. 1999).
Table 2.
The disulfide structure of PSI-1.2 as determined by enzymatic digestion and electron spray ionization mass spectrometry
| Molecular mass [Da] (S–S form) | Molcular mass [Da] after reduction | ||||||
| Enzyme | Native peptide fragment (predicted structure) | Observed | Predicted | Reduced fragment (predicted structure) | Observed | Predicted | S–S bridge |
| 2ACPR5 | 445.5 | 445.54 | 3–32 | ||||
| T + C | 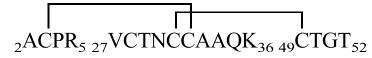 |
1860.6 | 1862.16 | ||||
| 27VCTNCCAAQK36 | 1040.3 | 1040.24 | 31–49 | ||||
| 2ACPR5 27VCTNCCAAQK36 49CTGT52 | 49CTGT52 | 380.4 | 380.42 | ||||
| T + C | 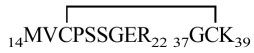 |
1270.2 | 1269.47 | 14MVCPSSGER22 | 965.2 | 965.12 | 16–38 |
| 14MVCPSSGER22 37GCK39 | 37GCK39 | 306.4 | 306.38 | ||||
| 3CPRNCDTDIA12 | 1107.6 | 1107.22 | 3–32 | ||||
| P | 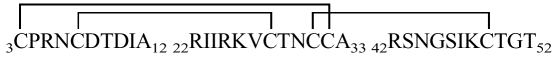 |
3603.3 | 3604.14 | 22RIIRKVCTNCCA33 | 1379.8 | 1379.72 | 7–28 |
| 3CPRNCDTDIA12 22RIIRKVCTNCCA33 42RSNGSIKCTGT52 | 42RSNGSIKCTGT52 | 1123.6 | 1123.25 | 31–49 | |||
| P | 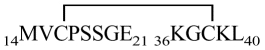 |
1354.6 | 1354.62 | 14MVCPSSGE21 | 808.3 | 808.92 | 16–38 |
| 14MVCPSSGE21 36KGCKL40 | 36KGCKL40 | 547.3 | 547.71 | ||||
T + C indicates trypsin/chymotrypsin; P, pepsin; at pH 3.0.
Fig. 4.
Ideication of dehydroalanine (DHA) during Edman sequencing by monitoring the high-performance liquid chromatography–effluent of the phenylhydantoin (PTH) degradation products at 313 nm. Appearance of PTH-DHA in position 32 indicates that this residue is disulfide bonded to a sequentially upstream cysteine, which by exclusion is Cys3. This arrangement is compatible with scheme A and not with scheme B.
Enzyme inhibitor assays
PSI-1.2B, the product with a fully known sequence was isolated for enzymatic analysis. It is a strong inhibitor of trypsin (Ki = 4.6 × 10−9 M) and a somewhat weaker inhibitor of α-chymotrypsin (Ki = 1.1 × 10−8 M), whereas elastase and subtilisin DY are not inhibited (Table 3). The enzymes thrombin and factor Xa, related to the blood clotting system, are only weakly inhibited by PSI-1.2 (Ki = 1.1 × 10−6 M and Ki = 2.6 × 10−5 M, respectively). As a whole, PSI-1.1 appears to be a stronger inhibitor of thrombin (100×), trypsin (10×), and factor Xa (10×) than the presently isolated PSI-1.2. Pepsin was found not to hydrolyze PSI-1.2 over a period of 30 min at pH 2.0. Heat treatment (100°C) at pH 4.0 for 10 min had no effect on the anti-trypsin activity of PSI-1.2B.
Table 3.
Inhibition constants of PSI–1.2
| Inhibitor | P4 | P3 | P2 | P1 | P`1 | P`2 | P`3 | Trypsin Ki (M) | α-chymotrypsin Ki (M) | Thrombin Ki (M) | Factor Xa Ki (M) |
| PSI–1.2 | A | C | P | R | N | C | D | 4.6 × 10−9 | 1.1 × 10−8 | 1.1 × 10−6 | 2.6 × 10−5 |
| PSI–1.1 | A | C | P | R | Y | C | D | 4.8 × 10−10 | 4.7 × 10−8 | 1.7 × 10−8 | 1.1 × 10−6 |
Inhibition effects were compared with PSI-1.1 isolated from C. annuum seeds (Antcheva et al. 1996). The numbering of residues in contact with the protease is taken from Bode and Huber (1992)
Sequence similarity searches
The sequence of PSI-1.2 is not identical with any known protein found in the protein and DNA databases. However, it shows a sequence similarity to various protein precursors of the PT-II family proteinase inhibitors. Comparing the sequence with the domain database SBASE, it becomes apparent that the sequence of PSI-1.2 corresponds to a complete IP-repeat (Fig. 3B ▶), the repeat unit of the PT-II family precursors (Murvai et al. 1999). A comparison with mature PT-II inhibitors reveals, on the other hand, that the sequence of PSI-1.2 is circularly permuted compared with that of the mature proteins, as if a domain swapping event had taken place (Bennett et al. 1995; Heringa and Taylor 1997). For example, of the 45 residues of the potato tuber inhibitor PCI-1 (PDB code 4SGB_I) that can be aligned with PSI-1.2, 25 are identical (56%). If we divide PSI-1.2 into three fragments, A-P-B, PCI-1 should be represented as B-A, with P denoting the putative processing site, which is missing in the mature PCI-1 protein (Fig. 3B ▶).
