Abstract
Spinach 5-phospho-d-ribosyl α-1-diphosphate (PRPP) synthase isozyme 4 was synthesized in Escherichia coli and purified to near homogeneity. The activity of the enzyme is independent of Pi; it is inhibited by ADP in a competitive manner, indicating a lack of an allosteric site; and it accepts ATP, dATP, GTP, CTP, and UTP as diphosphoryl donors. All of these properties are characteristic for class II PRPP synthases. Km values for ATP and ribose 5-phosphate are 77 and 48 μM, respectively. Gel filtration reveals a molecular mass of the native enzyme of ∼110 kD, which is consistent with a homotrimer. Secondary structure prediction shows that spinach PRPP synthase isozyme 4 has a general folding similar to that of Bacillus subtilis class I PRPP synthase, for which the three-dimensional structure has been solved, as the position and extent of helices and β-sheets of the two enzymes are essentially conserved. Amino acid sequence comparison reveals that residues of class I PRPP synthases interacting with allosteric inhibitors are not conserved in class II PRPP synthases. Similarly, residues important for oligomerization of the B. subtilis enzyme show little conservation in the spinach enzyme. In contrast, residues of the active site of B. subtilis PRPP synthase show extensive conservation in spinach PRPP synthase isozyme 4.
Keywords: Kinetics, nucleotide synthesis, oligomerization, phosphoribosylpyrophosphate (PRPP), secondary structure prediction
The enzyme 5-phospho-d-ribosyl α-1-diphosphate (PRPP) synthase (ATP: d-ribose-5-phosphate pyrophosphotransferase, EC 2.7.6.1) is encoded by a PRS gene and catalyzes the reaction: ribose 5-phosphate (Rib-5-P) + ATP → PRPP + AMP (Khorana et al. 1958). PRPP is an important precursor in the biosynthesis of purine, pyrimidine, and pyridine nucleotides and of the amino acids histidine and tryptophan (Hove-Jensen 1988, 1989). Prokaryotes contain one prs gene, whereas generally eukaryotes contain more than one PRS gene. Three genes encoding PRPP synthase have been ideied in mammalian organisms (Tatibana et al. 1995). Results of analysis of gene libraries of the flowering plants Arabidopsis thaliana and spinach (Spinacia oleracea) have ideied five and four PRS genes, respectively. Two of the spinach PRS gene products have been shown, or proposed, to be located in organelles, whereas a third is located in the cytosol (Krath et al. 1999; Krath and Hove-Jensen 1999).
Two classes of PRPP synthases exist. Class I, the "classical" PRPP synthases, is represented by the enzymes from Escherichia coli, Salmonella enterica serovar Typhimurium, Bacillus subtilis, and mammalian organisms. Class II consists of PRPP synthase isozymes 3 and 4 from spinach and A. thaliana and appears to be specific for plants. The distinction between the two classes is based on their enzymatic properties: dependence on Pi for activity, allosteric regulation, and specificity for diphosphoryl donor. Thus, the activity and stability of class I PRPP synthases is dependent on Pi, whereas class II PRPP synthases are independent of Pi. Enzymes of class I are inhibited allosterically by purine ribonucleoside diphosphates, whereas class II enzymes are not. Finally, class I enzymes use ATP or, in some instances, dATP as well as diphosphoryl donors, whereas class II enzymes have much broader specificity, accepting dATP, GTP, CTP, or UTP in addition to ATP. The dramatic differences in enzymatic properties are also reflected in the low amino acid sequence similarity between the two classes (Krath and Hove-Jensen 1999, 2001; Krath et al. 1999).
In the present work, we report the properties of recombinant spinach PRPP synthase isozyme 4 and show that this enzyme belongs to class II. A comparison of the predicted secondary structure of spinach PRPP synthase isozyme 4 with that of B. subtilis PRPP synthase is presented. This comparison, together with amino acid sequence comparison, reveals residues important for catalysis and regulation of PRPP synthases.
Results
Properties of spinach PRPP synthase isozyme 4
Recombinant spinach PRPP synthase isozyme 4 was produced in an E. coli strain in which the endogenous PRPP synthase gene was deleted. The enzyme was purified to near homogeneity as described in Materials and Methods. Amino acid sequencing of the purified enzyme showed the N-terminal amino acid sequence to be Met-Glu-Lys-Pro-Asn-Thr, as expected from the nucleotide sequence.
