Abstract
Haspin (haploid germ cell–specific nuclear protein kinase) is reported to be a serine/threonine kinase that may play a role in cell-cycle cessation and differentiation of haploid germ cells. In addition, Haspin mRNA can be detected in diploid cell lines and tissues. Here, Haspin-like proteins are identified in several major eukaryotic phyla—including yeasts, plants, flies, fish, and mammals—and an extended group in Caenorhabditis elegans. The Haspin-like proteins have a complete but divergent eukaryotic protein kinase domain sequence. Although clearly related to one another and to other eukaryotic protein kinases, the Haspin-related proteins lack conservation of a subset of residues that are almost invariant in known kinases and possess distinctive inserted regions. In fact, phylogenetic analysis indicates that the Haspin-like proteins form a novel eukaryotic protein kinase family distinct from those previously defined. The identification of related proteins in model organisms provides some initial insight into their functional properties and will provide new experimental avenues by which to determine the function of the Haspin proteins in mammalian cells.
Keywords: ALK-1, cell cycle, classification, eukaryotic protein kinase, Gsg2, haploid cells, Haspin, serine/threonine kinase
The protein kinases constitute a large group of enzymes that catalyze the transfer of a phosphate group from ATP to specific serine, threonine, or tyrosine residues in specific substrate proteins. This process of phosphorylation modulates the function of a huge variety of substrates, including metabolic enzymes, transcription factors, adapter molecules, and other kinases. Thus, protein kinases are critical in the regulation of many aspects of cellular metabolism. Most known protein kinases in eukaryotes are members of the so-called eukaryotic protein kinase superfamily (ePKs), although members of this class are also found in bacteria and archea (Hardie and Hanks 1995; Leonard et al. 1998). The crystal structures of a number of ePK domains reveal a common bi-lobed structure, and the conserved features of the 250–300 amino acid domain are well defined (Hanks and Hunter 1995). The smaller N-terminal lobe contains conserved residues required for nucleotide binding, whereas the larger C-terminal lobe is involved in substrate binding and phosphotransfer initiation. The cleft between the lobes contains further invariant amino acids that form the active site. This knowledge allows the identification of likely functional ePKs from primary sequence information. Often, these protein kinases can be identified as either serine/threonine kinases or tyrosine kinases by analysis of protein sequence, and they can be further classified into subfamilies based on amino acid sequence homology. Classification in a given subfamily has proven to be a good indicator of functional similarity (Hardie and Hanks 1995; Hunter and Plowman 1997).
In contrast, a number of proteins with protein kinase activity, including DNA-PK, ATM, and ATR, do not fall within the ePK superfamily but instead are related to the phosphatidylinositol kinases (lipid kinases or PIKs). The catalytic domains of PIKs possess only weak homology with some subdomains of the ePKs, yet the crystal structure of PI3Kγ reveals a bi-lobed structure with a fold reminiscent of the ePKs, particularly in the central ATP-binding core (Walker et al. 1999). It is even more striking that APH(3′)-IIIa and PIPKIIβ, members of the aminoglycoside phosphotransferase (APH) and phosphatidylinositol phosphate kinase (PIPK) superfamilies, respectively, also show similarity to the bi-lobed structure of the ePKs despite essentially undetectable protein sequence homology (Hon et al. 1997; Rao et al. 1998). Thus, a number of protein kinase and other kinase families share secondary and tertiary structural features, although they lack significant similarity in primary sequence.
The protein Haspin (haploid germ cell–specific nuclear protein kinase; encoded by germ cell–specific gene 2 [Gsg2]) was recently identified in mice and humans and reported to possess serine/threonine kinase activity. Haspin protein and its mRNA were found specifically in male germ cells, and expression appeared greatest in haploid spermatids (Tanaka et al. 1999, 2001). However, Haspin mRNA can also be detected in diploid cell lines and tissues (Higgins 2001). When ectopically expressed in HEK-293 cells, Haspin localized to the nucleus, had DNA-binding capacity, and led to reduced cell proliferation. It was suggested, therefore, that Haspin plays a critical role in cell-cycle cessation and differentiation of haploid germ cells (Tanaka et al. 1999). The mammalian Haspin proteins have been shown to possess homology limited to the N-terminal portion of the ePK domain consensus sequence and particularly to the Cdc2 kinase (Tanaka et al. 1999, 2001). Here, we show that Haspin-related proteins are found throughout the eukaryotes and that they contain a complete but divergent ePK domain. Furthermore, we find that the Haspin-like proteins comprise a new family of putative ePKs with distinctive structural features.