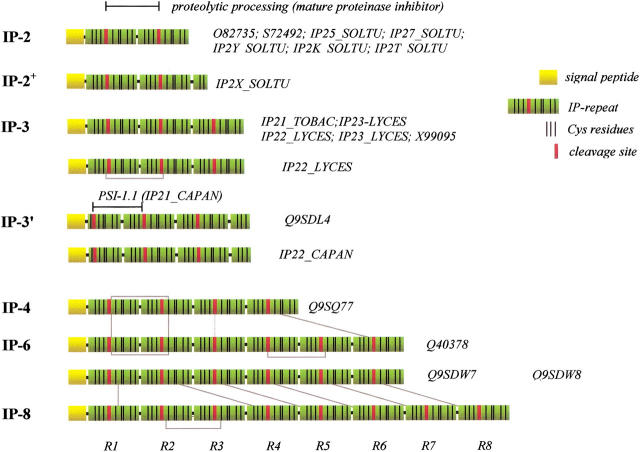
A database search for IP-repeats similar to PSI-1.2 in databases resulted in the ideication of several proteinase precursor proteins containing from two to eight IP-repeats (Table 1). In the precursor proteins from Nicotiana (tobacco), Solanum (potato), and Lycopersicon (tomato), the number of IP-repeats can vary from two to eight. In general, the precursor consists of an integer number of IP-repeats, the only exception being IP2X_SOLTU, for which a short C-terminal partial repeat containing two cysteines is present. The C. annuum precursors IP22_CAPAN and Q9SDL4 are unique in the sense that they contain two IP-repeats flanked by N- and C-terminal partial repeats. Their N-terminal partial repeat contains six cysteines, whereas the C-terminal partial repeat contains three cysteines only. In several cases, different IP-repeats share the same amino acid sequence. For example, five of the Q9SDW7 repeats (R2 to R6) are 100% identical to R4 through R8 from Q9SDW8. The previously isolated PSI-1.1 (IP21_CAPAN) protein (Antcheva et al. 1996) is identical to residues 36–90 of Q9SDL4, indicating that it derives from the proteolytic processing of the Q9SDL4 precursor.
The exon/intron structure was reported for a number of genes. In the known cases, all the IP-repeats (i.e., the entire precursor apart from the signal peptide) are coded by one single exon.
We performed a systematic sequence comparison on the individual IP-repeats found in the databases. For better ideication, the repeats are numbered as R1 through R8, starting from the N terminus of the precursors, and the precursors are denoted by the number of IP-repeats indicated in Table 1. For example, a precursor with two IP-repeats is denoted IP-2. The multiple alignment (deposited) revealed that only the cysteine pattern is completely conserved. The results of the comparison are displayed as a phylogenetic tree (Fig. 5 ▶) as well as in a graphical form (Fig. 6 ▶). One major cluster includes R1 and R2 repeats from precursors with two and three IP-repeats (IP-2 and IP-3), including IP2X_SOLTU, which has an additional short truncated repeat at the C terminus (IP-2+). This group includes all the known potato precursors as well as the one expressed in tobacco leaves. The second group contains all the IP-repeats of the N. alata and N. glutinosa precursors that consist of four to eight IP-repeats (IP-4, IP-6, and IP-8). These precursors are expressed in floral organs and young tissues of the plants.
Fig. 5.
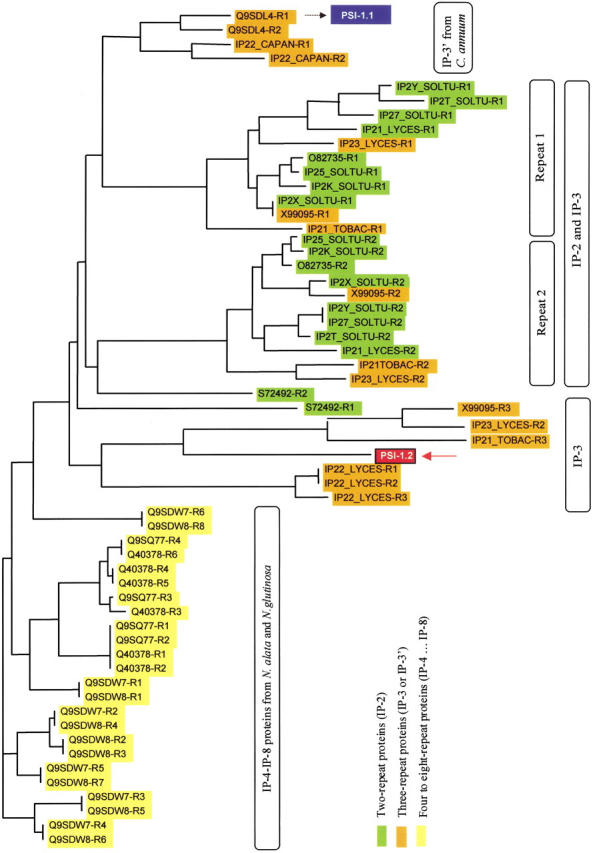
Phylogenetic tree of the proteinase inhibitor precursor IP-repeats. The repeat number is abbreviated as R1, R2, R3, etc., starting from the N terminus. The amino acid sequence of PSI-1.1 (Antcheva et al. 1996) exactly corresponds to the first putative mature product of the hypothetical 22.5-kD protein of Capsicum annuum (Q9SDL4).