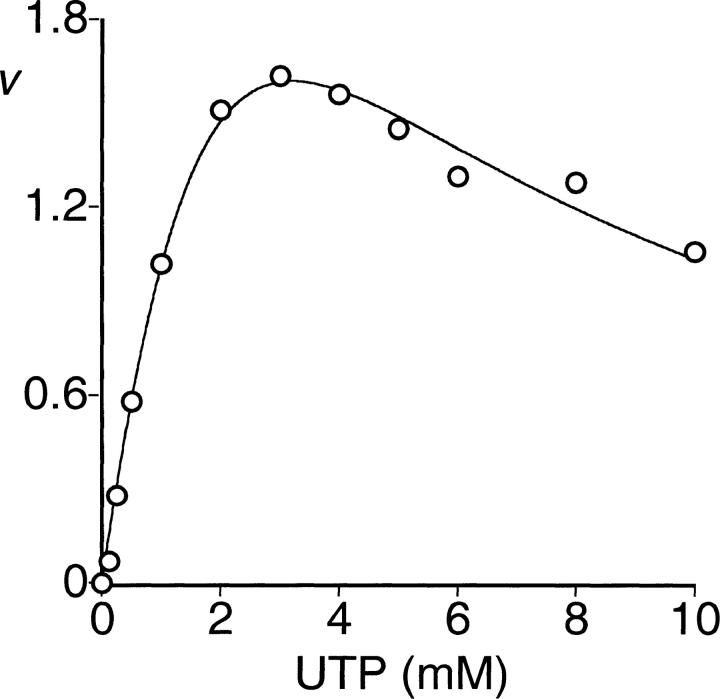
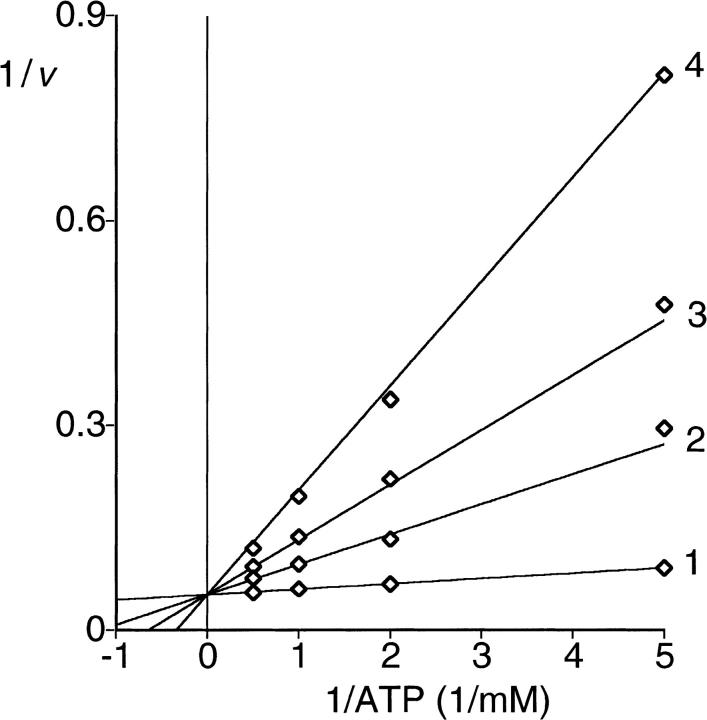
The activity of the enzyme was independent of Pi. Furthermore, the addition of Pi had no effect on the enzyme activity at least up to 80 mM of Pi (data not shown). The enzyme accepted ATP, dATP, GTP, CTP, or UTP as a diphosphoryl donor. With ATP, dATP, GTP, or CTP as substrate, hyperbolic saturation kinetics was observed, whereas with UTP, substrate inhibition was observed (Fig. 1 ▶). Kinetic constants are given in Table 1. Inhibition by ADP was linear competitive with respect to the substrate ATP (Fig. 2 ▶; Table 2). Addition of GDP had no effect with either ATP or GTP as substrate (data not shown).
Fig. 1.
Saturation of PRPP synthase activity by UTP. Activity was determined as described in Materials and Methods. The Rib-5-P concentration was 3.0 mM. The MgCl2 concentration was at least 2 mM in excess of the UTP concentration; v is expressed as μmol/(min × mg of protein). The data were fitted to Equation 4.
Table 1.
Kinetic constants of spinach PRPP synthase isozyme 4 a
| Kmb | Kappc | Vmax | Vapp | |
| Substrate | μM | μmol/(min × mg of protein) | ||
| Rib-5-P | 48 ± 12 | |||
| ATP | 77 ± 10 | 16.2 ± 0.5 | ||
| dATP | 84 ± 5 | 15 ± 0.2 | ||
| GTP | 490 ± 58 | 6.4 ± 0.2 | ||
| CTP | 680 ± 130 | 4.2 ± 0.2 | ||
| UTP | 500 ± 200 | 6.6 ± 2.1 | ||
a Kinetic constants were determined as described in Materials and Methods. Standard errors are those given by the computer program.
b The concentration of ATP was varied from 0.05 to 1.00 mM and the concentration of Rib-5-P was varied from 0.125 to 2.00 mM in the presence of 5.0 mM MgCl2.
c The concentration of dATP, GTP, CTP or UTP was varied from 0.125 to 8.0 mM at 3.0 mM Rib-5-P and MgCl2 in at least 2 mM excess of diphosphoryl donor concentration.
Fig. 2.
Kinetics of inhibition of PRPP synthase activity by ADP. Activity was determined as described in Materials and Methods. Double-reciprocal plots of initial velocity versus ATP at four concentrations of ADP are shown. The concentrations of Rib-5-P and MgCl2 were 1.0 and 5.0 mM, respectively. The concentration of ATP was varied from 0.2 to 2.0 mM in the presence of different concentrations of ADP: 1, 0.0 mM; 2, 0.5 mM; 3, 1.0 mM; 4, 2.0 mM. The lines represent fitting of the data set to Equation 2.
Table 2.
Inhibition of spinach PRPP synthase isozyme 4 a
| Kis | Kii | |||
| Substrate | Inhibitor | Mode of inhibition | μM | |
| Rib-5-P | AMPb | non competitive | 380 ± 63 | 3900 ± 570 |
| Rib-5-P | PRPPc | non competitive | 330 ± 36 | 2200 ± 140 |
| ATP | AMPd | non competitive | 430 ± 62 | 5600 ± 1600 |
| ATP | PRPPe | competitive | 160 ± 8 | |
| ATP | ADPf | competitive | 110 ± 7 |
a Inhibition constants were determined as described in Materials and Methods. Standard errors are those given by the computer program.
b The concentration of Rib-5-P was varied from 0.1 to 2.0 mM in the presence of 0.0 to 2.0 mM AMP, 1.0 mM ATP and 5.0 mM MgCl2.
c The concentration of Rib-5-P was varied from 0.1 to 2.0 mM in the presence of 0.0 to 2.0 mM PRPP, 1.0 mM ATP and 5.0 mM MgCl2.
d The concentration of ATP was varied from 0.05 to 1.0 mM in the presence of 0.0 to 2.0 mM AMP, 1.0 mM Rib-5-P and 5.0 mM MgCl2.
e The concentration of ATP was varied from 0.1 to 2.0 mM in the presence of 0.0 to 2.0 mM PRPP, 1.0 mM Rib-5-P and 5.0 mM MgCl2.
f The concentration of ATP was varied from 0.2 to 2.0 mM in the presence of 0.0 to 2.0 mM ADP, 1.0 mM Rib-5-P and 5.0 mM MgCl2.
