Abstract
Ribotoxins are a family of potent cytotoxic proteins from Aspergillus whose members display a high sequence identity (85% for about 150 amino acid residues). The three-dimensional structures of two of these proteins, α-sarcin and restrictocin, are known. They interact with phospholipid bilayers, according to their ability to enter cells, and cleave a specific phosphodiester bond in the large subunit of ribosome thus inhibiting protein biosynthesis. Two nonconservative sequence changes between these proteins are located at the amino-terminal β-hairpin of α-sarcin, a characteristic structure that is absent in other nontoxic structurally related microbial RNases. These two residues of α-sarcin, Lys 11 and Thr 20, have been substituted with the equivalent amino acids in restrictocin. The single mutants (K11L and T20D) and the corresponding K11L/T20D double mutant have been produced in Escherichia coli and purified to homogeneity. The spectroscopic characterization of the purified proteins reveals that the overall native structure is preserved. The ribonuclease and lipid-perturbing activities of the three mutants and restrictocin have been evaluated and compared with those of α-sarcin. These proteins exhibit the same ability to specifically inactivate ribosomes, although they show different activity against nonspecific substrate analogs such as poly(A). The mutant variant K11L and restrictocin display a lower phospholipid-interacting ability correlated with a decreased cytotoxicity. The results obtained are interpreted in terms of the involvement of the amino-terminal β-hairpin in the interaction with both membranes and polyadenylic acid.
Keywords: Cytotoxic protein, protein bilayer interaction, restrictocin, ribosome-inactivating protein, α-sarcin, site-directed mutagenesis
Ribotoxins are a group of secreted fungal ribonucleases (RNases) that inactivate ribosomes and inhibit protein biosynthesis by cleaving a single phosphodiester bond of the larger rRNA, releasing a characteristic RNA fragment (known as the α-fragment; Schindler and Davies 1977; Endo and Wool 1982). The target bond, located at a highly conserved rRNA sequence present in all known prokaryota and eukaryota, is known as the sarcin/ricin loop (SRL; Wool et al. 1992; Szewaczak and Moore 1995; Correll et al. 1998, 1999) recognized by ribosome-inactivating proteins such as ricin (Endo et al. 1990). Ribotoxins are among the most potent inhibitors of translation because the hydrolysis of the phosphodiester bond abolishes both the elongation factor 1-dependent binding of aminoacyl-tRNA and the GTP-dependent binding of elongation factor 2 to ribosomes. Ribotoxins inactivate ribosomes from all living species tested so far when assayed in a cell-free lysate system (Miller and Bodley 1988); however, they display selectivity when assayed against intact cells (Olson et al. 1965; Turnay et al. 1993). This selectivity is likely related to a differential recognition of membrane phospholipids, as observed for α-sarcin in lipid model systems, because no protein membrane receptors have been found for these toxins. In fact, α-sarcin interacts with membranes containing acid phospholipids promoting aggregation of vesicles, lipid-mixing between aggregated bilayers, and leakage of the intravesicular aqueous contents (Gasset et al. 1989, 1990, 1994; Mancheño et al. 1995b, 1998; Oñaderra et al. 1998). It gains access to cleave tRNA encapsulated in liposomes (Oñaderra et al. 1993). One of the initial steps in α-sarcin–vesicle interactions is the formation of a vesicle dimer maintained by protein molecules, as demonstrated by stopped-flow techniques (Mancheño et al.1994a), suggesting the involvement of different regions of the protein molecule in bridging together the vesicles.
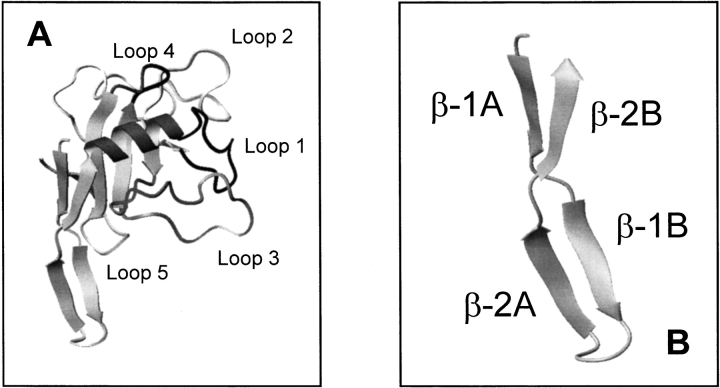
Ribotoxins show a high degree of sequence similarity (Wirth et al. 1997; Martínez-Ruiz et al. 1999a, b). α-Sarcin, and restrictocin (150 and 149 amino acid residues, respectively) are most extensively characterized and display 85% sequence identity. Their three-dimensional structures have been solved (Campos-Olivas et al. 1996; Yang and Moffat, 1996; Pérez-Cañadillas et al. 2000), revealing that their global shape and overall main-chain fold are closely matched. Both proteins display a core composed of a small α-helix packed against a five-stranded antiparallel β-sheet and connected by large loops (Fig. 1A ▶). These loops have been proposed to be involved in the cytotoxic activity—related to the ability for entering cells—because they are absent from other nontoxic microbial extracellular RNases (RNases U2 and T1) with a highly similar polypeptide fold (Martínez del Pozo et al. 1988; Kao and Davies 1995, Mancheño et al. 1995a). In fact, loop-2 of α-sarcin has been proposed to be one of the protein regions involved in the interaction with lipid vesicles (de Antonio et al. 2000). In this respect, preliminary studies with restrictocin have revealed a lower ability of this protein to perturb lipid membranes (L. García-Ortega, J.M. Mancheño, M. Oñaderra, R. Kao, A. Martínez del Pozo, and J.G. Gavilanes, unpubl.). α-Sarcin contains an amino-terminal β-hairpin (residues 2–26) that forms a solvent-exposed protuberance and shows a complex topology that can be considered as two consecutive minor β-sheets connected by a hinge region (Fig. 1B ▶; Pérez-Cañadillas et al. 2000). The second β-sheet of this amino-terminal hairpin is absent in the nontoxic RNases U2 and T1 (Pace et al. 1991; Noguchi et al. 1995). The region between residues 11 and 16 of this amino-terminal hairpin in restrictocin showed ill-defined electron density, which precluded its structural analysis (Yang and Moffat 1996). α-Sarcin and restrictocin sequences differ in only 20 residues. Nine of these changes are of conservative character, and only five result in charge modification (amino-terminal hairpin: K11L, T20D; loop 2: K84Q, T90D; loop 5: E140G; α-sarcin/restrictocin). Because restrictocin exhibits a decreased membrane-perturbing ability in comparison with α-sarcin, and the mentioned charge changes involve both loop-2 (one of the proposed bilayer-interacting regions of α-sarcin) and the amino-terminal β-hairpin, the latter appears to be a second region potentially required to maintain a vesicle dimer. In addition, the amino-terminal hairpin has been suggested to modulate the catalytic activity on the basis of results obtained with different mutants of mitogillin, a ribotoxin with a single substitution relative to restrictocin (Kao and Davies 1999, Kao and Davies 2000). Lys 11 and Thr 20 have relevant and specific structural roles in α-sarcin. Lys 11 is involved in a salt bridge with Glu 140, which displays unusual torsion backbone angles forced to adopt a conformation identical to the Gly located at the equivalent position in other ribotoxins. Thr 20 is very close to Asp 9, which is involved in a salt bridge with Lys 139 and hydrogen bonded to HN of Glu 140 (Pérez-Cañadillas et al. 2000). Considering the potential involvement of the amino-terminal hairpin in cytotoxicity and ribonuclease activity of ribotoxins and the specific structural role of Lys 11 and Thr 20 in α-sarcin, we prepared mutants of α-sarcin by substituting these two residues of the hairpin with their sequence counterparts in restrictocin: Lys 11 by Leu (variant K11L) and Thr 20 by Asp (variant T20D). These single variants and the corresponding K11L/T20D double mutant proteins have been purified and characterized to analyze the involvement of this protein region in the structure–function relationships of α-sarcin.
