Abstract
The energetic parameters for the folding of small globular proteins can be very different if derived from guanidine hydrochloride (GdnHCl) or urea denaturation experiments. A study of the equilibrium and kinetics of the refolding of wild-type (wt) cytochrome c551 (cyt c551) from Pseudomonas aeruginosa and of two site-directed mutants (E70Q and E70V) shows that the nonionic nature of urea reveals the role of a salt bridge between residues E70 and K10 on the transition state, which is otherwise completely masked in GdnHCl experiments. Mixed denaturant refolding experiments allow us to conclude that the masking effect of GdnHCl is complete at fairly low GdnHCl concentrations (≅0.1 M). The fact that potassium chloride is unable to reproduce this quenching effect, together with the results obtained on the mutants, suggests a specific binding of the Gdn+ cation, which involves the E70–K10 ion pair in wt cyt c551.We propose, therefore, a simple kinetic test to obtain a mechanistic interpretation of nonlinear dependences of ΔGw on GdnHCl concentration on the basis of kinetic refolding experiments in the presence of both denaturants.
Keywords: Protein folding, mutants, stability, kinetics, denaturants, guanidinium binding
Protein folding studies rely on experiments in which the equilibrium between the denatured (D) and the native (N) states of a protein is altered by changing solution conditions. Urea and guanidine hydrochloride (GdnHCl) are the most extensively used chemical denaturants, and they presumably exert their effect by interacting with peptide bonds (Creighton 1993; Mayo and Baldwin 1993). However, in some cases, estimates of protein stability derived from urea or GdnHCl experimental data may be considerably different (Pace 1990), and it has been suggested that these differences depend on the electrostatic and hydrophobic makeup of the protein (Monera et al. 1994).
The kinetics of folding is studied by rapidly mixing the protein, in the presence of a chemical denaturant, with a large volume of buffer. However, because the dilution factor is generally no more than 10-fold, denaturant concentrations below 0.2–0.3 M have rarely been explored. One of the aims of the present study is to highlight the observation that the specific role of some electrostatic interactions in the folding mechanism may become evident only at very low GdnHCl concentrations (below ≅0.1 M) (Shortle et al. 1989; Ahmad et al. 1994).
In the course of our studies on the folding mechanism of cytochrome c551 (cyt c551) from Pseudomonas aeruginosa (Travaglini-Allocatelli et al. 1999; Gianni et al. 2001), we found that the destabilizing effect of two site-directed mutants on the salt bridge interaction involving E70 and K10 at the interface between the N- and C-terminal helices (Fig. 1 ▶) is completely masked when GdnHCl, instead of urea, is used as a denaturant. We present evidence to support the contention (Monera et al. 1993; Gupta et al. 1996) that the overall stability of a protein may be better estimated by using urea rather than GdnHCl.
Fig. 1.
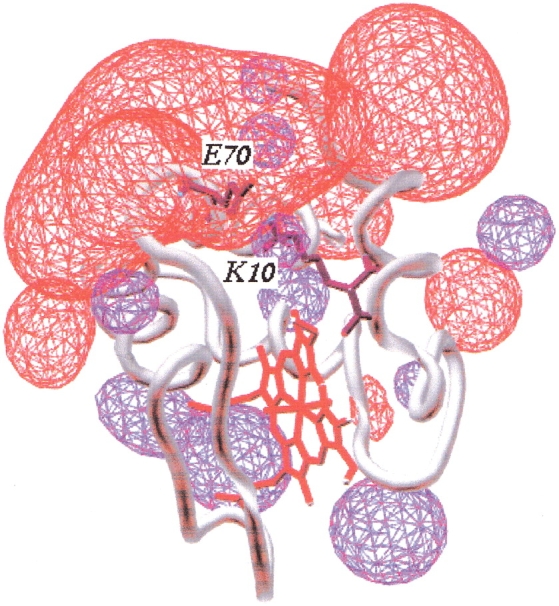
Schematic model of the three-dimensional structure of cytochrome c551. Electrostatic potential is calculated at pH 7.0 and ionic strength at 0.1 M. The red and blue contours represent the negative and the positive electric fields, respectively. The electrostatic potential was calculated by using gromos96 in the SPDBV package software. It is evident that the E70–K10 salt bridge (represented in black) is located at the bottom of an extensive negative field.