Results and Discussion
We recently cloned a human Haspin cDNA (Higgins 2001) and used the derived protein sequence to conduct BLAST searches of the translated nonredundant nucleotide sequence databases. In addition to murine Haspin, highly significant matches of the C-terminal domain of Haspin to many proteins in the database were revealed. All of these proteins were known or predicted eukaryotic protein kinases. Among such proteins, a subset of hypothetical gene products has significantly higher homology with Haspin and each other than with other kinases. This group includes translations of genomic sequences from Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Schizosaccharomyces pombe, and Saccharomyces cerevisiae (Fig. 1 ▶). Partial pig, bovine, and zebrafish sequences were also identified among expressed sequence tags (ESTs) that encode polypeptides with 81%, 77%, and 52% identity to human Haspin over segments of the kinase domain (GenBank accession numbers AW668743, AW478927, and AW343515).
Fig. 1.
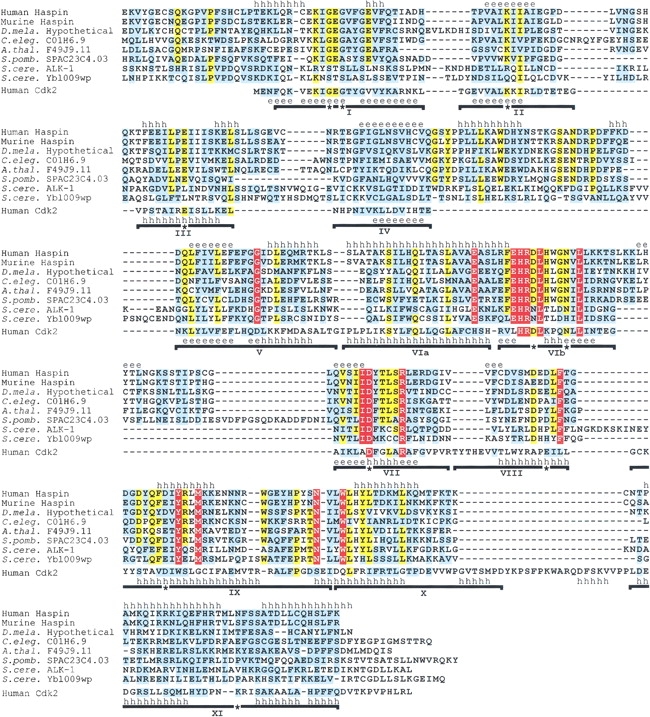
Multiple sequence alignment of Haspin kinase domains and human Cdk2. Residues that are completely conserved in all Haspin-like proteins are shown in white on a red background. Residues that are identical (yellow background) or similar (cyan background, based on Gonnet Pam250 matrix positive values) in ≥80% sequences are indicated. Introduced gaps are shown as dashes. Subdomains I to XI of the Cdk2 kinase fold are indicated with horizontal lines, and residues that are essentially invariant in previously identified kinases are labeled with asterisks (Hanks and Quinn 1991). The known secondary structure of Cdk2 is shown above its sequence (PDB code 1FIN). Above the alignment is a secondary structure prediction using PHDsec (Rost and Sander 1993), based on the alignment without Cdk2. Regions predicted with an expected accuracy of >82% to form α-helices (h) or β-strands (e) are labeled. The alignment includes residues 461–798 of human Haspin (GenBank AF289865); 417–754 of murine Haspin (GenBank AF289866, NP_034483); and the C-terminal regions of hypothetical proteins from D. melanogaster (see Materials and Methods), C. elegans (residues 564–920 of hypothetical protein C01H6.9, GenBank CAA95786), A. thaliana (residues 263–599 of hypothetical protein F14J9.11, GenBank AAC33205), S. pombe (residues 134–488 of hypothetical protein SPAC23C4.03, GenBank CAB16874), and S. cerevisiae (residues 408–759 of ALK-1/Ygl021w, GenBank P43633, CAA61012; and 324–676 of hypothetical protein Ybl009wp, GenBank NP_009544); and the entire sequence of human Cdk2 (residues 1–298, GenBank NP_001789). The PSI-BLAST E values after a single iteration when searching with human Haspin residues 461–798 were murine Haspin, 10−164; C01H6.9, 10 −103; F49J9.11, 10 −108; SPAC23C4.03, 7 × 10−97; ALK-1, 3 × 10−19; and Ybl009wp, 6 × 10−44.