Fig. 1.
Diagrams corresponding to the three-dimensional structure of α-sarcin (A) and its amino-terminal β-hairpin (B) constructed from the atomic coordinates deposited in PDB (Protein Data Bank; reference 1DE3). Images were generated by the molmol program (Koradi et al. 1996). Residues corresponding to loops in A are: loop 1 (black), 38–49; loop 2 (light grey), 53–93; loop 3 (grey), 98–119; loop 4 (black), 126–132; loop 5 (light grey), 139–143.
Results
Purification and structural characterization of the mutants
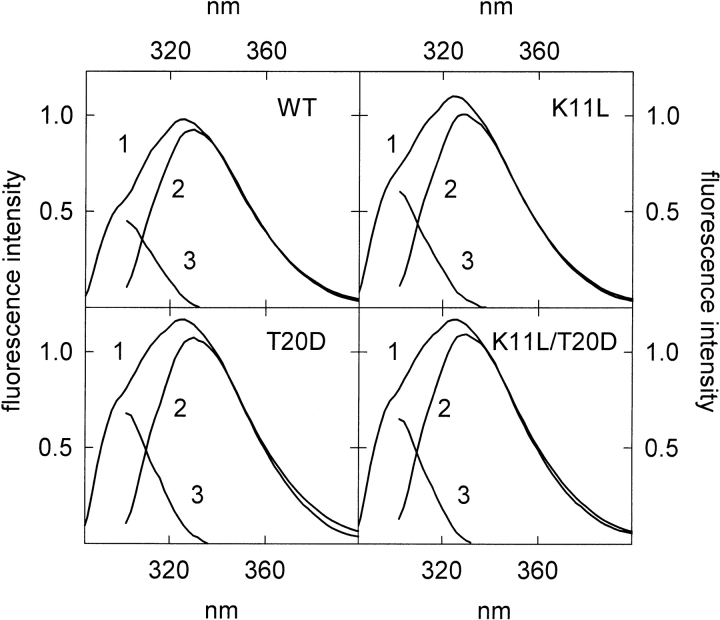
The three mutants were isolated with a yield ranging from 2 to 4 mg/L of original culture medium, slightly lower than that obtained for recombinant wild-type α-sarcin. The amino acid composition was in agreement with the mutations made in each case. The E0.1% (280 nm, 1 cm) calculated from the corresponding UV-absorption spectra showed only small variations (Table 1). Far- and near-ultraviolet (UV) circular dichroism (CD) spectra of the three variants were coincident with those already reported for the native fungal enzyme (Gavilanes et al. 1983; Martínez del Pozo et al. 1988) and some other mutants (Lacadena et al. 1995, 1999). The fluorescence quantum yield of the three variants was higher than that of the wild-type protein (Fig. 2 ▶). Differential analysis of the tyrosine and tryptophan contributions, upon excitation at 275 and 295 nm, revealed a larger increase for the Tyr quantum yield. T20D and K11L/T20D variants showed nearly the same Tyr and Trp emission increments, which were larger than for the K11L variant (Table 1). Restrictocin cannot be used for comparison because the counterpart of Tyr 18 in α-sarcin is Trp 17 in restrictocin.
Table 1.
Spectroscopic features of wild-type α-sarcin and the three mutants studieda
| Protein | E0.1%(280 nm, 1 cm) | QTyrb | QTrpb |
| Wild-type | 1.34 | 1 | 1 |
| K11L | 1.31 | 1.32 | 1.08 |
| T20D | 1.49 | 1.41 | 1.25 |
| K11L/T20D | 1.45 | 1.41 | 1.21 |
Values are the average of three different determinations.
a SD ± 0.05.
b Relative quantum yield of Tyr and Trp referred to the values of the wild-type protein.
Fig. 2.
Fluorescence emission spectra of wild-type α-sarcin (WT) and the K11L, T20D, and K11L/T20D mutant variants, at identical protein concentrations, for excitation at 275 nm (1) and 295 nm (2) (tryptophan contribution) normalized at wavelengths above 380 nm. (3) Calculated difference spectra 1 − 2 (tyrosine contribution). Fluorescence emission is expressed in arbitrary units considering the intensity at the wavelength of the emission maximum of the wild-type protein for excitation at 275 nm as 1.00.