Fig. 6.
The domain structure of the potato II inhibitor family precursors that show sequence similarity to PSI-1.2. IP-2 through IP-8 designate the number of inhibitor precursor IP-repeats in the group. Gray lines indicate sequence identity (>98%); dashed line, similarity. It is noted that the C-terminal truncated repeats in IP-2+ and IP-3` are not of the same length.
Finally, the precursors that contain three repeats form a further group that can be subdivided into two subgroups. One group, IP-3`, is represented by two bell pepper inhibitor precursors that have an unusual architecture, as they contain one repeat consisting of two partial repeats attached to the N and C terminus of two intact repeats. The other subgroup of three-repeat precursors, IP-3, consists of three regular IP-repeats. In these sequence comparisons, PSI-1.2 was found to be most similar to the R3 (the third IP-repeat) of the IP-3 precursors that form a separate cluster (Fig. 5 ▶).
Model building
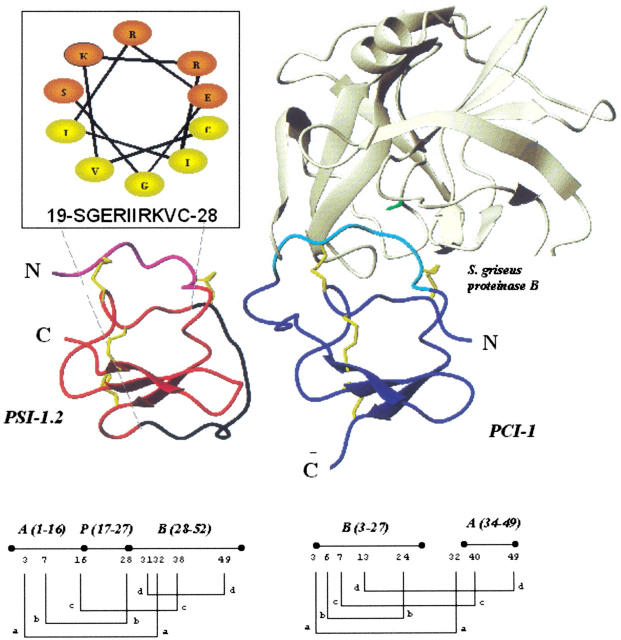
To idey the putative region responsible for proteinase inhibition, we built a three-dimensional model of PSI-1.2 based on the NMR structure of the proteinase inhibitor aPI1 (Scanlon et al. 1999). PSI-1.2 and aPI1 have a 53% sequence identity and the same disulfide bond pattern, so they are likely to adopt the same fold. Figure 7 ▶ compares the aPI1-based model of PSI-1.2 with the crystallographic structure of the potato chymotrypsin inhibitor-1 (PCI-1) complexed with the proteinase B from Streptomyces griseus (PDB code 4SGB). Of the available proteinase/inhibitor complexes, 4SGB_I shows the highest sequence similarity to PSI-1.2. Figure 7 ▶ clearly shows that despite the different positions of the N and C termini, the overall fold of the two proteins and the orientation of the disulfide bonds are very similar, with a conserved core composed of a triple-stranded anti-parallel β-sheet and a network of four disulfide bridges. The PCI-1 residues involved in contacts with the active site of the S. griseus proteinase are located in the long loop connecting strands 2 and 3. These residues (33-PKACPLNCDP-42) correspond to the N-terminal region in PSI-1.2. The central eight residues of PSI-1.2 (34-KACPRNCD-41) are well conserved, with the only exception of the L→R substitution. Although constrained by two disulfide bridges at positions 3 and 7, this region shows a high root mean square derived value in the NMR structure of aPI1 and is thus likely to be conformationally flexible.
Fig. 7.
(Left) The model of PSI-1.2 built by homology modeling. (Right) The structure of the complex between the polypeptide chymotrypsin inhibitor-1 from potato tubers (PCI-1, in blue) and the proteinase B from Streptomyces griseus (PDB code 4SGB; Greenblatt et al. 1989). The PCI-1 residues involved in contacts with the enzyme are shown in light blue; the corresponding residues of PSI-1.2, in magenta. The catalytically active residue (S195) of the enzyme is shown in green. Although the orientation of the disulfides (yellow) is identical in the two molecules, the disulfide topologies (below) are different: abcbdacd in PSI-1.2 and abcdbcad in PCI-1. (Inset) The helical wheel diagram of the central region of PSI-1.2. In the model, the same segment is shown in black.