The equilibrium denaturation curves of wild-type (wt) cyt c551 as a function of GdnHCl and urea compared with two site-directed mutants (E70Q and E70V) are shown in Figure 2 ▶ (panel A and panel B, respectively). The curve profiles show that the wt protein and the two mutants have approximately the same [GdnHCl]1/2 and meq values (Table 1); this observation would suggest that the E70–K10 salt bridge has no significant contribution to the stability of the native state of cyt c551. When urea is used as a denaturant, the mutants are less stable than wt cyt c551, although they show similar urea dependences (see Table 1). This observation leads us to conclude that the ionic nature of GdnHCl effectively masks the energetic contribution of the E70–K10 salt bridge to the stabilization of the native state.
Fig. 2.
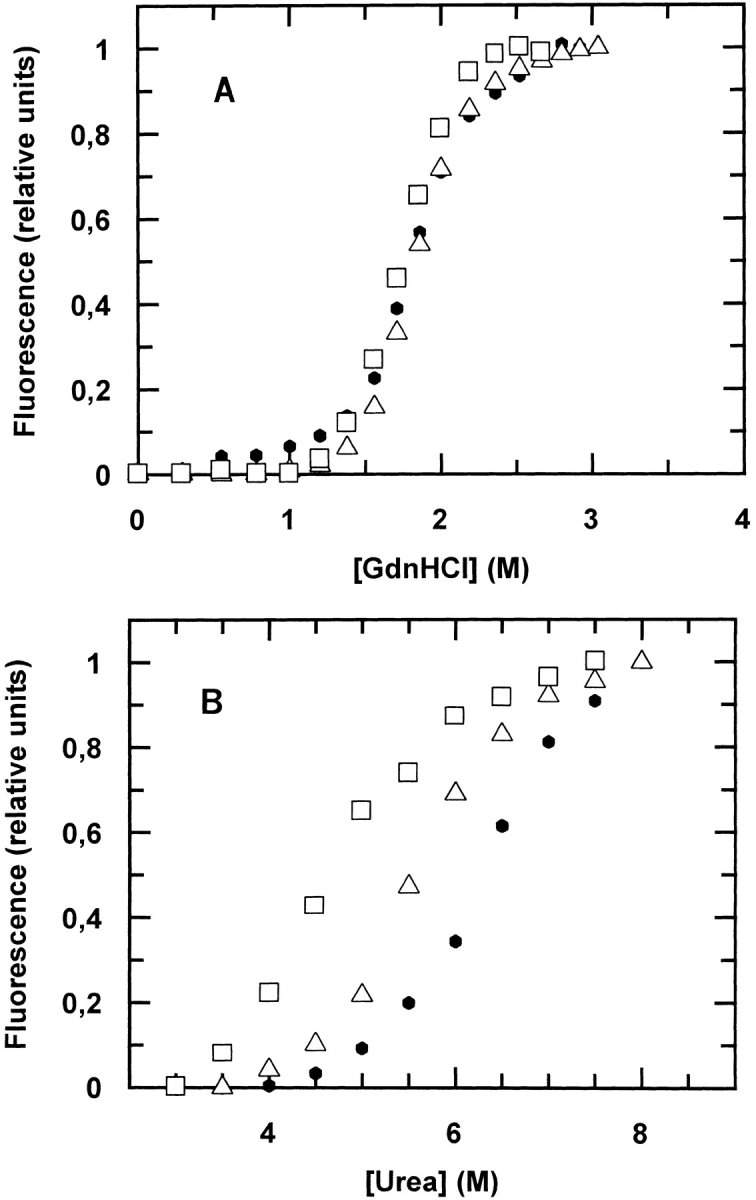
Equilibrium unfolding of wild-type cytochrome c551 (filled circles) and two site-directed mutants, E70Q (open triangles) and E70V (open squares), at pH 7.0 and 10°C. The two panels refer to experiments with two different chaotropic denaturants: guanidine hydrochloride (GdnHCl; panel A) and urea (panel B). It is evident that these three proteins unfold at approximately the same GdnHCl concentration but at different urea concentrations (see Table 1). This observation may be accounted for if GdnHCl masks the energetic contribution of the E70–K10 salt bridge to the stability of the native state.