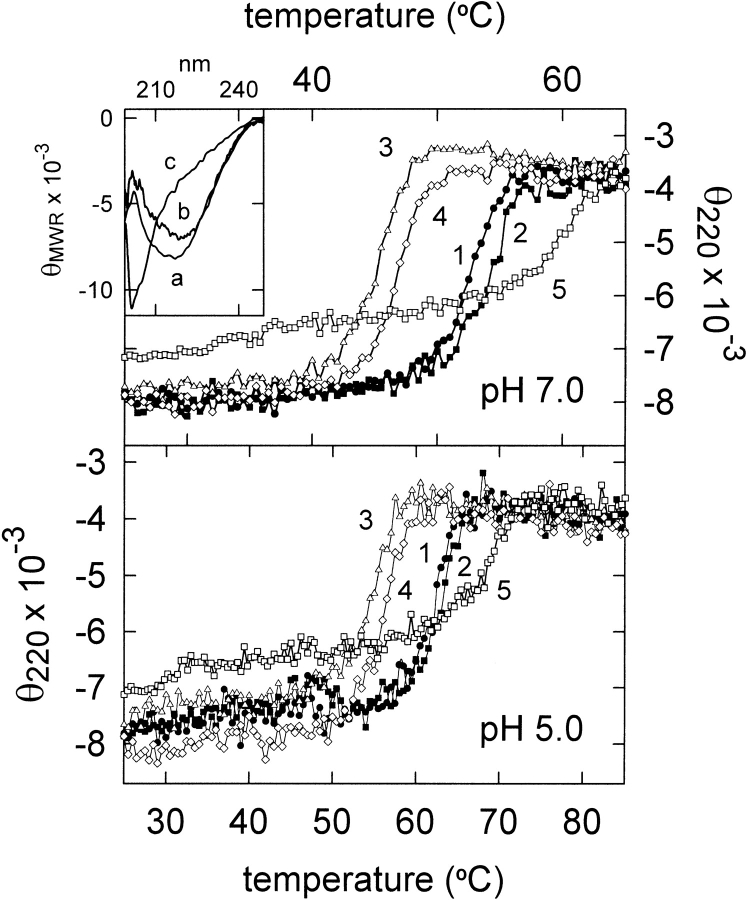
The optimum pH for the RNase activity of wild-type α-sarcin is 5.0 (Pérez-Cañadillas et al. 1998; Lacadena et al. 1999). Thus, thermal denaturation of fungal α-sarcin and the three mutants was studied at pH 5.0 and pH 7.0 by continuous recording of the thermal variation of the ellipticity at 220 nm (Fig. 3 ▶). The Tm value for the wild-type protein at pH 7.0 was 52°C, in good agreement with that obtained by differential scanning calorimetry (Gasset et al. 1995). This value increased up to 62°C at pH 5.0 (Table 2).The K11L variant showed a higher Tm at both pH values whereas the T20D and K11L/T20D variants displayed decreased values. The enthalpy changes calculated from these plots were in the range 140–145 kcal/mole. The ΔHcal value determined for the wild-type protein at pH 7.0 from differential scanning calorimetric measurements was 136 kcal/mole (Gasset et al. 1995). According to the calculated Δ(ΔG), the K11L substitution would involve a slight stabilization of the protein at both pH values, but replacement of Thr 20 by Asp would produce a decrease in stability (Table 2). Restrictocin was also studied for comparison (Table 2). Its Tm value was 7°C higher than that of α-sarcin at both pH values. It is striking that A. restrictus, the mold that produces restrictocin, can grow at 37°C whereas the growth of A. giganteus, the microorganism producing α-sarcin, shows impaired growth above 30°C.
Fig. 3.
Thermal denaturation profiles of wild-type α-sarcin (1), K11L (2), T20D (3), K11L/T20D (4), and restrictocin (5) at pH 7.0 and pH 5.0. The measurements were performed by continuous recording of the mean residue weight ellipticity at 220 nm (θ220). (Inset) Far-UV circular dichroism spectra of (a) wild-type: α-sarcin, K11L, T20D, and K11L/T20D mutant variants at both pH 5.0 and pH 7.0; (b) restrictocin at both pH 5.0 and pH 7.0; and (c) thermally denatured proteins at both pH 5.0 and pH 7.0. Mean residue weight ellipticity (θMRW) is expressed in units of °cm2/dmole.
Table 2.
Thermal denaturation parameters of wild-type α-sarcin and the three mutants studied calculated from ellipticity at 220 nm versus temperature profilesa
| Tm (°C) | Δ(ΔG)b (Kcal/mol) | |||
| Protein | pH 5.0 | pH 7.0 | pH 5.0 | pH 7.0 |
| Wild-type | 62 | 52 | — | — |
| K11L | 63 | 54 | 0.2 | 0.7 |
| T20D | 55 | 45 | −2.3 | −3.3 |
| K11L/T20D | 56 | 47 | −2.0 | −2.8 |
| Restrictocin | 69 | 59 | ||
Average values obtained from three independent denaturation profiles.
a SD ± 1°C.
b Δ(ΔG) ∼ (ΔH × ΔTm/Tm) is the stability change produced by the mutation [ΔH, enthalpy change for the wild-type protein; ΔTm = Tm (mutant) − Tm (wild-type); Tm, value obtained for the mutant variant] (Becktel and Schellman 1987).
Ribonucleolytic characterization
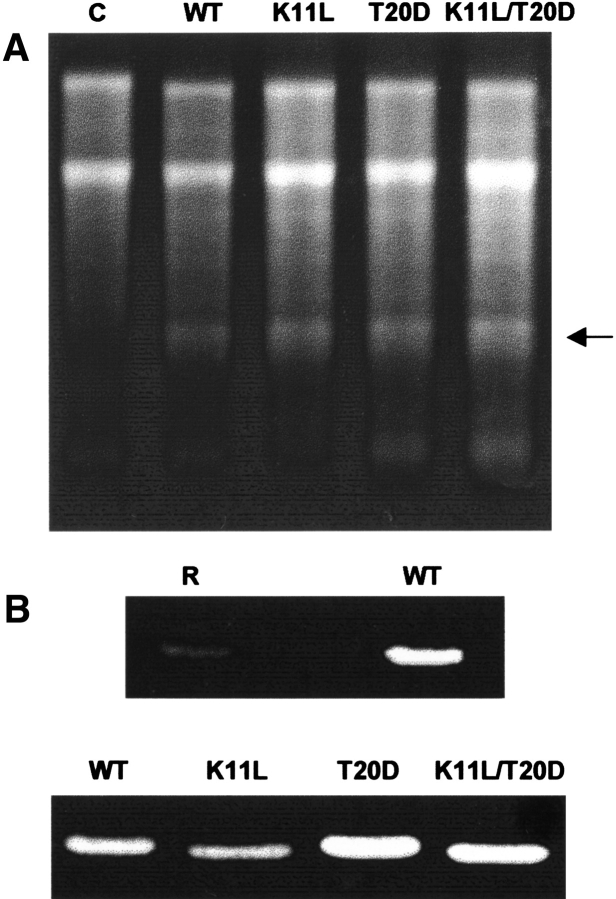
The three mutants of α-sarcin exhibited the same activity against ribosomes in a cell-free lysate (Fig. 4 ▶). The amounts of α-fragment produced by these protein forms were in the ±10% range of the wild-type protein. In this regard, α-sarcin and restrictocin produced identical extents of protein synthesis inhibition in a cell-free lysate (Fando et al. 1985). Ribotoxins also cleave homopolynucleotides, synthetic substrates lacking the characteristic structural element required for the exquisite specificity of ribotoxins against ribosomes (Endo et al. 1983, 1990) through a cyclizing mechanism (Lacadena et al. 1998). The K11L mutant showed a decreased activity against polyadenylic acid [poly(A); 70% of the wild-type activity] but the other variants, T20D and K11L/T20D, displayed higher activity, 126% and 130%, respectively. Restrictocin showed a very low activity against poly(A) (Fig. 4 ▶).