The segment 19–28 of PSI-1.2 is of particular interest. This segment is not part of any other mature protein isolated so far; moreover, the corresponding segment of aPI1 is not well defined. A helical wheel plot reveals, on the other hand, that the distribution of hydrophobic and hydrophilic amino acids in this segment of PSI-1.2 is characteristic of amphiphilic helices (Fig. 7 ▶, inset). In the hypothetical helix, residues S, E, R, R, and K are facing the solvent, whereas residues G, I, I, V, and C face the interior of the protein. This asymmetric distribution of hydrophilic and hydrophobic residues is unique to PSI-1.2 among the known IP-repeats.
Discussion
Of the bell pepper seed inhibitors described in this work, PSI-1.1 is apparently a proteolytic cleavage product of the Q9SDL4 precursor, whereas PSI-1.2 is different from any known protein. PSI-1.2 is the first naturally occurring inhibitor that shows the circularly permuted topology present in the hypothetical ancestral PT-II protein aPI1 (Scanlon et al. 1999). Circular permutation of sequences had been reported in other cases (for a review, see Heringa and Taylor 1997; Lindqvist and Schneider 1997). Until now, however, circular rearrangements were observed only between species, such as the β-lectin of Vicia fava and the lectin concanavalin A from Canavalia ensiformis (Edelman et al. 1972), or the plant aspartyl proteinases and human lung surfactant proteins (Ponting and Russell 1995). PSI-1.2 is apparently the first example in which circularly permuted members of a protein family are expressed within the same organism, moreover within the same organ. As both proteinase inhibitors and lectins are proteins that play roles in the defense mechanisms of plants, it is tempting to speculate that the underlying sequence rearrangements are part of a general scenario by which plants produce functional diversity against pathogenic attack. In fact, the specificity of PSI-1.2 is slightly different from that of the previously isolated PSI-1.1 (Table 3).
Homology modeling confirmed that the sequence can be built into the structure of aPI1 without steric clashes. In the region that corresponds to the in vivo proteolytic processing site in the aPI1 sequence, PSI-1.2 has the theoretical capability to form an amphiphilic helix that would have the hydrophobic face toward the interior of the protein and the hydrophilic face toward the solvent. As no other IP-repeat sequence shows similar distribution of hydrophobic and hydrophilic amino acids in the corresponding region, we speculate that this feature may be of significance and worth of structural studies. For example, the amphiphilic helix might contribute to the stability of PSI-1.2 in solution; moreover, it might provide protection against proteolytic cleavage as it might not be recognized by the proteases in the plant secretory pathway (Heath et al. 1995).
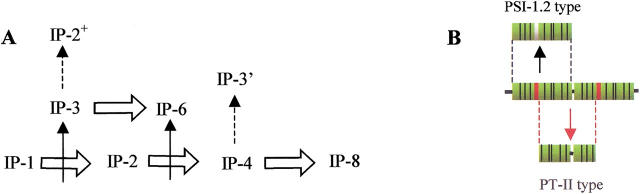
The similarities that exist between the individual IP-repeats of the various PT-II precursors (indicated by gray lines in Fig. 5 ▶) reveal a consistent pattern that suggests that insertion of double repeats may have taken place several times during the evolution of this protein family. For example, an insertion of two repeats between R3 and R4 of the N. alata precursor Q9SQ77 may have given rise to Q40378. In a similar way, the structure of the eight-repeat precursor Q9SDW8 can be deduced as a result of an analogous insertion N-terminal to R2 in Q9SDW7. This supposition has an interesting implication regarding the potential origins of PSI-1.2. The sequence of PSI-1.2 is most similar to R3 of the three-membered precursors IP-3. The similarity leads us to speculate that IP-3 proteins may in fact have evolved by insertion of a double repeat into a putative ancestral protein IP-1 (Fig. 8A ▶), and PSI-1.2 might in fact be the product of the ancestral IP-1 precursor as predicted by Scanlon and associates (1999). This hypothetical precursor might then give rise to PSI-1.2 by cleavage of the signal peptides without further proteolytic processing. In other terms, all members of the PT-II family might have emerged via a combination of duplications, insertions, and subsequent or concomitant modification events (e.g., emergence of proteolytic cleavage sites, sequence losses in the case of IP-3` precursors) from the same ancestral sequence, as schematically shown in Figure 8A ▶. This explanation is speculative and awaits experimental confirmation. It is equally possible that PSI-1.2 is derived via an alternative proteolytic processing scheme from an unknown precursor consisting of several IP-repeats (Fig. 8B ▶). Irrespective of the precise evolutionary route, it appears that the general mechanisms producing the permuted precursor genes and protein products in this family of Solanaceae genes are presumably similar to those invoked for the explanation of circular rearrangements in other species (Heringa and Taylor 1997; Lindqvist and Schneider 1997; Jeltsch 1999).
Fig. 8.