Table 1.
Equilibrium parameters for folding of wt and mutants of cyt c551 (pH 7.0, 10°C)
| ΔGw (urea) (kcal mol−1) | ΔGw (GdnHCl) (kcal mol−1) | meq (urea) (kcal mol−1 M−1) | meq (GdnHCl) (kcal mol−1 M−1) | [urea]1/2 (M) | [GdnHCl]1/2 (M) | |
| wt | −7.1 | −6.1 | −1.1 | −3.3 | 6.4 | 1.8 |
| E70Q | −6.2 | −6.3 | −1.1 | −3.4 | 5.7 | 1.8 |
| E70V | −5.1 | −5.8 | −1.2 | −3.3 | 4.3 | 1.8 |
We subsequently attempted to mimic a quenching effect of GdnHCl by performing urea denaturation experiments in the presence of potassium chloride (KCl), as proposed by Monera et al. (1993). These experiments (data not shown) showed a progressive destabilization of cyt c551 with increasing salt concentration, up to [KCl] = 0.5 M. However, increasing the salt concentration from 0.5 M to 3 M led to a recovery of the [urea]1/2 observed in the absence of salt. Moreover, the same behavior was observed for the wt cyt c551 and the two mutants (data not shown). These results clearly suggest that KCl is not as effective as GdnHCl in exerting specific electrostatic quenching (Monera et al. 1993; Gupta et al. 1996).
When the refolding rate constant of cyt c551, obtained in stopped-flow experiments, was plotted as a function of denaturant concentration (Chevron plot), we observed that the kFwt extrapolated to zero GdnHCl (kFwt[GdnHCl] ∼300 s−1) is more then 10-fold lower than that calculated from urea experiments (kFwt[urea] ∼4000 s−1) (Travaglini-Allocatelli et al. 1999; Gianni et al. 2001). Moreover, the ratio (kFwt/kFmut) calculated from urea experiments is ∼6 for E70Q and ∼14 for E70V, whereas that calculated from GdnHCl experiments is ∼2 in both cases.
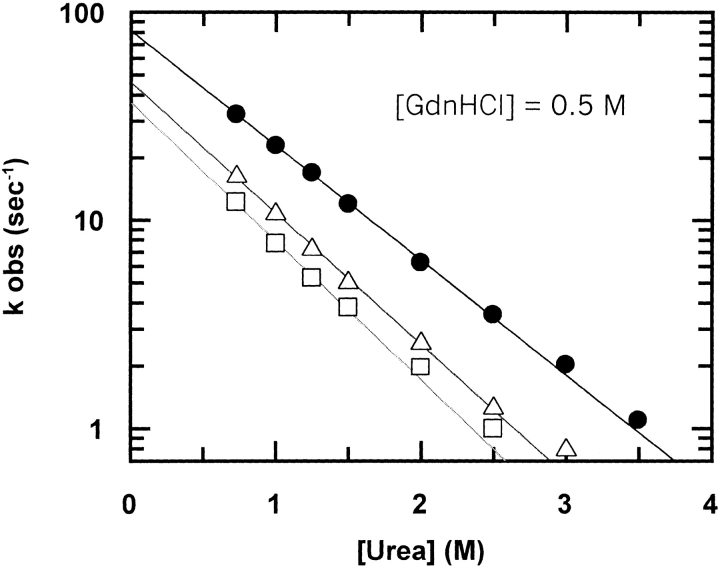
To analyze the masking effect of GdnHCl outlined above and to study its mechanistic and kinetic relevance, we performed urea refolding experiments in the presence of a constant concentration of GdnHCl (Johnson and Fersht 1995; Gupta et al. 1996). Figure 3 ▶ shows the dependence on urea of the refolding rates for wt cyt c551 and both mutants at [GdnHCl] = 0.5 M. Surprisingly, the ratio kFwt/kFmut is ∼2 for both mutants, that is, similar to that calculated from GdnHCl refolding experiments and different from those obtained in urea (Table 2). It is important to note that the calculated mU and mF values are the same as those calculated from "classical" urea experiments (without added GdnHCl; data not shown), confirming that mixing a constant low concentration of a chaotropic denaturant with another does not alter the refolding and unfolding mechanism.