Interestingly, the Haspin-like proteins do not appear to fall into previously described ePK families. The best match to a member of a known family identified by a PSI-BLAST search with the human Haspin kinase domain is Cdc2 from Bombyx mori, with an E value of 7 × 10−11 (after one iteration). A number of other cyclin-dependent kinases (Cdks) are identified with E < 10−4. However, members of several different ePK families, including Ste20/p21-activated kinases (Paks) and some protein tyrosine kinases, also have E < 10−4, whereas the Haspin-like proteins match with E < 3 × 10−19 (see Fig. 1 ▶). Furthermore, none of the Haspin-like sequences reported here were identified as members of the Cdk-related kinase family in a recent extensive survey of yeast and metazoan genomes (Liu and Kipreos 2000) or as members of other known ePK families (Hunter and Plowman 1997; Plowman et al. 1999). Thus, although the Haspins share a degree of homology with many ePKs (see below), they appear to form a unique subgroup with a subset of previously uncategorized kinase-like proteins from a variety of species.
Several specific features distinguish the Haspin-like proteins from Cdks and other kinases and further indicate their categorization as structurally distinct from known ePK families. Alignment of the Haspin-like proteins with an ePK of known structure (human Cdk2; Fig. 1 ▶; Tsai et al. 1991; De Bondt et al. 1993; Jeffrey et al. 1995) reveals that in most cases, residues known to be critical in forming the Mg2+-ATP–binding and catalytic sites are well conserved (the G-x-G-x-x-G-x-V motif in region I and the conserved lysine, glutamate, and aspartate in regions II, III, and VII). However, in the Haspin-like proteins, the motif surrounding the catalytic aspartate residue in region VIb often contains a histidine residue in place of the lysine typical of almost all serine/threonine kinases (H-x-D-[LIVA]-H-x-x-N, rather than [HY]-x-D-[LIVMFY]-K-x-x-N). At least one known ePK (the S. pombe cyclin-suppressing kinase, Csk1) has a histidine at this position, but it is more closely related to the Cdk family and is not otherwise related to the Haspin family (see below; Molz and Beach 1993; Liu and Kipreos 2000). Many members of the more recently described and divergent ABC1 family of putative ePKs also possess histidine at this position, but again, these ePKs are not otherwise closely related to the Haspin-like proteins (see below; Leonard et al. 1998). Notably, members of the PIK superfamily also have a histidine residue at this position (Walker et al. 1999). In contrast, the Haspin-related ALK-1/Ygl021w and Ybl009wp proteins of S. cerevisiae contain a threonine at this position, a residue normally associated with tyrosine kinase activity. However, these two proteins also have an S-x-S-x-x-S-x-[NS] motif that replaces the G-x-G-x-x-G-x-V motif in region I, and they lack invariant lysine, glutamate, and aspartate residues in regions II, III, and VIb, respectively. Whether these two molecules have protein kinase activity clearly will require experimental determination.
The C-terminal regions VIII through XI in the Haspin-like proteins appear to lack some of the highly conserved residues found in other ePKs, and there is a relative lack of sequence similarity to other ePKs in regions that may play a role in substrate recognition (particularly regions VIII and IX, Fig. 1 ▶). However, the similarity of the predicted secondary structure of the Haspin-like proteins to the known structure of human Cdk2 in regions IX through XI, and the coincidence of their C termini with the end of the ePK consensus (PROSITE PS50011) support the validity of the sequence alignment. Therefore, it is likely that the Haspin-like proteins contain an entire kinase domain that would adopt a bi-lobed conformation similar to that of other ePKs. However, given that known kinases of the APH, PIPK, and PIK superfamilies have structural similarities to the ePKs in regions I through VII but diverge in more C-terminal regions, it remains possible that the Haspin-like proteins might also adopt a conformation somewhat different from classical ePKs in regions VIII through XI. Residues conserved within the Cdk family, such as the cyclin-binding PSTAIRE motif (region III) and GDSEID motif (region IX and X), are not found in the Haspin-like proteins. The Haspins also do not have conserved threonine, serine or tyrosine residues in the activation-loop or T-loop between regions VII and VIII. Phosphorylation of residues in this region regulates kinase activity in a number of kinase families. Overall, the sequence between regions VII and IX is unusually short, and it is difficult to identify an APE motif in region VIII, a very widespread feature of ePKs.