Fig. 4.
(A) Ribosome-inactivating activity assay of (WT) wild-type α-sarcin, and its K11L, T20D, and K11L/T20D mutant variants (C, control in the absence of ribotoxin). The highly specific ribonuclease activity of the ribotoxins is shown by the release of the 400-nt α-fragment (arrow) from the 28S rRNA of eukaryotic ribosomes. Cell-free reticulocyte lysates were incubated in the presence of 100 ng of each protein. The reaction mixture was analyzed on 2.4% agarose gels and stained with ethidium bromide. (B) Zymograms corresponding to the ribonucleolytic activity of (WT) wild-type α-sarcin and its mutant variants, and (R) restrictocin. The amount of protein analyzed was 500 ng. The poly(A)-degrading activity of the proteins produced a nonstained region in this negative-staining analysis.
Protein–lipid interaction
It is well documented that α-sarcin interacts with lipid model vesicles containing acid phospholipids through electrostatic and hydrophobic interactions. Regarding this specific requirement for acid phospholipids, it is important to mention that some tumor cells express much more acid phospholipids in the membrane outer leaflet than their nontumor counterparts (Utsugi et al. 1991), and α-sarcin was discovered because of its antitumor action (Olson et al. 1965). Although α-sarcin is a basic protein, its effect on membranes cannot be explained simply by charge interaction. This charge interaction has been also observed for other basic proteins like bovine seminal ribonuclease, a homodimeric basic protein (pI, 10.4). The native and the monomer form are active ribonucleases, but only the dimeric protein presents antitumor activity and destabilizes negatively charged membranes (Mancheño et al. 1994b). The interaction of α-sarcin with membranes occurs through a complex process that can be dissected in three steps: vesicle aggregation, mixing of phospholipids from different bilayers, and leakage of intravesicular aqueous contents. Stopped-flow analysis (Mancheño et al. 1994a) has revealed that one of the first steps of vesicle aggregation is the formation of a vesicle dimer maintained by protein molecules through essentially electrostatic interactions. This initial step occurs on a scale of milliseconds, but vesicle aggregation continues to form large aggregates of complex structure on a scale of minutes. Vesicle aggregates scatter more light than unaggregated vesicles, and the process can be followed by measurement of the resulting apparent absorbance at 360 nm of a reaction mixture. The membranes are perturbed by the protein within the vesicle aggregates, and further mixing of phospholipids from different bilayers occurs. This process can be followed by a resonance energy transfer assay by use of two fluorescent phospholipid probes (donor and acceptor) incorporated into the membranes. The assay is performed with a mixture of fluorescence-labeled and unlabeled vesicles (1 : 9 ratio, labeled to unlabeled, respectively). The lipid mixing results in dilution of the two phospholipid probes and consequently in decrease of the fluorescence energy transfer. This decrease can reach a maximum value (infinite dilution of the probes in the assay) although mixing of lipids continues. As consequence of this perturbation promoted by the protein in the membrane, its permeability is altered, and leakage of the intravesicular aqueous content occurs. This effect can be measured by co-encapsulation of a fluorescence probe and its specific collisional quencher. Leakage results in dilution of both molecules into the extravesicular medium, and the fluorescence quenching decreases.
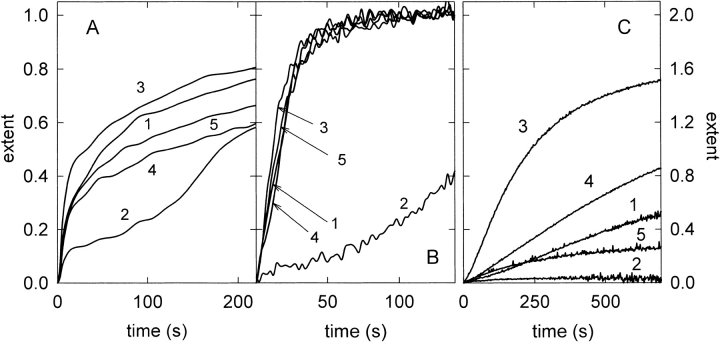
From a general point of view, vesicle-interacting proteins can produce either simply vesicle aggregation or leakage without vesicle aggregation (e.g., proteins promoting the formation of membrane pores) or lipid mixing between aggregated vesicles without or with leakage of aqueous contents. α-Sarcin promotes aggregation, lipid mixing and leakage of aqueous content of its target vesicles. The native protein and its three mutants bound to phosphatidylglycerol-containing vesicles to the same extent, saturation being reached at the same protein/lipid ratio in all the cases. However, significant differences in their vesicle-interacting ability were observed. All three promoted vesicle aggregation and lipid-mixing but only T20D and K11L/T20D produced leakage of intravesicular contents (Fig. 5 ▶). Regarding vesicle aggregation, all the studied proteins promoted a similar final variation of the apparent absorbance at 360 nm. However, significant differences were observed on comparison of the kinetic traces (Fig. 5 ▶) and initial rates (Table 3) of the absorbance variation. T20D displayed a higher initial rate of vesicle aggregation than that observed for the wild-type protein, whereas K11L showed a lower initial rate; the double mutant form behaved essentially as the average of the two single mutants. The variation of the apparent absorbance promoted by K11L displayed a biphasic behavior (Fig. 5A ▶) that may be related to the formation of a critical vesicle aggregate that further evolves to larger aggregates. The different behavior of the K11L mutant was also observed when lipid mixing was analyzed (Fig. 5B ▶). The measured lipid mixing reached the same final extent for all the studied proteins, although the process was slower for the K11L mutant, suggesting that phospholipid mixing occurs only within the above-mentioned critical vesicle aggregate. Furthermore, the K11L mutant did not perturb the permeability barrier of the bilayer enough to produce leakage of intravesicular contents (Fig. 5C ▶). The T20D mutant otherwise displayed a higher initial rate for lipid mixing and leakage, and the final extent of this later process was double that produced by the wild-type protein (Table 3; Fig. 5B,C ▶). Although restrictocin and wild-type α-sarcin showed similar behavior regarding vesicle aggregation and lipid-mixing, distinct differences were observed in their permeabilizing abilities (Fig. 5C ▶ and Table 3).