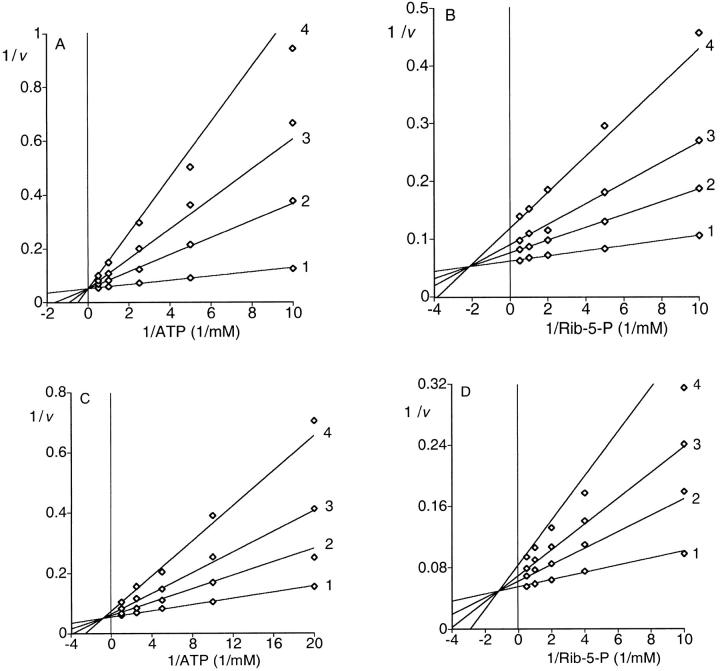
The effect of the ATP concentration on initial velocity was measured at different concentrations of Rib-5-P. Double-reciprocal plots of the data resulted in intersecting lines characteristic of a sequential mechanism (data not shown). The Km and Vmax values for ATP and Rib-5-P are given in Table 1. Product inhibition analysis was used to distinguish between an ordered and a random mechanism. Rib-5-P and ATP were varied at different fixed concentrations of the products PRPP or AMP. Inhibition by PRPP was competitive with respect to ATP and noncompetitive with respect to Rib-5-P. Inhibition by AMP was noncompetitive with respect to both ATP and Rib-5-P (Fig. 3A ▶–D). The mode of inhibition and the calculated inhibition constants are listed in Table 2. These data indicate a steady state ordered Bi Bi mechanism.
Fig. 3.
Product inhibition of PRPP synthase. Activity was determined as described in Materials and Methods. The concentration of MgCl2 was 5.0 mM. (A) Double-reciprocal plot of initial velocity versus ATP at four concentrations of PRPP. The concentration of Rib-5-P was 1.0 mM. The concentration of ATP was varied from 0.1 to 2.0 mM in the presence of different concentrations of PRPP: 1, 0.0 mM; 2, 0.5 mM; 3, 1.0 mM and 4, 2.0 mM. The lines represent fitting of the data set to Equation 2. (B) Double-reciprocal plot of initial velocity versus Rib-5-P at four concentrations of PRPP. The concentration of ATP was 1.0 mM. The concentration of Rib-5-P was varied from 0.1 to 2.0 mM in the presence of different concentrations of PRPP: 1, 0.0 mM; 2, 0.5 mM; 3, 1.0 mM and 4, 2.0 mM. The lines represent fitting of the data set to Equation 3. (C) Double-reciprocal plot of initial velocity versus ATP at four concentrations of AMP. The concentration of Rib-5-P was 1.0 mM. The concentration of ATP was varied from 0.05 to 1.0 mM in the presence of different concentrations of AMP: 1, 0.0 mM; 2, 0.5 mM; 3, 1.0 mM and 4, 2.0 mM. The lines represent fitting of the data set to Equation 3. (D) Double-reciprocal plot of initial velocity versus Rib-5-P at four concentrations of AMP. The concentration of ATP was 2.0 mM. The concentration of Rib-5-P was varied from 0.1 to 2.0 mM in the presence of different concentrations of AMP: 1, 0.0 mM; 2, 0.5 mM; 3, 1.0 mM and 4, 2.0 mM. The lines represent fitting of the data set to Equation 3.
Spinach PRPP synthase isozyme 4 required an Mg2+ concentration in excess of the ATP concentration for maximal activity. Therefore, the enzyme required free Mg2+ in addition to ATP-bound Mg2+, a requirement similar to that observed for other PRPP synthases. With Mn2+ the activity was 25% of that obtained with Mg2+, whereas the enzyme was inactive with Ca2+, Cd2+, Co2+, Cu2+, Fe2+, Ni2+, or Zn2+ (data not shown). Maximal enzyme activity was observed between pH 8.5 and 9.3, determined with Tris/HCl, Na+-K+/phosphate, or glycine/HCl buffer systems. Maximal activity was at 37°C in Tris/HCl at pH 8.5 (data not shown).