Notable features that are conserved within the Haspin-like proteins include an inserted segment between regions IV and V that is predicted to contain an α-helical region and another insert between regions VIb and VII that may adopt a β-strand conformation (except in the S. cerevisiae proteins, where this insert is missing). A G-G-x-x-L motif is present in region V, an S-I-hydrophobic-x-Q-x-[VT]-x-x-L-x-[IV]-[AV]-[EK] motif is found in region VIa, and an I-I-D-[FY]-[TSG]-x-[AS] motif surrounds the invariant aspartate residue in region VII. A D-x-x-L-F motif replaces the common APE sequence in region VIII, and invariant tyrosine, methionine, asparagine, and tryptophan residues are found within conserved motifs in regions IX and X (see Fig. 1 ▶). A leucine zipper motif in mammalian Haspins (Tanaka et al. 1999) is incompletely conserved in the other Haspin-like proteins, and it is questionable whether this motif could function as a dimerization domain because it spans regions VIa and VIb, which straddle the predicted catalytic loop region.
Strikingly, C. elegans appears to have an extended family of Haspin-related proteins. Besides C01H6.9 (included in Fig. 1 ▶), the C. elegans genome encodes a closely related protein, F22H10.5, and several other somewhat more distantly related kinase-like proteins that have homology with the mammalian Haspins with statistically significant E values (Fig. 2 ▶). Like the proteins in Figure 1 ▶, these sequences contain an inserted element between regions IV and V and the conserved motifs described above in regions V and VIa. The sequence of the VIb to VII insert, [LIV]-x-[YF]-[TS]-hydrophobic-x-G-[KQRN]-x-[LIV]-x-[LIV]-[TS]-x-G, is particularly well conserved in all the Haspin-like proteins (except in S. cerevisiae, see above). Again, the fifth position of the H-x-D-[LIVA]-H-x-x-N motif in region VIb is often histidine, and the motif surrounding the invariant aspartate of region VII is maintained. In addition, several other partial Haspin-kinase–like domains were identified among hypothetical C. elegans genome translations (Fig. 2 ▶, legend), indicating that approximately 15 Haspin-like proteins are present in this species.
Fig. 2.
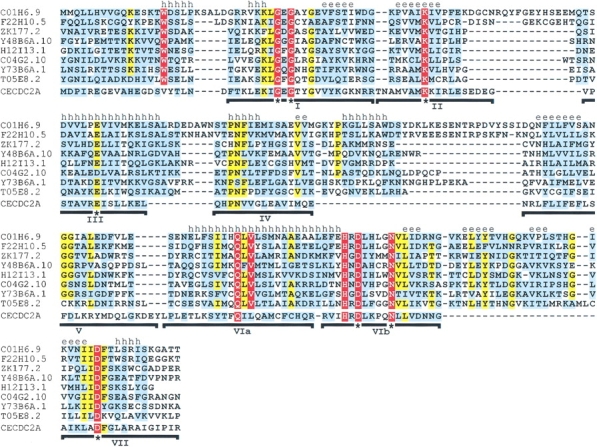
Multiple sequence alignment of Haspin-related proteins and Cdc2 (CECDC2A) from C. elegans. The alignment was generated and is colored and annotated as described for Figure 1 ▶. Above the alignment is a secondary structure prediction using PHDsec (Rost and Sander 1993), based on the alignment without CECDC2A. Only regions I through VII of the kinase domain are shown because many of the hypothetical translations of the C. elegans genome lack homology with kinase regions VIII and XI, probably because of incorrect prediction of C-terminal exons by gene prediction algorithms. The alignment includes residues 563–810 of C01H6.9 (GenBank CAA95786), 71–309 of F22H10.5 (GenBank AAB09102), 37–238 of ZK177.2 (GenBank AAG00045), 169–377 of Y48B6A.10 (GenBank CAB54446), 395–592 of H12I13.1 (GenBank AAF39881), 71–281 of C04G2.10 (GenBank CAA94675), 309–525 of Y73B6A.1 (GenBank AAF36040), 144–352 of T05E8.2 (GenBank AAB52426), and residues 1–176 of C. elegans CECDC2A (GenBank AAC60520, CAA48455). The PSI-BLAST E values after a single iteration when searching with human Haspin residues 461–798 (to allow comparison with Fig. 1 ▶) were C01H6.9, 10−103; F22H10.5, 2 × 10−97; ZK177.2, 10−21; Y48B6A.10, 3 × 10−21; H12I13.1, 2 × 10−25; C04G2.10, 2 × 10−30; Y73B6A.1, 6 × 10−63; and T05E8.2, 2 × 10−48. More severely truncated Haspin-like kinase domains not shown were also identified in C. elegans: F12A10.2, F12A10.6, F59E12.6 (all closely related to ZK177.2), C06E4.5, C26E6.1, C50H2.7, and K08B4.5 (GenBank accession numbers AAA68295, AAA68299, AAB54256, AAA82472, AAA21155, CAA98255, and AAC68982).