Fig. 5.
Effect of wild-type α-sarcin (1), K11L (2), T20D (3), K11L/T20D (4), and restrictocin (5) on phospholipid vesicle aggregation (A), lipid-mixing (B), and leakage of aqueous contents (C). The extent is referred to the final effect of wild-type α-sarcin considered as unit in all the cases. Assays were performed at 20 : 1 phospholipid/protein saturating molar ratio.
Table 3.
Relative initial rate (rin) and final extent of vesicle aggregation, lipid-mixing and leakage of intravesicular aqueous contents promoted by wild-type α-sarcin (WT), its three studied mutant variants and restrictocin on acid vesicles composed of phosphatidylglycerol, at saturating lipid/protein molar ratiosa
| Aggregation | Lipid-mixing | Leakage | ||||
| Protein | (rin) | Extent | (rin) | Extent | (rin) | Extent |
| WT | 1 | 1 | 1 | 1 | 1 | 1 |
| K11L | 0.4 | 0.8 | 0.1 | 1 | 0 | 0 |
| T20D | 1.5 | 1 | 1.5 | 1 | 4.9 | 2 |
| K11L/T20D | 0.9 | 0.9 | 0.7 | 1 | 1.4 | 1.7 |
| Restrictocin | 0.8 | 1 | 1.2 | 1 | 1.2 | 0.5 |
Results are expressed as relative values of those corresponding to WT considered as unit.
Average values from three different determinations.
a SD ± 0.07 for rin and SD ± 0.05 for extent.
Cytotoxicity activity
α-Sarcin has been reported to be cytotoxic for different human tumor cell lines including RD cells (from human rhabdomyosarcoma; Turnay et al. 1993). These cells were used to assay the cytotoxic activity of the studied proteins. Wild-type α-sarcin and its T20D and K11L/T20D variants exhibited similar IC50 values (concentration required for 50% protein biosynthesis inhibition), 0.6, 0.7, and 0.8 μM, respectively, whereas the values obtained for K11L and restrictocin were two- (1.5 μM) and threefold (2.0 μM) increased, respectively.
Discussion
Structural and spectroscopical characterization
Wild-type α-sarcin and the three mutants showed the same secondary and tertiary structural arrangement according to their near- and far-UV CD spectra. The substitutions engineered produce small, although significant, variations in the local environment of the aromatic chromophores as deduced from the spectroscopical properties of the mutant variants (Table 1). This result could be explained on the basis of the known structure of α-sarcin. In fact, the fluorescence properties of native α-sarcin are strongly dominated by the Trp 4 emission (de Antonio et al. 2000), a residue located at the amino-terminal β-hairpin. Moreover, Tyr 18 and Tyr 25 are also in the vicinity of the changed residues. In addition, Lys 11 is involved in the formation of a salt bridge with Glu 140 (loop 5), Tyr 18 displaying a π-cation interaction with Lys 139 (loop5), which also forms a salt bridge with Asp 9 (Pérez-Cañadillas et al. 2000).
It is also clear from the results presented that the Tm of wild-type α-sarcin is much higher (about 10°C) at pH 5.0 than at pH 7.0 (Table 2). The individual pKa values of most of the α-sarcin ionizable groups in the pH range of 3.0–8.5 have been determined (Pérez-Cañadillas et al. 1998). Therefore, it is tempting to assign this Tm increment to certain residues titrating within the pH 5.0 to pH 7.0 range. Inspection of the three-dimensional structure of α-sarcin reveals that His 36 (pKa of 6.8) and His 104 (pKa of 6.5) form surface salt bridges with Asp 102 and Asp 105, which display altered pKa values (Pérez-Cañadillas et al. 1998, 2000). It has even been proposed that these salt bridges would contribute to the global stability of the protein (Pérez-Cañadillas et al. 2000). Indeed, Glu 31, His 35, and His 36, located in the single α-helix of α-sarcin, form a group of titrable amino acids on the basis of their spatial proximity. This group of residues also titrate in the pH 5.0 to pH 7.0 range (Pérez-Cañadillas et al. 1998). The increased Tm at pH 5.0 for wild-type α-sarcin can be explained in terms of a higher degree of protonation for these His residues, which favors the formation of the salt bridges. All the residues mentioned are located in loop 3 or in the helix, in regions far away from the amino-terminal β-hairpin, which explains why this pH-dependent Tm increment is very similar for the three mutants; that is, the mutations do not affect the ionization equilibrium of the groups responsible for the increased stability at acid pH.
The additive character of the effects of the mutations studied on the Tm values suggests these changes are independent, presumably promoting only local structural changes. Loop 5 of α-sarcin (residues 139–143) connects the last two strands of the central β-sheet and establishes many interactions with other parts of the protein (Fig. 6 ▶). In particular, Glu 140 has unusual backbone torsional angles to maintain the unique conformation adopted by this loop (Pérez-Cañadillas et al. 2000). It has been proposed that mutating this Glu to Gly would stabilize α-sarcin (Pérez-Cañadillas et al. 2000). The salt bridge between Lys 11 and Glu 140 mentioned above would contribute to maintaining the unusual conformation of Glu 140. This salt bridge is not present in the K11L variant, and its increased stability would reflect the release of conformational tension. The T20D substitution has a destabilizing effect in both single and double mutant variants. In this regard, positions Glu 9 and Asp 20 are very close in the three-dimensional structure of α-sarcin (4.4 Å is the distance between the two Cαs; Fig. 6 ▶). Therefore, the presence of the negative charge in the T20D mutant might result in destabilization of the protein structure by charge repulsion between Glu 9 and Asp 20. In addition, the sequence around this position is highly charged (17-Lys-Tyr-Glu-Thr-Lys-Arg-22) in α-sarcin and the presence of Asp 20 in the mutants may also explain the decreased stability of the corresponding variants. As mentioned above, the structure of the amino-terminal region of restrictocin is not known and, therefore, it is not possible to assign this effect to a particular residue or group of residues.
Fig. 6.
Diagrams corresponding to the three-dimensional structure of α-sarcin (A) and details of the amino-terminal β-hairpin (B, C). Positively charged residues of loop 2 (from Lys 64 to Lys 89) and amino-terminal β-hairpin (from Lys 11 to Arg 22) are shown in the diagram in D. Images were generated by the molmol program (Koradi et al. 1996).