(A) Comparison of PSI-1.2 with the inhibitor precursor IP-repeats and hypothetical evolutionary pathways capable of producing the current proteinase inhibitor precursor genes of the PT-II family. Fat arrow indicates duplication events; thin arrow, insertion event; and dashed arrow, modification events (loss of N- and/or C-terminal sequence). (B) The possible products arising from alternative proteolytic processing pathways are also shown.
The emergence of the processing sites (red in Figs. 5, 7 ▶ ▶) may have had an interesting implication specific for this family of proteins. The ancestral repeat unit must have formed intra-repeat disulfides only, such as those in aPI1 and in PSI-1.2. In other terms, the ancestral repeat must have been an independent folding unit—as indirectly proved by Scanlon et al. (1999). In contrast, those repeats that are cleaved in their central sections form inter-repeat disulfides (e.g., PCI-1), so that the folding units are now formed from segments of two adjacent sequence repeats. The two disulfide topologies are different, as shown in Figure 7 ▶, even though the positions and the spatial orientation of the disulfide bridges are in good agreement. The emergence of an alternative folding ability, followed by the formation of a new disulfide topology, might have been a prerequisite for the new proteolytic processing to appear.
We can conclude that PSI-1.2 represents a novel type of plant serine proteinase inhibitor, which is related to the mature PT-II inhibitors by a circular permutation in the amino acid sequence. It corresponds to a complete IP-repeat unit of the precursor proteins and is derived from a hitherto unideied member of PT-II precursors. In the members of this protein family present in Solanaceae, rearrangements have apparently taken place both at the DNA level (duplications, insertions, loss of flanking sequences) as well as on the protein level (proteolytical processing variants, single-chain versus two-chain analogs). Nevertheless, the resulting protein products all seem to fold into the same overall three-dimensional structure, which may be required for binding to proteinases. The case of PSI-1.2 is apparently the first example in which circularly permuted members of a protein family are expressed within the same organism.
Materials and methods
Materials
Bell pepper (C. annuum) seeds were from the Canning Factory, Pazardjik, Bulgaria. Bovine trypsin (L-1-p-tosylamino-2-phenylethyl chloromethyl ketone-treated), α-chymotrypsin, elastase, and human plasma thrombin were purchased from Sigma; human plasma coagulation factor Xa, from Calbiochem; and pepsin, from Merck. Subtilisin DY was isolated according to Nedkov and Bobatinov (1976). H-D-Phe-Pec-Arg-pNA•AcOH was purchased from Bachem; N-methoxycarbonyl-Nle-Gly-Arg-pNA•AcOH (Chromozym X), from Boehringer Mannheim; Suc-Ala-Ala-Pro-Phe-pNA (Suc-X-pNA) and N-benzoyl-l-arginine-pNA hydrochloride (L-BApNA), from Fluka; Casein Hammarsten Servabacter, from Serva; and p-nitrophenyl-p`-guanidino-benzoate hydrochloride (p-NPGB) and N-trans-cinnamoyl-imidazole (NTCI), from Sigma.
Inhibitor isolation
The isolation of the inhibitors was performed essentially as described previously (Antcheva et al. 1996). The final purification was performed by RP-HPLC on a semi-preparative Delta Pak 300 15RP18 column (250 × 10 mm) using a Millipore Waters HPLC system. The two solvents used were 0.1% (v/v) TFA in water (solvent A) and 0.1% (v/v) trifluoroacetic acid in acetonitrile (solvent B). A linear gradient was used for elution (flow rate, 6.0 mL/min) in which the solvent composition changed from 0% to 20% solvent B in 45 min. The purity of the isolated proteins was checked on an analytical HPLC column (LiChrosphere 5RP18, 4 × 250 mm, Merck); chromatography was performed at a flow rate of 1 mL/min and gradient from 0% to 45% solvent B in 40 min, using the same buffer system as for the preparative runs. The molecular weight of inhibitors were determined by electron spray mass spectrometry on a Perkin Elmer Sciex API 150EX.
Enzyme inhibition assays
Inhibition of serine proteinases (except subtilisin) were followed by monitoring the rate of hydrolysis of p-NA substrates spectrophotometrically at 405 nm using a Ultrospec 3000 UV/Visible spectrophotometer (Pharmacia Biotech) using L-BApNA (trypsin), Suc-Ala-Ala-Pro-Phe-pNA (α-chymotrypsin), Suc-(Ala)3-pNA (elastase), H-D-Phe-Pec-Arg-pNA•AcOH (thrombin), and N-methoxycarbonyl-Nle-Gly-Arg-pNA•AcOH (factor Xa). Precise concentrations of the active enzymes were determined by standard active site titration procedures (Chase and Shaw 1970). In the inhibition assays, stoichiometric concentrations in the10−7 M range of the enzymes (trypsin, α-chymotrypsin) and the inhibitors (PSI-1.1 and PSI-1.2) were used. The residual enzyme activity was measured after 5-min incubation of the enzymes with the inhibitors at 22°C in 50 mM Tris-HCl buffer at pH 7.8), containing 0.02 M CaCl2. The substrates (10−4M) were added, and the reaction was followed at 405 nm (Geiger and Fritz 1984). In the inhibition assays of human plasma thrombin and blood coagulation factor Xa, concentrations in the 10−9 M range of the enzymes, 10−7M of the inhibitors, and 10−4 M of the substrates were used. The measurements were performed in 50 mM Tris/HCl, 100 mM NaCl, 0.5 mg/mL BSA, 20 mM EDTA at pH 7.8 and 22°C (van Dam-Mieras et al. 1984). PSI-1.2 was assayed for possible inhibitory activity against subtilisin DY using 1.2% casein as a substrate. The dissociation constants (Ki) of the enzyme-inhibitor complexes were determined by the method of Cha (1975).