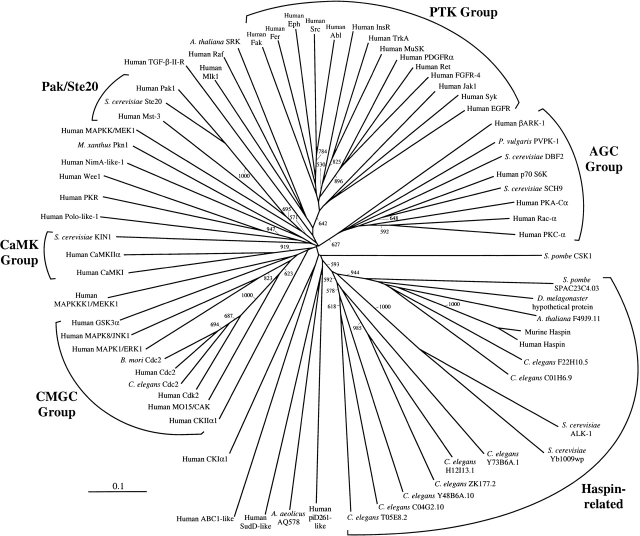
To confirm whether or not the Haspin-related proteins represent a distinct ePK subfamily, a phylogenetic analysis using the neighbor-joining method (Saitou and Nei 1987) was performed. Because of incomplete sequence information for some of the C. elegans Haspin-like domains (see legend of Fig. 2 ▶), the analysis was limited to kinase subdomains I through VII and included only residue positions for which there were no gaps in any sequence in the alignment. Kinase domains that were identified as most related to human Haspin by BLAST and PSI-BLAST searches were included, such as members of the Cdk, Ste20/Pak, and tyrosine kinase (PTK) families. Also included were sequences of multiple members of the CMGC, AGC, CaMK serine/threonine kinase families and representatives of the major other protein kinase families (OPKs; Hardie and Hanks 1995). The S. pombe kinase Csk1 mentioned above and representatives of the recently identified divergent ABC1, RIO1, piD261, and AQ578 putative ePK families (Leonard et al. 1998) were also incorporated. The results (Fig. 3 ▶) support the idea that the Haspin-like proteins form a family distinct from the Cdk family and other ePK subgroups. Note that this is the case even though the C-terminal portions of the Haspin kinase domains that diverge most significantly from other ePKs were excluded from the analysis. As indicated by the BLAST scores, the mammalian, Drosophila, Arabidopsis, S. pombe, and C. elegans C01H6.9 and F22H10.5 proteins form a subgrouping and may represent essentially orthologous proteins in different species. The S. cerevisiae ALK-1 and Ybl009wp proteins are rather divergent members of this first subgroup, whereas the remaining C. elegans proteins form a second somewhat more distantly related subfamily. In this work, human ePKs were included in the phylogeny where possible, but a recent analysis by parsimony of protein kinases in the C. elegans genome also indicates that the Haspin-like proteins identified here form a kinase family quite distinct from previously identified groups (http://www.kinase.com). Interestingly, in this study of C. elegans protein kinases, five subfamilies were tentatively described as worm-specific. Three of these subfamilies had C04G2.10, K08B4.5, and ZK177.2 as prototypical members (Plowman et al. 1999). The analysis presented here indicates that these putative kinases are all members of the Haspin-like family (see Figs. 2, 3 ▶ ▶) and that related proteins are found in many eukaryotic phyla.