Ribonucleolytic activity
Resolution of the three-dimensional structures of α-sarcin and restrictocin has allowed modeling of the docking of these proteins and their SRL substrate (Yang and Moffat 1996; Campos-Olivas et al. 1996, Pérez-Cañadillas et al. 2000). According to these analyses, the extraordinary high substrate specificity of α-sarcin would be based on the existence of two regions interacting with SRL RNA that are separated by >11 Å (Pérez-Cañadillas et al. 2000). One would be the lysine-rich region in loop 3 (residues 110–114) responsible for the interaction with the bulged G. This prediction seemed to be confirmed by the fact that deletion of the corresponding loop in mitogillin created a mutant that failed to recognize and cleave the SRL RNA (Kao and Davies 1999). The other region would be loop 5, which, in the model, is in close proximity to the RNA GAGA loop that is actually cleaved by the protein. The fact that the three α-sarcin mutants studied retain their capacity to specifically cleave the ribosomes suggests that the regions involved in SRL RNA recognition are not affected by the mutations, in agreement with the docking model, which suggested that loops 3 and 5 are responsible for such recognition (Fig. 4A ▶).
The nonspecific activity of α-sarcin seems to be slightly affected by the mutations (Fig. 4B ▶). Substitution of Lys 11 with Leu yielded an enzyme displaying a lower activity against poly(A) whereas the T20D activity was increased. These results agree with the fact that α-sarcin and restrictocin display different activity against poly(A) although they behave very similarly in ribosome cleavage (Fando et al. 1985). The enzymatic efficiency of α-sarcin would be dependent on the interactions between the catalytic His residue, loop 5, and the amino-terminal β-hairpin on the basis of comparison with the nontoxic RNase T1 (Pace et al. 1991; Noguchi et al. 1995; Pérez-Cañadillas et al. 2000). α-Sarcin His 137 is one of the two catalytic residues playing a general acid–base role during the ribonucleolytic action (Pérez-Cañadillas et al. 1998; Lacadena et al. 1999). It is deeply buried in a special environment as revealed by its anomalously low pKa of 5.8, because loop 5 folds toward the active center (Pérez-Cañadillas et al. 1998). In RNase T1, the loop folds in the opposite direction, and the catalytic His 92 is in an exposed environment exhibiting a higher pKa (Inagaki et al. 1981; Steyaert et al. 1990). In addition, the side chain of α-sarcin His 137 is hydrogen bonded to the carbonyl oxygen of Gly 143 (loop 5) although this hydrogen bond is not formed in RNase T1. The active site of α-sarcin is connected with the amino-terminal β-hairpin via the salt bridge established between Lys 11 and Glu 140 (Pérez-Cañadillas et al. 2000). Thus, the catalytic properties of ribotoxins against nonspecific substrates would be dependent on interactions involving some residues of the amino-terminal hairpin of these proteins. Disrupting these interactions by introducing single mutations in the amino-terminal region could affect the environment and/or accessibility of His 137, and consequently the nonspecific activity of α-sarcin. A similar proposal was made (Kao and Davies 1999, Kao and Davies 2000) on the basis of analysis of deletion mutants in different regions of mitogillin. Deletions comprising stretches between residues 13 and 23 (corresponding to 14 and 24 in α-sarcin) gave rise to mitogillin variants with elevated nonspecific ribonucleolytic activity, which is in good agreement with the higher activity of T20D against poly(A) observed in our study (Fig. 4B ▶), even considering that, in our case, only single amino acid changes have been made.
Interaction with lipids
α-Sarcin is a highly polar (Martínez del Pozo et al. 1988; Pérez-Cañadillas et al. 2000), water soluble protein that binds to and crosses membranes. The effect of this protein on phospholipid vesicles as well as the conditions required for this interaction to be established have been well documented (Gasset et al. 1989, 1990, 1994; Mancheño et al. 1994a, b, 1998; Oñaderra et al. 1993, 1998). It was predicted that the α-sarcin central β-sheet defined a hydrophobic core that could contribute to the interaction with membranes (Mancheño et al. 1995a). This prediction seemed to be confirmed in the light of the results obtained with different peptides containing sequences corresponding to some of the β-strands that conform this region (Mancheño et al. 1995b, 1998). Apart from these experiments, no other data have been reported concerning the participation of any ribotoxin structural element in their interaction with phospholipids, and very little is known about the protein structural determinants that would mediate this interaction. All the ribotoxin mutants studied so far either behaved like the wild-type protein (Lacadena et al. 1995; de Antonio et al. 2000) or had not been tested against phospholipid vesicles (Kao and Davies 1995, Kao and Davies 1999; Nayak and Batra 1997; Kao et al. 1998; Lacadena et al. 1999).
Inspection of the α-sarcin structure reveals that its surface is highly charged, with some regions within which positive charges concentrate (Fig. 6D ▶; Pérez-Cañadillas et al. 2000). This feature could explain its preference for negative phospholipids (Gasset et al. 1989, 1990). But, it is not easy to draw any other conclusion about the regions of the molecule that could be responsible for its specific passage across membranes. Yang and Moffat (1996) suggested that loop L3 (corresponding to loop 2 in α-sarcin nomenclature) would be involved in the membrane fusion process induced by α-sarcin (Gasset et al. 1990), but no experimental confirmation of this hypothesis has been obtained.
The results presented here suggest that the region around the 11th and 20th residues might be involved in the interaction of α-sarcin with lipid vesicles (Fig. 6A–C ▶). Substitution of K11 with Leu resulted in a decreased ability of the protein to perturb the bilayers whereas substitution of Thr 20 with Asp resulted in increased initial rates of perturbation in spite of the net positive charge decreases. The side chains of Thr 20 and Asp 9 are near in the three-dimensional structure of wild-type α-sarcin (4.4 Å between Cαs). The presence of Asp at position 20 in the T20D variant would result in electrostatic repulsion, that may change the orientation of the side chains of Lys 21 and Arg 22, favoring potential electrostatic interactions with the acidic phospholipids through Lys 11, Lys 14, and Lys 17 (which are located in the highly exposed protuberance, residues 9–20, of the amino-terminal β-hairpin), thus explaining the increased initial rates of perturbation of this mutant variant. Substitution of Lys 11 with Leu would reduce this effect.
Vesicle aggregation induced by the K11L mutant follows a biphasic kinetics (Fig. 5A ▶). This observation suggests the formation of an intermediate protein–vesicle complex that further evolves to a large aggregate. The initial step of vesicle aggregation is the formation of a vesicle dimer maintained by electrostatic interactions. This dimer is favored when the vesicle-interacting protein displays two lipid binding regions. A decreased affinity in one of these regions requires a higher concentration of an intermediate protein–vesicle complex to form large aggregates. One of the lipid-interacting regions of α-sarcin has been related to Lys and Arg residues of loop 2 (surface accessibilities >50%). In fact, Trp 51, close to this loop, is located in the hydrophobic core of the bilayer upon protein–vesicle interaction as its fluorescence is quenched by anthracene incorporated in the core of the membrane (de Antonio et al. 2000). Residues from the amino-terminal β-hairpin, located in the opposite side of the α-sarcin molecule with respect to loop 2, might contribute to the second lipid-binding region. The K11L change would decrease the affinity for electrostatic interaction with vesicles thus explaining the observed vesicle aggregation kinetics (Fig. 5A ▶).