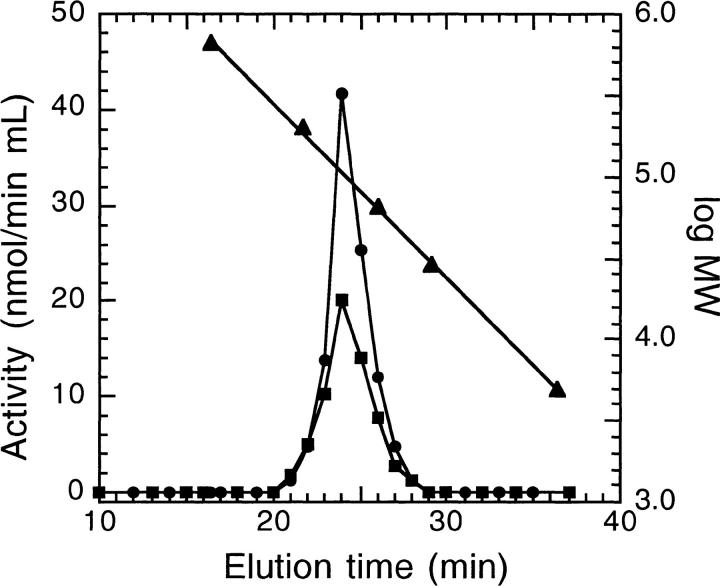
The quaternary structure of spinach PRPP synthase isozyme 4 was analyzed by gel filtration. The enzyme eluted as a single symmetrical peak at a position equivalent to a molecular mass of 115 kD. The molecular mass of the subunit calculated from the deduced amino acid sequence is 35.4 kD, indicating that the enzyme eluted as a trimer. Identical results were obtained in Tris- or phosphate-based buffers. To see if the presence of Mg • ATP had an influence on the molecular architecture of the enzyme, gel filtration was performed in the presence of ATP and Mg2+. Under these conditions, spinach PRPP synthase isozyme 4 eluted with a molecular mass of 106 kD (i.e, again as a trimer; Fig. 4 ▶). Spinach PRPP synthase isozyme 3 eluted at a similar position. The size of the subunit of the latter enzyme is identical to that of spinach PRPP synthase isozyme 4; thus, this enzyme also eluted as a trimer.
Fig. 4.
Determination of the molecular mass of PRPP synthase by gel filtration. Chromatography was performed as described in Materials and Methods with 50 mM Tris/HCl (pH 7.6) containing 2.0 mM ATP, 5.0 mM MgCl2 as the solvent. PRPP synthase activity was determined as described in Materials and Methods. Symbols: •, activity of spinach PRPP synthase isozyme 4; ▪, activity of spinach PRPP synthase isozyme 3; ▴, calibration of the column with the following compounds: thyroglobulin (molecular mass 670 kD), β-amylase (200 kD), bovine serum albumin (66 kD), carbonic anhydrase (29 kD), and ADP (0.4 kD) (Sigma Chemical Co., St. Louis, MO).
Implications of amino acid sequence on enzyme function
The secondary structure of spinach PRPP synthase isozyme 4 was predicted as described in Materials and Methods. Application of a combination of various prediction methods was used to improve the reliability of the prediction (King et al. 2000). The resulting secondary structural elements were aligned with the secondary structure of B. subtilis PRPP synthase (Eriksen et al. 2000). The position and extent of helices and β-sheets of the two enzymes were conserved except for the positions of the secondary structural elements corresponding to β4C, β5C, and α2N (Fig. 5 ▶).
Fig. 5.
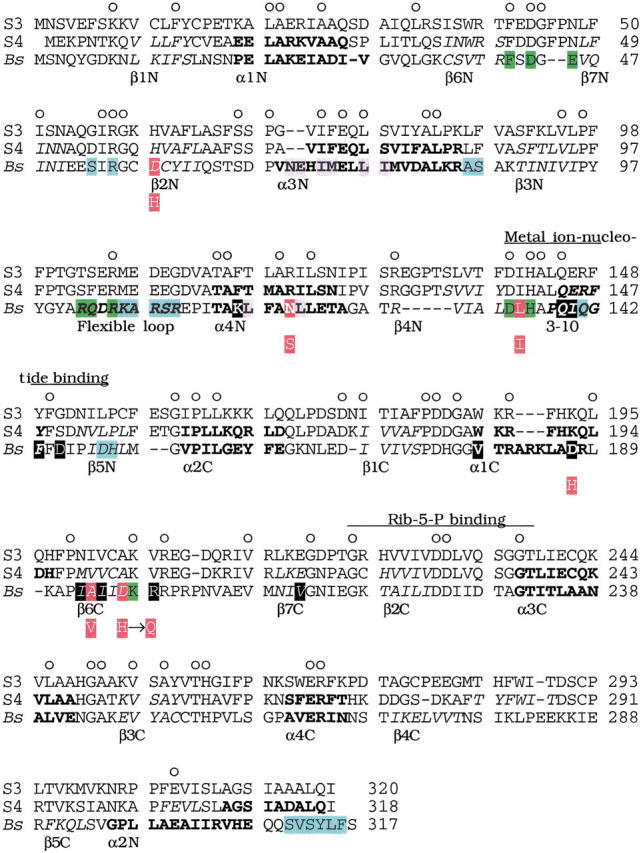
Alignment of amino acid sequences of spinach PRPP synthase isozyme 3 and 4 with B. subtilis PRPP synthase. Abbreviations: S3, spinach PRPP synthase isozyme 3; S4, spinach PRPP synthase isozyme 4 (Krath and Hove-Jensen 1999); Bs, B. subtilis PRPP synthase (Nilsson et al. 1989). Numbers (right) indicate amino acid position. Circles above the spinach PRPP synthase isozyme 3 sequence indicate positions with identical amino acids in the three sequences. Elements of secondary structure of B. subtilis PRPP synthase are indicated by amino acids in italics (β-sheet), bold (α-helix), or italics and bold (a 3–10 helix and a flexible loop) (Eriksen et al. 2000). The designation of each structural element is given below the B. subtilis PRPP synthase amino acid sequence. Similarly, predicted secondary structural elements of spinach PRPP synthase isozyme 4 are indicated in italics and bold as for the B. subtilis sequence. Green shading indicates amino acid residues of the active site of B. subtilis PRPP synthase. These residues were ideied by analysis of the crystal structure obtained as α,β-methylene ADP in complex with the enzyme, by analysis of the E. coli prs-1 specified mutant PRPP synthase altered in Asp129 (homologous with Asp134 of the B. subtilis sequence; Bower et al. 1989), or by analysis of affinity labeling of S. enterica PRPP synthase His131 (homologous with His136 of the B. subtilis sequence; Harlow and Switzer 1990), or of E. coli PRPP synthase Lys194 (homologous with Lys198 of the B. subtilis enzyme; Hilden et al. 1995). Blue shading indicates amino acid residues of the allosteric site of B. subtilis PRPP synthase ideied by analysis of the binding of α,β-methylene ADP in the crystal structure (Eriksen et al. 2000). The amino acid sequence ms ideied previously and designated by divalent metal ion-nucleotide binding (Bower et al. 1989) and by Rib-5-P binding (Hove-Jensen et al. 1986; Willemoës et al. 1996) are indicated by lines above the spinach PRPP synthase isozyme 3 amino acid sequence. Grey shading indicates amino acid residues ideied as involved in the formation of the so-called bent dimer of B. subtilis PRPP synthase. Black shading (white letters) indicates amino acid residues ideied as involved in the formation of the so-called parallel dimer of B. subtilis PRPP synthase (Eriksen et al. 2000). Red shading (white letters) of the B. subtilis PRPP synthase sequence indicates amino acids homologous with amino acids altered in the human PRPP synthase 1 owing to point mutations in the PRSP1 gene. Asp187, which belongs to this group of amino acids, is shaded in black. The amino acid of the mutant variant of human PRPP synthase 1 is shown below the B. subtilis sequence as a white letter with red shading. The actual alterations of human PRPP synthase 1, with the position of the homologous amino acid of B. subtilis PRPP synthase shown in parentheses, were: Asp51 → His (Asp58), Asn113 → Ser (Asn120), Leu128 → Ile (Leu135), Asp182 → His (Asp187), Ala189 → Val (Ala194). In addition, the alteration His192 → Gln was found in human PRPP synthase 1 (Becker et al. 1995). B. subtilis has aspartate at this position.