Protein sequence determination
Reduced and pyridylethylated samples (Hampton et al. 1992) were digested separately either with CNBr in 70% HCOOH overnight at room temperature or with trypsin for 3 h at 37°C (E:S, 1:20). The resulting peptides were isolated by narrow-bore RP-HPLC on an analytical Aquapore OD300 column (220 × 2.1 mm, 7-mm particle size, Applied Biosystems) using gradients of acetonitrile in 0.1% (v/v) aqueous TFA. The pyridylethylated protein and the peptides were sequenced by automated Edman degradation using an Applied Biosystems protein sequencer (model 471 A) equipped with a 120A PTH analyzer (Hunkapiller et al. 1983).
Determination of disulfide bridges
The disulfide topology of PSI-1.2B was deduced from mass spectrometric and N-terminal sequencing analysis of enzymatic digests of the protein using a combination of methods described previously (Chagolla-Lopez et al. 1994; Lu et al. 1999). Peptide samples corresponding to 75 mu;g PSI-1.2B were dissolved in 150 mu;L of 0.2M Tris-HCl buffer at pH 7.3 and digested with a mixture of trypsin (3mu;g) and chymotrypsin (3mu;g) for 15 h at 37°C. The mixture was separated by RP-HPLC on a Vydac C18 column (2.1 × 250mm) and subjected to mass spectrometry analysis, and the molecular weights were compared with those predicted for the expected peptides. Selected peaks were collected, liophylised, and subjected to automated Edman degradation. The released amino acid phenylthiohydantoins were detected at 269 and 313 nm (Li and Liang 1999).
Sequence similarity searches
The sequence homology search in the SWISS-PROT, trEMBL (Bairoch and Apweiler 2000), SBASE (Murvai et al. 2000), and PIR (Barker et al. 2000) protein databases for proteinase inhibitor precursor proteins was performed using BLAST (Altschul et al. 1997), FASTA (Pearson and Lipman 1988), and BLITZ (Collins and Coulson 1990). Full-length precursors only were retained. The precursors were dissected into repeats homologous to the PSI-1.2 sequence and clustered using CLUSTALW (Thompson et al. 1994). The phylogenetic tree was built using the PHYLIP package and drawn using DRAWGRAM according to Felsenstein (1981) using a neighbor-joining algorithm (Saitou and Nei 1987).
Modeling
The three-dimensional model of PSI-1.2 was built from the NMR structure of the N. alata aPI1 putative inhibitor. PSI-1.2 and aPI1 were aligned manually, and the first five structures of the NMR ensemble (PDB code 1CE3) were used as templates. The model was built by satisfaction of spatial restraints and energy minimization using MODELLER (Sali and Blundell 1993). The molecules were displayed using MOLMOL (Koradi et al. 1996).
Electronic supplemental material
The multiple sequence alignment of the IP-repeats used in this work is available.
Acknowledgments
The authors thank Prof. Francisco E. Baralle (ICGEB) for his advice and Dr. Kristian Vlahovicek (ICGEB) for his help with sequence comparison.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.21701.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antcheva, N., Patthy, A., Athanasiadis, A., Tchorbanov, B., Zakhariev, S., and Pongor, S. 1996. Primary structure and specificity of a serine proteinase inhibitor from paprika (Capsicum annuum) seeds. Biochim. Biophys. Acta 1298 95–101. [DOI] [PubMed] [Google Scholar]
- Atkinson, A.H., Heath, R.L., Simpson, R.J., Clarke, A.E., and Anderson, M.A. 1993. Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandin, T., van der Does, C., Albert, J.M., Bol, J.F., and Linthorst, H.J. 1995. Structure and induction pattern of a novel proteinase inhibitor class II gene of tobacco. Plant Mol. Biol. 27 1197–1204. [DOI] [PubMed] [Google Scholar]
- Barker, W.C., Garavelli, J.S., Huang, H., McGarvey, P.B., Orcutt, B.C., Srinivasarao, G.Y., Xiao, C., Yeh, L.S., Ledley, R.S., Janda, J.F., Pfeiffer, F., Mewes, H.W., Tsugita, A., and Wu, C. 2000. The protein information resource (PIR). Nucleic Acids Res. 28 41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Schlunegger, M.P., and Eisenberg, D. 1995. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 4 2455–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, W. and Huber, R. 1992. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 204 433–451. [DOI] [PubMed] [Google Scholar]