Fig. 3.
Refolding kinetics of wild-type cytochrome c551 (filled circles), E70Q (open triangles), and E70V (open squares) mutants measured by fluorescence stopped-flow (Applied Photophysics SX18, Letherhead, UK) urea dilution experiments at pH 7.0 (phosphate buffer 50 mM) and 10°C in the presence of [GdnHCl] = 0.5 M. The lines are the best fit to the experimental data by using the equation
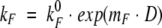 |
The ratio kFwt/kFmut obtained from the extrapolated values is approximately the same as that obtained in "classical" GdnHCl refolding experiments (see Table 2). This observation suggests that the E70–K10 salt bridge is already broken at 0.5 M GdnHCl.
Table 2.
Kinetic folding parameters of wt and mutants of cyt c551, measured by stopped-flow from urea dilution experiments (pH 7.0, 10°C)
| kF (sec−1) | mF (M−1) | kFwt/kFmut | kF (sec−1)a | mF (M−1)a | kFwt/kFmuta | |
| wt | 3900 | 1.4 | — | 81 | 1.3 | — |
| E70Q | 640 | 1.3 | 6.1 | 46 | 1.4 | 1.8 |
| E70V | 280 | 1.5 | 14 | 37 | 1.5 | 2.2 |
a In the presence of GdnHCl 0.5 M.
The fact that, for both mutants, the values of kFwt/kFmut obtained from urea experiments in the presence of [GdnHCl] = 0.5 M are very similar to those obtained from dilution of GdnHCl alone strongly suggests that the E70–K10 salt bridge is fully broken at 0.5 M GdnHCl.
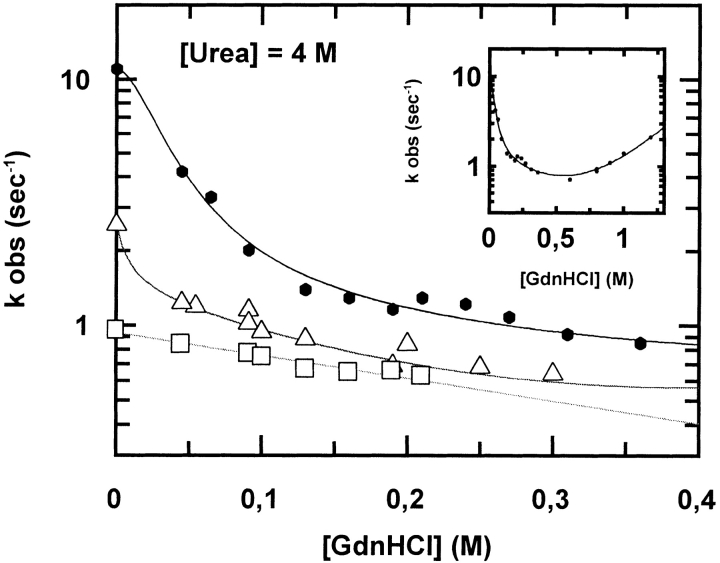
To further test this hypothesis and to investigate the folding pathway of these proteins below 0.5 M GdnHCl, we decided to measure the refolding rate constants of the wt and mutants of cyt c551 as a function of [GdnHCl] in the presence of 4 M urea. At the latter concentration, the three proteins are still native, as judged from far ultraviolet-circular dichroism and fluorescence spectroscopy, but destabilized. It is evident from the results reported in Figure 4 ▶ that the refolding branch of the wt protein has an atypical and surprising "upper curvature"; this curvature is seen only at very low [GdnHCl] (at or below 0.1 M); it is still detectable, though less evident, in the E70Q mutant, but it is absent in the E70V mutant.