Eighteen amino acids have been ideied as involved in the binding of α,β-methylene ADP to the allosteric site of B. subtilis PRPP synthase (Fig. 5 ▶). Of these, only Arg55 of B. subtilis PRPP synthase is conserved in spinach PRPP synthase isozyme 3 and 4. Six point mutations of the human PRPS1 gene causing PRPP synthase 1 superactivity have been ideied as responsible for uric acid overproduction and gout (Fig. 5 ▶). The six recombinant enzymes have been characterized (Becker et al. 1995). The substitutions result in enzymes with less sensitivity to allosteric inhibition by ADP compared to normal human PRPP synthase 1. In addition, less Pi is required for activation of the mutant forms of human PRPP synthase 1 compared to the normal enzyme. Two of the mutant forms of human PRPP synthase 1 have the amino acid substitutions, Asp52(58) → His and Leu128(135) → Ile (the homologous positions for the B. subtilis PRPP synthase are shown in parentheses). Spinach PRPP synthase isozyme 3 and 4 have histidine and isoleucine, respectively, at these positions. A third amino acid substitution of human PRPP synthase 1 is Ala189(194) → Val. Similarly, spinach PRPP synthase isozyme 4 has valine at this position (Fig. 5 ▶). None of these six amino acids are among the 18 residues ideied as involved in the binding of α,β-methylene ADP at the allosteric site described above. The mutant forms of human PRPP synthase 1 resemble the spinach PRPP synthase isozyme 3 and 4 by their lack of inhibition by ADP and reduced requirement or independence of Pi for activity.
Seven amino acid residues have been ideied as involved in binding of the 5`-AMP moiety of α,β-methylene ADP to the active site of B. subtilis PRPP synthase. These residues are Phe41, Asp43, Glu45, Arg102, Gln103, Arg105, and His136 (Eriksen et al. 2000). Two other residues have been ideied as involved in ATP binding, Asp134 and Lys198 (Fig. 5 ▶). Six of these nine amino acids were conserved in spinach PRPP synthase isozyme 3 and 4. The conserved residues were Phe41, Asp43, Arg105, Asp134, His136, and Lys198.
Analysis of the crystal structure of B. subtilis PRPP synthase also ideied amino acids involved in the association of subunits. Two types of associations were found (Fig. 5 ▶), with a total of 19 residues involved. Of these, only five were conserved in the spinach sequences (Ile73, Leu77, Leu121, Gln139, and Phe143 of the B. subtilis PRPP synthase sequence). Several of the remaining residues were quite different in the spinach and B. subtilis PRPP synthase sequences. Examples of these were (numbering refers to the B. subtilis sequence; the corresponding amino acid of spinach PRPP synthase isozyme 4 is given in parentheses): Ile78 (Ser), Leu117 (Thr), Ile140 (Glu), Asp145 (Ser), Asp187 (Lys), and Arg199 (Val) (Eriksen et al. 2000).
The amino acids of the Rib-5-P binding m were also conserved in spinach PRPP synthase isozyme 4; as before, the folding in this region was conserved (Hove-Jensen et al. 1986; Willemoës et al. 1996).
Discussion
We have shown that spinach PRPP synthase isozyme 4 belongs to class II PRPP synthases. This conclusion was based on the enzyme's Pi-independent activity, its relaxed specificity towards nucleoside triphosphates as diphosphoryl donor, and its simple competitive inhibition by ADP. Currently, Class II contains one additional well-characterized member, spinach PRPP synthase isozyme 3 (Krath and Hove-Jensen 2001). Despite their similar properties, the two enzymes also differ. Spinach PRPP synthase isozyme 3 shows substrate inhibition with GTP and hyperbolic saturation kinetics with UTP, a reciprocal result of that observed for spinach PRPP synthase isozyme 4. Although both spinach PRPP synthase isozyme 3 and 4 were inhibited in a competitive manner by ADP, only spinach PRPP synthase isozyme 3 was inhibited by GDP.