Fig. 3.
Neighbor-joining phylogeny of subdomains I through VII of the Haspin-related proteins and known eukaryotic protein kinases. An unrooted dendrogram is shown. Bootstrap values >500 from 1000 trials are given on branches, and the scale bar indicates distance of divergence. Members of ePK families as previously defined (Hardie and Hanks 1995) are indicated. GenBank accession numbers for those proteins (human unless otherwise stated) not identified in Figures 1 and 2 ▶ ▶ are piD261-like protein dJ28H20.2, CAC00561; Aquifex aeolicus AQ578, AAC06806; SudD-(Aspergillus)-like, AAC26080; ABC1-like protein, NP_064632; casein kinase I α1 (CKIα1), NP_001883; CKIIα1, NP_001886; MO15/CAK, CAA55785; Cdc2, A29539; B. mori Cdc2, BAA21483; mitogen-activated protein kinase-1 (MAPK1/ERK1), NP_002736; MAPK8/JNK1, NP_002741; glycogen synthase kinase 3α (GSK3α), P49840; MAPKKK1/MEKK1, Q13233; Ca2+/calmodulin kinase-1 (CaMKI), NP_003647; CaMKIIα, AAD55815; S. cerevisiae KIN1, AAA34722; Polo-(Drosophila)-like kinase, NP_005021; Protein Kinase RNA–dependent (PKR), AAA36409; Wee1, CAA43979; NimA-like protein kinase 1, G01452; Myxococcus xanthus Pkn1, AAA25402; MAPKK1/MEK1, AAA36318; Mst-3, AAB82560; S. cerevisiae Ste20, NP_011856; Pak1, NP_002567; TGF-β type II receptor, AAA61164; mixed lineage kinase-1 (Mlk1), P80192; Raf proto-oncogene, P04049; A. thaliana S-receptor kinase (SRK), T00534; focal adhesion kinase (Fak), Q05397; Fer, AAA61190; Eph, AAA36747; Src proto-oncogene, P22455; Abl proto-oncogene, P00519; insulin receptor (InsR), P06213; high affinity nerve growth factor receptor (TrkA), P04629; muscle, skeletal, receptor tyrosine kinase (MuSK), NP_005583; platelet-derived growth factor receptor α (PDGFRα), AAA60048; Ret proto-oncogene, P07949; fibroblast growth factor receptor 4 (FGFR4), P22455; Janus kinase-1 (JAK1), AAA36527; spleen tyrosine kinase (Syk), NP_003168; epidermal growth factor receptor (EGFR), P00533; β-adrenergic receptor kinase-1 (βARK1), NP_001610; Phaseolus vulgaris PvPK1, AAA33772; S. cerevisiae DBF2, AAA34559; ribosomal protein S6 kinase (p70 S6K), P23443; S. cerevisiae cAMP-dependent protein kinase SCH9, P11792; cAMP-dependent kinase catalytic subunit-α (PKA-Cα), P17612; Rac-α, AAA36539; protein kinase C-α (PKC-α), P17252; and S. pombe Csk1, AAB26194.
The overall structure of the Haspin-like proteins is similar: The conserved kinase domain at the C terminus is preceded by an N-terminal region that lacks significant homology in the group. For example, human and murine Haspin are 83% identical in the C-terminal kinase domain (human amino acids 481–798), but only 53% identical in the N-terminal portion. The N-terminal regions of the proteins shown in Figure 1 ▶ share certain general features, such as a preponderance of serine and lysine residues, but otherwise appear unrelated (data not shown). It is likely that the DNA-binding capacity of the mammalian Haspins lies in the N-terminal domain (Tanaka et al. 1999).
Conclusions
In summary, the data define a group of Haspin-related proteins that, based on distinctive primary structural features, form a new family of ePKs that extends previous classification (Hanks and Quinn 1991; Hanks and Hunter 1995; Hardie and Hanks 1995). If, in addition to the mammalian Haspins, other Haspin-like proteins (or ABC1 family members) are confirmed to be serine/threonine kinases, it may be useful to relax definition of the serine/threonine kinase signature (PROSITE PS00108). Specifically, histidine might be allowed at the fifth position of the [HY]-x-D-[LIVMFY]-K motif, as this may aid identification of more divergent protein kinase domains in the growing genome databases. Likely Haspin orthologs are found in several major eukaryotic phyla, including yeasts, plants, flies, fish, and mammals, and an extended group is found in C. elegans. In fact, the Haspin-like proteins can be added to the list of kinase families that are expanded in C. elegans, including the casein kinase I, FER, and KIN-15 families (Plowman et al. 1999).