Cytotoxicity
The observed differences in lipid-interacting ability of the studied proteins correlate well with their cytotoxic effect. Restrictocin and the K11L mutant variant are the least effective in inhibiting both cellular protein biosynthesis and ability to perturb the phospholipid bilayer. These observations strongly suggest the involvement of the amino-terminal β-hairpin in the interaction with membranes and the notion that the cytotoxicity of ribotoxins is related to both ribonuclease and lipid-interacting activities.
Conclusions
The results show that a single amino acid substitution in α-sarcin can alter its cytotoxicity without altering the overall structural folding or the activity against ribosomes. It is clear from these experiments that the amino-terminal β-hairpin of ribotoxins plays a role in their ability to penetrate different cells. Its absence from other nontoxic RNases, such as T1 and U2, could explain, in part, why they are not cytotoxic. Furthermore, this amino-terminal stretch is one of the regions of the molecule that shows a high degree of sequence variation among all known ribotoxins (Wirth et al. 1997; Martínez-Ruiz et al. 1999a, b), resulting in varying cytotoxicity.
Materials and methods
DNA manipulations
All materials and reagents were molecular biology grade. Cloning procedures and bacteria manipulations were carried out according to standard methods (Sambrook et al. 1989) as described previously (Lacadena et al. 1994, 1995, 1999). Oligonucleotide site-directed mutagenesis was used to replace Lys 11 by Leu (K11L) and Thr 20 by Asp (T20D) as described previously (Kunkel et al. 1987; Lacadena et al. 1994, 1995, 1999). To obtain the double mutant, a second mutagenic round was performed with T20D DNA as template. The mutagenic primers used were 5′-TTGAACGACCAGCTGAACCCCAAGACC-3′ for K11L and 5′-AACAAGTATGAGGACAAACGCCTCCTC-3′ for T20D (the site of mutation is underlined). The E. coli strains used were BW313 {(HfrKL16 pol45 [LysA (61–62)] dut1 ung1 thi1 relA } to obtain the uridine-rich ssDNA, DH5αF′({[F′] endA1 hsdR17 (r−K m−K) supE44 thi-1 recA1 gyrA (NaIR) relA1 Δ(lacZYA-argF) U169 deoR [∅80 dlac Δ(lacZ) M15]}) for the expression constructs, and BL21(DE3)(F′ ompT [lon] hsdB (r−B m−B)) for protein production. The thioredoxin-producing plasmid (pT-Trx; Yasukawa et al. 1995) was a generous gift of Dr. S. Ishii, from the Riken Tsukuba Life Science Center (Japan).
Protein production and purification
BL21(DE3) cotransformed with pT-Trx and the corresponding α-sarcin mutant plasmid were used to produce and purify the mutants, as described for the wild-type protein (Lacadena et al. 1994; García-Ortega et al. 2000). Wild-type α-sarcin and restrictocin were produced and purified according to methods reported previously (Olson and Goerner 1965; Gavilanes et al. 1983; Lacadena et al. 1994). Polyacrylamide gel electrophoresis of proteins, protein hydrolysis, and amino acid analysis were also performed according to standard procedures (Gavilanes et al. 1983; Lacadena et al. 1994).
Spectroscopic characterization
Absorbance measurements were carried out with a Uvikon 930 spectrophotometer at 100 nm/min scanning speed, at room temperature, and in 1-cm optical-path cells. CD spectra were obtained on a Jasco 715 spectropolarimeter at 0.2 nm/sec scanning speed; 0.1- and 1.0-cm optical-path cells were used for the far- and near-UV, respectively. Mean residue weight ellipticities were expressed in units of degree ×cm2/dmole. Extinction coefficients E(0.1%, 1 cm, 280 nm) were calculated from their absorbance spectra and amino acid analyses. Thermal denaturation profiles were obtained by measurement of the temperature dependence of the ellipticity at 220 nm in the range of 25°C–85°C; the temperature was continuously changed at a rate of 0.5°C/min. Tm values were calculated assuming a two-state unfolding mechanism. Fluorescence emission spectra were obtained on an SLM Aminco 8000 spectrofluorimeter at 25°C and in 0.2-cm optical-path cells, as described previously (Gavilanes et al. 1983; Lacadena et al. 1999).
Ribonucleolytic activity
The specific ribonucleolytic activity of α-sarcin was followed by detection of the release of the 400-nt α-fragment (Schindler and Davies 1977; Endo and Wool 1982; Endo et al. 1983) from a cell-free reticulocyte lysate (Promega) (Lacadena et al. 1994, 1995, 1999). The production of the 400-nt α-fragment was visualized by ethidium bromide staining after electrophoresis on a 2.4% agarose gel. The amount of α-fragment was estimated from the ethidium bromide-stained gel with the photo documentation system UVI-Tec and the software facility UVIsoft UVI band Windows Application V97.04. The residual 16S rRNA was used as reference for normalization. The activity of the purified proteins against poly(A) was assayed at pH 4.5 in 15% polyacrylamide gels containing 0.1% SDS and 0.3 mg/mL of the homopolynucleotide. This zymogram method was based on one described previously (Blank et al. 1982; Lacadena et al. 1994, 1995; Kao and Davies 1995, Kao and Davies 1999, Kao and Davies 2000). Proteins exhibiting ribonuclease activity appear as colorless bands after appropriate destaining. Volumograms (density or quantity of a spot calculated from its volume made of the sum of all pixel intensities composing the spot) of these bands (obtained with the photo documentation system described above) were also used to quantify the activity. All assays were performed with controls to test potential nonspecific degradation of the substrates, which does not occur under the conditions used.
Protein–lipid interactions
All phospholipids used were purchased from Avanti Polar Lipids Inc. Vesicles were formed by hydration of a dry lipid film with Tris buffer (15 mM Tris [pH 7.0] containing 0.1 M NaCl and 1 mM EDTA) for 60 min at 37°C. The lipid suspension was subjected to five cycles of extrusion through two stacked 0.1 μm (pore diameter) polycarbonate membranes (Mancheño et al. 1994a). The average diameter of the vesicle population was 100 nm (85% of the vesicles in the range 75–125 nm), as determined by electron microscopy studies (Mancheño et al. 1994a). Phospholipid concentration was determined as described (Barlett 1959).