- Bowles, D. 1998. Signal transduction in the wound response of tomato plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353 1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstadter, J., Rossbach, C., and Theres, K. 1996. Expression of genes for a defensin and a proteinase inhibitor in specific areas of the shoot apex and the developing flower in tomato. Mol. Gen. Genet. 252 146–154. [DOI] [PubMed] [Google Scholar]
- Casaretto, J.A. and Corcuera, L.J. 1995. Plant proteinase inhibitors: A defensive response against insects. Biol. Res. 28 239–249. [PubMed] [Google Scholar]
- Cha, S. 1975. Tight-binding inhibitors-I: Kinetic behavior. Biochem. Pharmacol. 24 2177–2185. [DOI] [PubMed] [Google Scholar]
- Chagolla-Lopez, A., Blanco-Labra, A., Patthy, A., Sanchez, R., and Pongor, S. 1994. A novel α-amylase inhibitor from amaranth (Amaranthus hypocondriacus) seeds. J. Biol. Chem. 269 23675–23780. [PubMed] [Google Scholar]
- Chase, I. and Shaw, E. 1970. Titration of trypsin, plasmin, and thrombin with p-nitrophenyl p`-guanidinobenzoate HCl. Methods Enzymol. 19 20–27. [Google Scholar]
- Choi, Y., Moon, Y., and Lee, J.S. 1990. Primary structure of two proteinase inhibitor II genes closely linked in the potato genome. Korean J. Biochem. 23 214–220. [Google Scholar]
- Choi, D., Park, J., Seo, Y.S., Chun, Y.J., and Kim, W.T. 2000. Structure and stress-related expression of two cDNAs encoding proteinase inhibitor II of Nicotiana glutinosa. Biochim. Biophys. Acta 1492 211–215. [DOI] [PubMed] [Google Scholar]
- Collins, J.F. and Coulson, A.F. 1990. Significance of protein sequence similarities. Methods Enzymol. 183 474–487. [DOI] [PubMed] [Google Scholar]
- Dammann, C., Rojo, E., and Sanchez-Serrano, J.J. 1997. Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J. 11 773–782. [DOI] [PubMed] [Google Scholar]
- Edelman, G.M., Cunningham, B.A., Reeke, G.N., Jr., Becker, J.W., Waxdal, M.J., and Wang, J.L. 1972. The covalent and three-dimensional structure of concanavalin A. Proc. Natl. Acad. Sci. 69 2580–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. 1981. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17 368–376. [DOI] [PubMed] [Google Scholar]
- Gadea, J., Mayda, M.E., Conejero, V., and Vera, P. 1996. Characterization of defense-related genes ectopically expressed in viroid-infected tomato plants. Mol. Plant Microbe Interact. 9 409–415 [DOI] [PubMed] [Google Scholar]
- Geiger, R. and Fritz, H. 1984. Proteinases and their inhibitors. In Methods of enzymatic analysis (ed. H. Bergmeyer), Vol. 5, pp. 99–129. Wiley-VCH, Weinheim.
- Graham, J.S., Pearce, G., Merryweather, J., Titani, K., Ericsson, L., and Ryan, C.A. 1985a. Wound-induced proteinase inhibitors from tomato leaves, I: The cDNA-deduced primary structure of pre-inhibitor I and its post-translational processing. J. Biol. Chem. 260 6555–6560. [PubMed] [Google Scholar]
- ———. 1985b. Wound-induced proteinase inhibitors from tomato leaves, II: The cDNA-deduced primary structure of pre-inhibitor II. J. Biol. Chem. 260 6561–6564. [PubMed] [Google Scholar]
- Greenblatt, H.M., Ryan, C.A., and James, M.N. 1989. Structure of the complex of Streptomyces griseus proteinase B and polypeptide chymotrypsin inhibitor-1 from Russet Burbank potato tubers at 2.1 Å resolution. J. Mol. Biol. 205 201–228. [DOI] [PubMed] [Google Scholar]
- Hampton, B.S., Marshak, D.R., and Burgess, W.H. 1992. Structural and functional characterization of full-length heparin-binding growth associated molecule. Mol. Biol. Cell 3 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, R.L., Barton, P.A., Simpson, R.J., Reid, G.E., Lim, G., and Anderson, M.A. 1995. Characterization of the protease processing sites in a multidomain proteinase inhibitor precursor from Nicotiana alata. Eur. J. Biochem. 230 250–257. [DOI] [PubMed] [Google Scholar]
- Heringa, J. and Taylor, W.R. 1997. Three-dimensional domain duplication, swapping and stealing. Curr. Opin. Struct. Biol. 7 416–421. [DOI] [PubMed] [Google Scholar]
- Hunkapiller, M.W., Hewick, R.M., Dreyer, W.J., and Hood, L.E. 1983. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 91 399–413. [DOI] [PubMed] [Google Scholar]
- Jeltsch, A. 1999. Circular permutations in the molecular evolution of DNA methyltransferases. J. Mol. Evol. 49 161–164. [DOI] [PubMed] [Google Scholar]
- Johnson, R. and Ryan, C.A. 1990. Wound-inducible potato inhibitor II genes: Enhancement of expression by sucrose. Plant Mol. Biol. 14 527–536. [DOI] [PubMed] [Google Scholar]
- Jongsma, M.A., Bakker, P.L., Stiekema, W.J., and Bosch, D. 1995. Phage display of a double-headed proteinase inhibitor: Analysis of the binding domains of potato proteinase inhibitor II. Mol. Breed. 1 181–191. [Google Scholar]