The identity of the amino acid sequence of the two spinach enzymes with that of the B. subtilis enzyme is 23%, whereas the identity of the amino acid sequence of spinach PRPP synthase isozyme 4 with that of isozyme 3 is 75% (Krath and Hove-Jensen 1999). We observed a remarkable difference between spinach PRPP synthase isozyme 4 and B. subtilis PRPP synthase in conservation of amino acids participating in the binding of nucleotide to the active site versus amino acids participating in the binding of nucleotide to the allosteric site. Thus, 67% (six out of nine) of the residues of the active site of B. subtilis PRPP synthase were conserved in spinach PRPP synthase isozyme 4, whereas only 6% (one out of 18) of the residues of the allosteric site of B. subtilis PRPP synthase were conserved in spinach PRPP synthase isozyme 4. Prominent residues ligating the adenine moiety to the active site of B. subtilis PRPP synthase are Phe41, Asp43, Glu45, and Arg102. The corresponding residues of spinach PRPP synthase isozyme 4 are Phe41, Asp43, Asn47, and Ser102. Other important residues ligate to the phosphate chain of the nucleoside triphosphate; these are Arg105 and His136 of the B. subtilis enzyme corresponding to Arg105 and His141 of spinach PRPP synthase isozyme 4.
Contrary to the conservation of amino acids of the active site, very little, if any, conservation was found among the amino acids ideied as important in forming the allosteric site of B. subtilis PRPP synthase or among residues of human PRPP synthase 1 ideied as involved in modulation of enzyme activity by ribonucleoside diphosphates. The only conserved residue is Arg57 of spinach PRPP synthase isozyme 4 (Arg55 of B. subtilis PRPP synthase). An SO42− ion, which is an analog of Pi, occupied a position similar to the β-phosphorus of α,β-methylene ADP in one complex of crystals of B. subtilis PRPP synthase. It has been postulated that class I PRPP synthases must have either ADP or Pi at this position at any time (Eriksen et al. 2000; Willemoës et al. 2000). This lack of an allosteric binding site seems to be the basis for the Pi-independence of class II PRPP synthases.
The quaternary structure of PRPP synthase is a matter of constant debate. The enzyme from rat liver appears to consist of two different but related subunits (Sonoda et al. 1998). The precise number of subunits in the native enzyme is unknown, but a structure with higher states of aggregation, 16 and 32 subunits, has been suggested for the human enzyme (Becker et al. 1977; Meyer and Becker 1977). The structure of the enzyme from B. subtilis has been defined as a homohexamer by crystallographic analysis. Oligomerization of B. subtilis PRPP synthase occurs primarily by hydrophobic interaction of the α3N with the α4N helix and by interaction of the α1C helix with the β6C strand. Additional interactions occur among 3–10 helices and by hydrogen bonding (Asn70 with Glu71) or salt bridges (Lys116 with Asp145 and Asp187 with Arg199). Although the secondary structure alignment revealed that most of the elements are conserved among B. subtilis PRPP synthase and spinach PRPP synthase isozyme 4, only five of the 19 amino acid residues shown to interact with each other were conserved. In particular, no conservation was found among the residues responsible for stabilization by formation of hydrogen bonds or salt bridges. This low conservation in spinach PRPP synthase isozyme 4 of residues involved in oligomerization of B. subtilis PRPP synthase is consistent with the proposed trimeric, rather than a hexameric, structure of spinach PRPP synthase isozyme 4. The question of oligomerization is particularly important in view of the participation of amino acids from two subunits in the formation of the active site of B. subtilis PRPP synthase. Among the amino acids of B. subtilis PRPP synthase mentioned above, Phe41, Asp43, and Glu45 are donated by one subunit, whereas Arg102, Arg105, and His136 are donated by a second subunit (Eriksen et al. 2000). This arrangement may be retained in spinach PRPP synthase isozyme 4. The allosteric site of B. subtilis PRPP synthase is formed by amino acids from three subunits. It is of interest to note that five of the six amino acid residues ideied as involved in regulating human PRPP synthase 1 are located in or very close to the secondary-structural elements that are involved in subunit interactions. Therefore, it is possible that these amino acids take part in allosteric regulation by altering subunit–subunit interactions. Thus, the difference between spinach PRPP synthase isozyme 4 and B. subtilis PRPP synthase in the oligomeric structures is consistent with differences in allosteric properties and vice versa.
Materials and methods
Bacterial strain and growth medium
The E. coli strain used was HO1088 (araCam araD Δ(lac)U169 trpam malam rpsL relA thi supF deoD gsk-3 udp Δprs-4/F lacIq zzf::Tn10) (Krath and Hove-Jensen 2001). Cells were grown at 37°C in NZY broth (Hove-Jensen and Maigaard 1993) supplemented when necessary with ampicillin (100 mg/L), tetracycline (10 mg/L), isopropyl 1-thio-β-d-galactopyranoside (50 μM) or NAD (40 mg/L).
Manipulation of PRPP synthase isozyme 4-specifying cDNA
A plasmid containing an inducible PRS4 allele was constructed as follows. PCR with DNA of pHO304 as the template (Krath and Hove-Jensen 1999), the oligodeoxyribonucleotides 5`-CAGACG TTAACCCAAGCTTTTAAGAGGAGAAATTAACTATGGAGA AACCCAACACG(So4Hinc) and 5`-CCATGCCATGGTTCATA TCTGAAGAGCGTC (So4Nco) (Hobolth DNA Syntese, Hillerød, Denmark) as primers, Pyrococcus furiosus DNA polymerase (Gibco Brl, Paisley, UK), and the four deoxyribonucleoside triphosphates was performed by standard procedures in a Trio-Thermoblock (Biometra, Göttingen, Germany). The underlined nucleotides of the So4Hinc oligodeoxyribonucleotide indicate the codons specifying the N-terminal end of spinach PRPP synthase isozyme 4 polypeptide. Italicized nucleotides indicate recognition sites of the restriction endonucleases HindIII of So4Hinc or NcoI of So4Nco. The amplified DNA was purified (Qiagen, Hilden, Germany) and digested by restriction endonucleases HindIII and NcoI, ligated by T4 DNA ligase to HindIII, NcoI-digested DNA of pUHE23-2 (H. Bujard, pers. comm.), and transformed to E. coli strain HO1088 followed by a selection for Prs+ (Mandel and Higa 1970; Hove-Jensen 1989). The nucleotide sequence of the resulting plasmid, pBK869, was determined by using an Abi Prism 310 DNA Sequencer as recommended by the supplier (Perkin-Elmer, Foster City, CA); the expected sequence was revealed.