Fig. 4.
Refolding kinetics of wt cyt c551 (closed circles), E70Q (open triangles), and E70V (open squares) mutants measured by fluorescence stopped-flow GdnHCl dilution experiments at pH 7.0 and 10°C in the presence of [urea] = 4 M. Inset: complete Chevron plot of wt cyt c551. For wt cyt c551, the refolding branch has an atypical "upper curvature" that becomes evident at very low [GdnHCl]. This curvature is possibly still present for the E70Q mutant, but is not seen in the E70V mutant.
This atypical Chevron plot has been fitted by using the equation
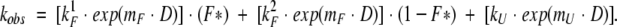 |
Because we have shown that the E70–K10 salt bridge is fully formed in the refolding transition state (Gianni et al. 2001), F* represents the fractional population that refolds (with rate constant kF1) via a transition state stabilized by this ionic interaction, and kF2 is the refolding rate constant in the absence of the E70–K10 salt bridge interaction. mF and mU are the denaturant dependencies of the refolding and unfolding rate constants, respectively. D is the denaturant concentration.
Following a binding model
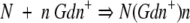 |
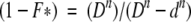 |
in which D represents the [Gdn+], N is native cyt c551, n the number of Gdn+ cations that bind to the protein, and d the concentration of denaturant at which F* is half populated. Because we have shown that mutation of residue E70 does not affect the dependence of the observed refolding and unfolding rate constants on denaturant concentration (Gianni et al. 2001), the equation postulates that the two alternative pathways have the same urea dependence (mF).
Because we have shown that the E70–K10 salt bridge is fully formed in the refolding transition state (Gianni et al. 2001), we suggest that this unusual "upper-curved" Chevron behavior is linked to the breakage of this salt bridge, which occurs at [GdnHCl] ≅0.1 M; it can be kinetically detected in refolding experiments only below this critical concentration, resulting in an increase of the apparent refolding rate. This behavior may be described with a model involving a two-state transition, as detailed in the legend to Figure 4 ▶. This hypothesis is consistent with the observation that, in these experiments (i.e., at 4 M urea), kFwt/kFmut values extrapolated to zero [GdnHCl] are the same as those calculated from "classical" urea dilution experiments. It should also be noted that the apparent kFwt/kFmut in the right limb of the refolding branch (0.1 M < [GdnHCl] < 0.4 M) is similar to that observed in classical GdnHCl experiments, in which, we presume, the refolding reaction leads to a native-like state (N*) that does not involve the formation of an E70–K10 salt bridge. We believe that the persistence of some curvature in the E70Q mutant is due to the formation of a hydrogen bond between the Q70 and K10 side chains, which is also broken by GdnHCl > 0.1 M; this hydrogen bonding capability is absent in the E70V mutant and, as expected, its refolding branch displays the usual linear profile in the refolding limb of the Chevron plot.
The results of our study are significant in at least three ways. First, we have unequivocal evidence that GdnHCl denaturation experiments may fail to detect the thermodynamic and kinetic contribution of a specific salt bridge in cyt c551 and possibly other proteins, in agreement with Monera et al. (1994) for coiled-coil synthetic peptides. This masking effect seems to be complete at fairly low [GdnHCl] (≅0.1 M), that is, over a range that is rarely explored in classical stopped-flow dilution experiments. On the other hand, because the equilibrium estimate of ΔGw has relatively high intrinsic errors that depend on the distance of the transition region from the extrapolated value at zero denaturant (Pace 1990), these masking effects are very difficult to detect from differences in the ΔGw values obtained by urea or GdnHCl.