- Keil, M., Sanchez-Serrano, J., Schell, J., and Willmitzer, L. 1986. Primary structure of a proteinase inhibitor II gene from potato (Solanum tuberosum). Nucleic Acids Res. 14 5641–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- Li, F. and Liang, S. 1999. Assignment of the three disulfide bonds of Selenocosmia huwena lectin-I from the venom of spider Selenocosmia huwena. Peptides 20 1027–1034. [DOI] [PubMed] [Google Scholar]
- Lindqvist, Y. and Schneider, G. 1997. Circular permutation of natural protein sequences: Structural evidence. Curr. Opin. Struct. Biol. 7 422–427. [DOI] [PubMed] [Google Scholar]
- Lu, S., Deng, P., Liu, X., Luo, J., Han, R., Gu, X., Liang, S., Wang, X., Li, F., Lozanov, V., Patthy, A., and Pongor, S. 1999. Solution structure of the major α-amylase inhibitor of the crop plant amaranth. J. Biol. Chem. 274 20473–20478. [DOI] [PubMed] [Google Scholar]
- Miller, E.A., Lee, M.C., Atkinson, A.H., and Anderson, M.A. 2000. Ideication of a novel four-domain member of the proteinase inhibitor II family from the stigmas of Nicotiana alata. Plant Mol. Biol. 42 329–333. [DOI] [PubMed] [Google Scholar]
- Murray, C. and Christeller, J.T. 1994. Genomic nucleotide sequence of a proteinase inhibitor II gene. Plant. Physiol. 106 1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murvai, J., Vlahovicek, K., Barta, E., Szepesvári, C., Acatrinei, C., and Pongor, S. 1999. The SBASE protein domain library, release 6.0: A collection of annotated protein sequence segments. Nucleic Acids Res. 27 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murvai, J., Vlahovicek, K., Barta, E., Cataletto, B., and Pongor, S. 2000. The SBASE protein domain library, release 7.0: A collection of annotated protein sequence segments. Nucleic Acids Res. 28 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedkov, P. and Bobatinov, M. 1976. Rapid method for isolation of purified alkaline protease and partial characterization of the obtained product. Bulg. Chem. Commun. 11: 443–450. [Google Scholar]
- Nielsen, K.J., Heath, R.L., Anderson, M.A., and Craik, D.J. 1994. The three-dimensional solution structure by 1H-NMR of a 6-kDa proteinase inhibitor isolated from the stigma of Nicotiana alata. J. Mol. Biol. 242 231–243. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Structures of a series of 6-kDa trypsin inhibitors isolated from the stigma of Nicotiana alata. Biochemistry 34 14304–14311. [DOI] [PubMed] [Google Scholar]
- Park, S. and Thornburg, R.W. 1996. Isolation and characterization of a proteinase inhibitor II gene that is not wound-inducible. Plant Gene Register PGR96-007 (ed. Plant Physiol.).
- Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 85 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P. and Russell, R.B. 1995. Swaposins: Circular permutations within genes encoding saposin homologues. Trends Biochem. Sci. 20 179–180. [DOI] [PubMed] [Google Scholar]
- Richardson, M. 1977. The proteinases inhibitors of plants and microorganisms. Phytochemistry 16 159–169. [Google Scholar]
- Ryan, C.A. 1980. Wound-regulated synthesis and vacuolar compartmentation of proteinase inhibitors in plant leaves. Curr. Top. Cell Regul. 17 1–23. [DOI] [PubMed] [Google Scholar]
- ———. 1989. Proteinase inhibitor gene families: Strategies for transformation to improve plant defenses against herbivores. Bioessays 10 20–24. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sali, A. and Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Sanchez-Serrano, J., Schmidt, R., Schell J., and Willmitzer, L. 1986. Nucleotide sequence of proteinase inhibitor II encoding cDNA of potato (Solanum tuberosum) and its mode of expression. Mol. Gen. Genet. 203 15–20. [Google Scholar]
- Scanlon, M.J., Lee, M.C., Anderson, M.A., and Craik, D.J. 1999. Structure of a putative ancestral protein encoded by a single sequence repeat from a multidomain proteinase inhibitor gene from Nicotiana alata. Structure Fold. Des. 7 793–802. [DOI] [PubMed] [Google Scholar]
- Taylor, B.H., Young, R.J., and Scheuring, C.F. 1993. Induction of a proteinase inhibitor II-class gene by auxin in tomato roots. Plant Mol. Biol. 23 1005–1014. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam-Mieras, M.C.E., Muller, A.D., van Dieijen, G., and Coenraad Hemker, H. 1984. In Methods of enzymatic analysis (ed. H. Bergmeyer), Blood coagulation factors. Vol. 5, pp. 365–394. Wiley-VCH, Weinheim.