Purification of recombinant spinach PRPP synthase isozyme 4
4 L NZY broth supplemented with ampicillin, tetracycline, and isopropyl 1-thio-β-d-galactopyranoside were inoculated with 200 mL of an overnight culture of strain HO1088/pBK869 and incubated with shaking for 20 h. The following procedures were carried out at 4°C. Cells (16 g of wet weight) were harvested by centrifugation (Sorvall, Wilmington, DE, GS3 rotor) at 6000 rpm for 12 min, resuspended in 0.9% NaCl, collected by centrifugation (Sorvall, SS34 rotor) at 10,000 rpm for 15 min, resuspended in 60 mL of 50 mM Tris/HCl (pH 7.6) and homogenized for 6 × 1 min in a Soniprep ultrasonic disintegrator (model 150; Measuring and Scieic Equipment, London, UK). Debris was removed by centrifugation at 10,000 rpm for 15 min. Streptomycin sulfate (10% (w/v) in 50 mM Tris/HCl at pH 7.6) was added to a final concentration of 1% and the precipitate was removed by centrifugation at 10,000 rpm for 15 min. Protein precipitating between 45% and 65% saturation of (NH4)2SO4 was recovered and dissolved in 50 mM Tris/HCl (pH 7.6). Protein precipitating between 5% and 15% (w/v) polyethyleneglycol 6000 was recovered and dissolved in 50 mM Tris/HCl (pH 7.6) and applied to a column (1.0 × 25 cm) of Dyematrex Gel Blue B (Millipore, Lexington, MA). After washing the column with three volumes of 50 mM Tris/HCl (pH 7.6) protein was eluted with a gradient of KCl (0.0–0.5 M) in 50 mM Tris/HCl (pH 7.6) at a rate of 1 mL/min. Fractions containing PRPP synthase activity were combined, dialyzed against 50 mM Tris/HCl (pH 7.6) and applied to a column (1.0 × 30 cm) of DE52 (Whatman, Maidstone, UK). Protein was eluted with a gradient of NaCl (0.0–0.5 M) in 50 mM Tris/HCl (pH 7.6) at a rate of 1 mL/min. Fractions containing PRPP synthase activity were combined, concentrated in a Centriprep-10 concentrator (Amicon, Bedford, MA), dialyzed against 50 mM Tris/HCl (pH 7.6) containing 50% glycerol, and stored at −20°C. The yield was 15 mg of >95% pure enzyme as evaluated by SDS-PAGE and Coomassie Brilliant Blue-staining (Laemmli 1970). The specific activity was 16.2 μmol/(min × mg of protein). Automated Edman degradation was performed by the Department of Biochemistry and Nutrition, the Technical University of Denmark. Protein content was determined by the bicinchoninic acid procedure with chemicals provided by Pierce Chemical Co. (Rockfort, IL) using bovine serum albumin as the standard (Smith et al. 1985). Activity of purified PRPP synthase was assayed at 37°C as described previously (Krath and Hove-Jensen 2001). Fractions from column chromatography were assayed for PRPP synthase activity by a one-point assay.
Gel filtration
Fast protein liquid chromatography was performed using a Bio Logic system with UV detection at 280 nm at room temperature (Bio-Rad, Hercules, CA). Enzyme (300 μg) was applied to a gel filtration column (1.0 × 30 cm) of Superose 12, which has a lower cut off limit of 5 kD (Pharmacia, Uppsala, Sweden). Protein was eluted isocratically in 50 mM Tris/HCl (pH 7.6) in 50 mM Na+-K+/phosphate buffer (pH 7.6) or in 50 mM Tris/HCl (pH 7.6) containing 2.0 mM ATP, 5.0 mM MgCl2; in either case the rate was 0.5 mL/min.
Analysis of kinetic data
Initial velocities are average values of determinations at three enzyme concentrations. Results of initial velocity experiments and of product inhibition studies were fitted to the following equations using the UltraFit program (version 3.01; Biosoft, Cambridge, UK). Equation 1 is the rate equation for a sequential Bi Bi mechanism. For competitive, noncompetitive and substrate inhibition the initial velocities were fitted to Equations 2, 3, and 4, respectively. Equation 5 is the Michaelis-Menten equation for hyperbolic substrate saturation kinetics (Cleland 1963).
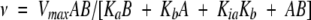 |
(1) |
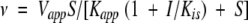 |
(2) |
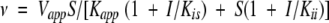 |
(3) |
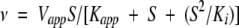 |
(4) |
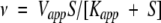 |
(5) |
in which v is the initial velocity, Vmax is the maximal velocity, Ka and Kb are the Michaelis-Menten constants for the varied substrate A or B, respectively, and Kia is the dissociation constant for substrate A. Vapp is the apparent maximal velocity, Kapp is the apparent Michaelis-Menten constant for the varied substrate S, Kii and Kis are the inhibitor constants for inhibitor I obtained from the effect on intercept and slopes, respectively, and Ki is the inhibitor constant for the substrate S (Cleland 1963).
Computer-assisted analysis of amino acid sequences
Amino acid sequences were aligned with the ClustalX program (Thompson et al. 1997). Secondary structure was predicted with PSI-PRED (Jones 1999), JPRED (Cuff et al. 1998), SAMT99 (Karplus et al. 1998), PHD (Rost and Sander 1993), Prof (Ouali and King 2000), and DSSP algorithms (Kabsch and Sander 1983).
Acknowledgments
Financial support was obtained from the Danish Natural Science Research Council and from the Center for Enzyme Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.11801.