What are the functions of the Haspin-like proteins In its GenBank entry, the S. cerevisiae gene for ALK-1 is annotated as a novel DNA damage response gene, and ALK1 gene expression is reported to be responsive to pheromones, although no effect on yeast mating differentiation was observed when the gene was disrupted (Erdman et al. 1998). Disruption of the ALK1 or Ybl009w gene does not prevent vegetative growth of S. cerevisiae (http://genome-www. stanford.edu/cgi-bin/sgd), but expression of both genes appears to be regulated in a periodic manner during the mitotic cell cycle, with ALK1 and Ybl009w expression peaking during M and G1 phases, respectively (Cho et al. 1998; Spellman et al. 1998). Characterization of murine Haspin indicates that the family may play a role in regulating the cell cycle of haploid cells (Tanaka et al. 1999). Consistent with a role in meiosis, Ybl009w gene transcription also is induced during the early–mid phase of sporulation in budding yeast, whereas ALK1 induction was below that considered significant in this study (Chu et al. 1998). However, Haspin mRNA can also be detected in diploid human and mouse cells, so the Haspin-related proteins are likely to have a wider functional role than previously anticipated (Higgins 2001). The presence of multiple Haspin-like proteins in C. elegans may complicate analysis of their function in this organism. Indeed, the Haspin-related C. elegans gene C26E6.1 was among a large panel of genes targeted by RNA interference analysis in a recent study able to detect subtle variations in meiotic and mitotic cell division, but no phenotype was apparent (Gönczy et al. 2000). Among other explanations, it is possible that the C. elegans Haspin-like proteins show functional redundancy that obscures the effect of single-gene targeting. However, the identification of related proteins in model organisms such as yeasts and Drosophila will provide experimental avenues by which to determine the function of the Haspins in mammalian cells.
Materials and methods
Sequences related to the kinase domain of human Haspin were identified by TBLASTN and PSI-BLAST searches (Altschul et al. 1997) of the nonredundant database at NCBI (http://www.ncbi.nlm.nih.gov/blast/). The sequence of a Haspin-related Drosophila hypothetical protein was reconstructed by translation of nucleotides 29203–29406 and 29464–29759 of GenBank AE003029 and 4468–4669 of GenBank AC005334. The validity of this join was confirmed by the Berkeley Drosophila Genome Project cDNA clone LD07633, which links the two genomic regions (http://www.flybase.org). Alignments of the kinases shown in Figures 1 and 2 ▶ ▶ were produced by CLUSTAL W (Thompson et al. 1994), with default settings at the University of California San Diego Supercomputer Center Biology Workbench (http://workbench.sdsc.edu) and adjusted manually according to Hanks and Quinn (1991) and by comparison to protein kinase alignments at the Protein Kinase Resource (http://www.sdsc.edu/Kinases). Alignments were colored using BOXSHADE (K. Hofmann, and M.D. Baron, ISREC, Epalinges s/Lausanne, Switzerland). Secondary structure predictions were performed with PHDsec (http://cubic.bioc.columbia.edu/predictprotein; Rost and Sander 1993). Phylogenetic analysis was performed using the neighbor-joining method of Saitou and Nei (1987) as implemented by CLUSTAL X. In this case, alignments were limited to regions I through VII. When calculating the distance matrix, residue positions in the alignment containing gaps were excluded, and no correction was made for multiple substitutions. Phylogenetic trees were displayed using TREEVIEW (1996).