The fact that KCl cannot reproduce the masking effect seen with GdnHCl suggests some specificity in the effect of the Gdn+ cation in cyt c551 (Wu and Wang 1999), with binding to the E70–K10 ion pair. Inspection of the calculated electrostatic potential of wt cyt c551, reported in Figure 1 ▶, shows a large negative electric field around the N- and C-terminal helices where the E70–K10 salt bridge is located. We propose that the Gdn+ ion may enter this interface between the two helices and displace the K10 side chain from its ionic partner E70; this effect takes place at lower concentration than the solvation of hydrophobic groups that leads to complete unfolding.
Finally, to the best of our knowledge, this is the first kinetic test that gives a mechanistic explanation of nonlinear dependences of ΔGw on GdnHCl concentration. We suggest that this test may be helpful in elucidating complex denaturant dependences already noticed at equilibrium for other proteins such as metmyoglobin (Gupta et al. 1996), ribonuclease A, lysozyme, and mammalian cytochrome c (Ahmad et al. 1994). In this context, this kinetic test may have general significance in the interpretation of refolding experiments performed with different denaturants.
Acknowledgments
This work was partially supported by the MURST of Italy (PRIN 1999, Dinamica strutturale di emoproteine). We express our thanks to Prof. Francesca Cutruzzolà (Rome, Italy) for stimulating discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.5101
References
- Ahmad, F., Taneja, S., Yadav, S., and Haque, S.E. 1994. A new method for testing the functional dependence of unfolding free energy changes on denaturant concentration. J. Biochem. (Tokyo) 115 322–327. [DOI] [PubMed] [Google Scholar]
- Creighton, T.E. 1993. Proteins: Structures and molecular properties, 2nd ed. W.H. Freeman, New York.
- Gianni, S., Travaglini-Allocatelli, C., Cutruzzolà, F., Bigotti, M.G., and Brunori, M. 2001. Snapshots of protein folding. A study on the multiple transition state pathway of cytochrome c551 from Pseudomonas aeruginosa. J. Mol. Biol. 309 1177–1187. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Yadav, S., and Ahmad, F. 1996. Protein stability: Urea-induced versus guanidine-induced unfolding of metmyoglobin. Biochemistry 36 11925–11930. [DOI] [PubMed] [Google Scholar]
- Johnson, C.M. and Fersht, A.R. 1995. Protein stability as a function of denaturant concentration: Thermal stability of barnase in the presence of urea. Biochemistry 34 6795–6804. [DOI] [PubMed] [Google Scholar]
- Mayo, S.L. and Baldwin, R.L. 1993. Guanidinium chloride induction of partial unfolding in amide proton exchange in RNase A. Science 262 873–876. [DOI] [PubMed] [Google Scholar]
- Monera, O.D., Zhou, N.E., Kay, C.M., and Hodges, R.S. 1993. Comparison of antiparallel and parallel two-stranded α-helical chains in two stranded α-helical coiled-coils: Design, synthesis, and characterization. J. Biol. Chem. 268 19218–19227. [PubMed] [Google Scholar]
- Monera, O.D., Kay, C.M., and Hodges, R.S. 1994. Protein denaturation with guanidine hydrocloride or urea provides a different estimate of stability depending on the contribution of electrostatic interactions. Protein Sci. 3 1984–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, C.N. 1990. Measuring and increasing protein stability. Trends Biotechnol. 8 93–98. [DOI] [PubMed] [Google Scholar]
- Shortle, D., Meeker, A.K., and Gerring, S.L. 1989. Effects of denaturants at low concentrations on the reversible denaturation of staphylococcal nuclease. Arch. Biochem. Biophys. 272 103–113. [DOI] [PubMed] [Google Scholar]
- Travaglini-Allocatelli, C., CutruzzolÀ, F., Bigotti, M.G., Staniforth, R.A., and Brunori, M. 1999. Folding mechanism of Pseudomonas aeruginosa cytochrome c551: Role of electrostatic interactions on the hydrophobic collapse and transition state properties. J. Mol. Biol. 289 1459–1467. [DOI] [PubMed] [Google Scholar]
- Wu, J.W. and Wang, Z.X. 1999. New evidence for the denaturant binding model. Protein Sci. 10 2090–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]