References
- Becker, M.A., Meyer, L.J., Huisman, W.H., Lazar, C., and Adams, W.B. 1977. Human erythrocyte phosphoribosylpyrophosphate synthetase. Subunit analysis and states of subunit association. J. Biol. Chem. 252 3911–3918. [PubMed] [Google Scholar]
- Becker, M.A., Smith, P.R., Taylor, W., Mustafi, R., and Switzer, R.L. 1995. The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J. Clin. Invest. 96 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower, S.G., Harlow, K.W., Switzer, R.L., and Hove-Jensen, B. 1989. Characterization of the Escherichia coli prsA1-encoded mutant phosphoribosylpyrophosphate synthetase ideies a divalent cation-nucleotide binding site. J. Biol. Chem. 264 10287–10291. [PubMed] [Google Scholar]
- Cleland, W.W. 1963. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. II. Inhibition: Nomenclature and theory. Biochim. Biophys. Acta 67 173–187. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M., and Barton, G.J. 1998. JPred: A consensus secondary structure prediction server. Bioinformatics 14 892–893. [DOI] [PubMed] [Google Scholar]
- Eriksen, T.A., Kadziola, A., Bentsen, A.-K., Harlow, K.W., and Larsen, S. 2000. Structural basis for the function of Bacillus subtilis phosphoribosylpyrophosphate synthetase. Nat. Struct. Biol. 7 303–308. [DOI] [PubMed] [Google Scholar]
- Harlow, K.W. and Switzer, R. L. 1990. Chemical modification of Salmonella typhimurium phosphoribosylpyrophosphate synthetase with 5`-(p-fluorosulfonylbenzoyl)adenosine. Ideication of an active site histidine. J. Biol. Chem. 265 5487–5493. [PubMed] [Google Scholar]
- Hilden, I., Hove-Jensen, B., and Harlow, K.W. 1995. Inactivation of Escherichia coli phosphoribosylpyrophosphate synthetase by the 2`,3`-dialdehyde derivative of ATP. Ideication of active site lysines. J. Biol. Chem. 270 20730–20736. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen, B., Harlow, K.W., King, C.J., and Switzer, R.L. 1986. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J. Biol. Chem. 261 6765–6771. [PubMed] [Google Scholar]
- Hove-Jensen, B. 1988. Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J. Bacteriol. 170 1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hove-Jensen, B. 1989. Phosphoribosylpyrophosphate (PRPP)-less mutants of Escherichia coli. Mol. Microbiol. 3 1487–1492. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen, B. and Maigaard, M. 1993. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J. Bacteriol. 174 6852–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Karplus, K., Barrett, C., and Hughey, R. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14 846–856. [DOI] [PubMed] [Google Scholar]
- Khorana, H.G., Fernandes, J.F., and Kornberg, A. 1958. Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J. Biol. Chem. 230 941–948. [PubMed] [Google Scholar]
- King, R.D., Ouali, M., Strong, A.T., Aly, A., Elmaghraby, A., Kantardzic, M., and Page, D. 2000. Is it better to combine predictions? Protein Eng. 13 15–19. [DOI] [PubMed] [Google Scholar]
- Krath, B.N. and Hove-Jensen, B. 1999. Organellar and cytosolic localization of four phosphoribosyl diphosphate synthase isozymes in spinach. Plant Physiol. 119 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krath, B.N., Eriksen, T.A., Poulsen, T.S., and Hove-Jensen, B. 1999. Cloning and sequencing of cDNAs specifying a novel class of phosphoribosyl diphosphate synthase in Arabidopsis thaliana. Biochim. Biophys. Acta 1430 403–408. [DOI] [PubMed] [Google Scholar]
- Krath, B.N. and Hove-Jensen, B. 2001. Class II recombinant phosphoribosyl diphosphate synthase from spinach: Phosphate-independence and phosphoryl donor specificity. J. Biol. Chem. 276 17851–17856. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Mandel, M. and Higa, A. 1970. Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53 159–162. [DOI] [PubMed] [Google Scholar]
- Meyer, L.J. and Becker, M.A. 1977. Human erythrocyte phosphoribosylpyrophosphate synthetase. Dependence of activity on state of subunit association. J. Biol. Chem. 252 3919–3925. [PubMed] [Google Scholar]
- Nilsson, D., Hove-Jensen, B., and Arnvig, K. 1989. Primary structure of the tms and prs genes of Bacillus subtilis. Mol. Gen. Genet. 218 565–571. [DOI] [PubMed] [Google Scholar]
- Ouali, M. and King, R.D. 2000. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9 1162–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B. and Sander, C. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232 584–599. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 76–85. [DOI] [PubMed] [Google Scholar]
- Sonoda, T., Ishiharu, T., Ishijima, S., Kita, K., Ahmad, I., and Tatibana, M. 1998. Rat liver phosphoribosylpyrophosphate synthetase is activated by free Mg2+ in a manner that overcomes its inhibition by nucleotides. Biochim. Biophys. Acta 1387 32–40. [DOI] [PubMed] [Google Scholar]
- Tatibana, M., Kita, K., Taira, M., Ishijima, S., Sonoda, T., Ishizuka, T., Iizasa, T., and Ahmad, I. 1995. Mammalian phosphoribosyl-pyrophosphate synthetase. Adv. Enzyme Regul. 35 229–249. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemoës, M., Nilsson, D., and Hove-Jensen, B. 1996. Effects of mutagenesis of aspartic acid residues in the putative phosphoribosyl diphosphate binding site of Escherichia coli phosphoribosyl diphosphate synthetase on metal ion specificity and ribose 5-phosphate binding. Biochemistry 35 8181–8186. [DOI] [PubMed] [Google Scholar]
- Willemoës, M., Hove-Jensen, B., and Larsen, S. 2000. Steady state kinetic model for the binding of substrates and allosteric effectors to Escherichia coli phosphoribosyl-diphosphate synthase. J. Biol. Chem. 275 35408–35412. [DOI] [PubMed] [Google Scholar]