Acknowledgments
This work was supported by the Harvard Digestive Diseases Center National Institutes of Health grant P30 DK34845. I thank Drs. Michael Brenner and Christopher Dascher for useful discussions, and Dr David Lee and the reviewers for comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.49901
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller,W., and Lipman, D.J. 1997. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, R.J., Campbell, M.J., Winzeler, E.A., Steinmetz, L., Conway, A., Wodicka, L., Wolfsberg, T.G., Gabrielian, A.E., Landsman, D., Lockhart, D.J., et al. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2 65–73. [DOI] [PubMed] [Google Scholar]
- Chu, S., DeRis, J., Mulholland, J., Botstein, D., Brown, P.O., and Herskowitz, I. 1998. The transcriptional program of sporulation in budding yeast. Science 282 699–705. [DOI] [PubMed] [Google Scholar]
- De Bondt, H.L., Rosenblatt, J., Jancarik, J., Jones, H.D., Morgan, D.O., and Kim, S.-H. 1993. Crystal structure of cyclin-dependent kinase 2. Nature 363 595–602. [DOI] [PubMed] [Google Scholar]
- Erdman, S., Lin, L., Malczynski, M., and Snyder, M. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J.M., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408 331–336. [DOI] [PubMed] [Google Scholar]
- Hanks, S. and Quinn, A.M. 1991. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Meth. Enzymol. 200 38–62. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K. and Hunter, T. 1995. The eukaryotic protein kinase family: Kinase (catalytic) domain structure and classification. FASEB J. 9 576–596. [PubMed] [Google Scholar]
- Hardie, G. and Hanks, S. 1995. The Protein Kinase FactsBook. Academic Press Ltd, London, UK.
- Higgins, J.M.G. 2001. The Haspin gene: Location in an intron of the Integrin αE gene, associated transcription of an Integrin αE-derived RNA and expression in diploid as well as haploid cells. Gene 267 55–69. [DOI] [PubMed] [Google Scholar]
- Hon, W.C., McKay, G.A., Thompson, P.R., Sweet, R.M., Yang, D.S.C., Wright, G.D., and Berghuis, A.M. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89 887–895. [DOI] [PubMed] [Google Scholar]
- Hunter, T. and Plowman, G.D. 1997. The protein kinases of budding yeast: Six score and more. Trends Biochem. Sci. 22 18–22. [DOI] [PubMed] [Google Scholar]
- Jeffrey, P.D., Russo, A.A., Polyak, K., Gibbs, E., Hurwitz, J., Massague, J., and Pavletich, N.P. 1995. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature 376 313–320. [DOI] [PubMed] [Google Scholar]
- Leonard, C.J., Avarind, L., and Koonin, E.V. 1998. Novel families of putative protein kinases in bacteria and archea: Evolution of the "eukaryotic" protein kinase superfamily. Genome Res. 8 1038–1047. [DOI] [PubMed] [Google Scholar]
- Liu, J. and Kipreos, E.T. 2000. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): Differential conservation of CAKs in yeast and metozoa. Mol. Biol. Evol. 17 1061–1074. [DOI] [PubMed] [Google Scholar]
- Molz, L. and Beach, D. 1993. Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J. 12 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. 1996. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Plowman, G.D., Sudarsanam, S., Bingham, J., Whyte, D., and Hunter, D. 1999. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. 96 13603–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, V.D., Misra, S., Boronenkov, I.V., Anderson, R.A., and Hurley, J.H. 1998. Structure of type IIβ phosphatidylinositol phosphate kinase: A protein kinase fold flattened for interfacial phosphorylation. Cell 94 829–839. [DOI] [PubMed] [Google Scholar]
- Rost, B. and Sander, C. 1993. Prediction of protein secondary structure at better than 72% accuracy. J. Mol. Biol. 232 584–599. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle–regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Yoshimura, Y., Nozaki, M., Yomogida, K., Tsuchida, J., Tosaka, Y., Habu, T., Nakanishi, T., Okada, M., Nojima, H., et al. 1999. Identification and characterization of a haploid germ cell–specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J. Biol. Chem. 274 17049–17057. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Iguchi, N., Nakamura, Y., Kohroki, J., Egydio de Carvalho, C., and Nishimune, Y. 2001. Cloning and characterization of human haspin gene encoding haploid germ cell–specific nuclear protein kinase. Mol. Hum. Reprod. 7 211–218. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, L.H., Harlow, E., and Meyerson, M. 1991. Isolation of the human cdk2 gene that encodes the cyclin A– and adenovirus E1A–associated p33 kinase. Nature 353 174–177. [DOI] [PubMed] [Google Scholar]
- Walker, E.H., Perisic, O., Ried, C., Stephens, L., and Williams, R.L. 1999. Structural insights into phosphoinositol 3-kinase catalysis and signalling. Nature 402 313–320. [DOI] [PubMed] [Google Scholar]