Aggregation of phospholipid vesicles was monitored as described before (Gasset et al. 1989) by measurement of the increase in the absorbance at 360 nm of a suspension of phosphatidylglycerol vesicles in Tris buffer (30 μM final lipid concentration) after addition of a small aliquot of a freshly prepared solution of protein. Intermixing of membrane lipids was analyzed by considering fluorescence energy transfer assays (Struck et al. 1981) as described (Mancheño et al. 1994a, b). A decrease in the donor-to-acceptor fluorescence energy transfer indicates lipid-mixing between membranes. Leakage of vesicle aqueous contents was measured by use of the 8-aminonaphthalene-1,3,6-trisulfonic acid/N,N′-p-xylenebispyridinium bromide assay also as described previously (Mancheño et al. 1995b, 1998). Binding of the proteins to the lipid vesicles was analyzed by measurement of the free protein concentration in the supernatant obtained by centrifugation (160000g/60 min, Beckman Airfuge) of protein–vesicle mixtures at different protein/lipid ratios. This protein concentration was calculated by analysis with a photo documentation system of the Coomassie-stained SDS gels resulting from subjecting supernatant aliquots to SDS–PAGE. A calibration plot, volumogram of the protein band (obtained as described above) versus protein concentration (determined by amino acid analysis of acid-hydrolyzed protein samples) was used for these calculations. Other experimental details were as reported previously (Gasset et al. 1989, 1990, 1994; Mancheño et al. 1994a, b, 1998; Oñaderra et al. 1993, 1998).
Cytotoxicity assay
This assay was performed essentially as described (Turnay et al. 1993) by use of RD (human rhabdomyosarcoma) cells. Protein synthesis was analyzed by measurement of the incorporation of l-[4,5–3H]leucine (166 Ci/mmole). The radioactivity was measured in a Beckman LS 3801 liquid scintillation counter. The results are expressed as percentage of incorporation in control samples. A plot of these percentage values versus toxic protein concentration in the cytotoxicity assay allows the calculation of the IC50 values. Those values reported correspond to the average of triplicate experiments.
Acknowledgments
This work was supported by grants BMC2000–0551 from the Ministerio de Ciencia y Tecnología and PB98–0083 from the Ministerio de Educación y Cultura (MEC) (Spain) and Natural Sciences and Engineering Research Council (Canada; R.K., J.D.). L.G.-O. is recipient of a fellowship from the MEC (Spain).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.9601
References
- Barlett, G.R. 1959. Colorimetric assay methods for free and phosphorylated glyceric acids. J. Biol. Chem. 234 466–468. [PubMed] [Google Scholar]
- Becktel, W.J. and Schellman, J.A. 1987. Protein stability curves. Biopolymers 26 1859–1877. [DOI] [PubMed] [Google Scholar]
- Blank, A., Sugiyama, R.H., and Dekker, C.A. 1982. Activity staining of nucleolytic enzymes after sodium dodecyl sulfate-polyacrylamide gel electrophoresis: Use of aqueous isopropanol to remove detergent from gels. Anal. Biochem. 120 267–275. [DOI] [PubMed] [Google Scholar]
- Campos-Olivas, R., Bruix, M., Santoro, J., Martínez del Pozo, A., Lacadena, J., Gavilanes, J.G., and Rico, M. 1996. Structural basis for the catalytic mechanism and substrate specificity of the ribonuclease α-sarcin. FEBS Lett. 399 163–165. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Munishkin, A., Chan, Y.L., Ren, Z., Wool, I.G., and Steitz, T.A. 1998. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. 95 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll, C.C., Wool, I.G., and Munishkin, A. 1999. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: The structure at 1.11 Å resolution. J. Mol. Biol. 292 275–287. [DOI] [PubMed] [Google Scholar]
- De Antonio, C., Martínez del Pozo, A., Macheño, J.M., Oñaderra, M., Lacadena, J., Martínez-Ruiz, A., Pérez-Cañadillas, J.M., Bruix, M., and Gavilanes, J.G.. 2000. Assignment of the contribution of the tryptophan residues to the spectroscopic and functional properties of the ribotoxin α-sarcin. Proteins 41 350–361. [DOI] [PubMed] [Google Scholar]
- Endo, Y. and Wool, I.G. 1982. The site of action of α-sarcin on eukaryotic ribosomes: The sequence at the α-sarcin cleavage-site in 28S ribosomal ribonucleic acid. J. Biol. Chem. 257 9054–9060. [PubMed] [Google Scholar]
- Endo, Y., Hubert, P.W., and Wool, I.G. 1983. The ribonuclease activity of the cytotoxin α-sarcin: The characteristics of the enzymatic activity of α-sarcin with ribosomes and ribonucleic acids as substrates. J. Biol. Chem. 258 2662–2667. [PubMed] [Google Scholar]
- Endo, Y., Glück, A., Chan, Y.-L., Tsurugi, K., and Wool, I.G. 1990. RNA–protein interaction: An analysis with RNA oligonucleotides of the recognition by α-sarcin of a ribosomal domain critical for function. J. Biol. Chem. 265 2216–2222. [PubMed] [Google Scholar]
- Fando, J.L., Alaba, I., Escarmis, C., Fernández-Luna, J.L., Méndez, E., and Salinas, M. 1985. The mode of action of restrictocin and mitogillin on eukaryotic ribosomes. Eur. J. Biochem. 149 29–34. [DOI] [PubMed] [Google Scholar]
- García-Ortega, L., Lacadena, J., Lacadena, V., Masip, M., De Antonio, C., Martínez-Ruiz, A., and Martínez del Pozo, A. 2000. The solubility of the ribotoxin α-sarcin, produced as a recombinant protein in Escherichia coli, is significantly increased in the presence of thioredoxin. Lett. Applied Microbiol. 30 298–302. [DOI] [PubMed] [Google Scholar]
- Gasset, M., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1989. Study of the interaction between the antitumor protein α-sarcin and phospholipid vesicles. Biochem. J. 258 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset, M., Oñaderra, M., Thomas, P.G., and Gavilanes, J.G. 1990. Fusion of phospholipid vesicles produced by the anti-tumour protein α-sarcin. Biochem. J. 265 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset, M., Mancheño, J.M., Lacadena, J., Turnay, J., Olmo, N., Lizarbe, M.A., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1994. α-Sarcin, a ribosome-inactivating protein that translocates across the membrane of phospholipid vesicles. Curr. Topics Peptide Protein Res. 1 99–104. [Google Scholar]
- Gasset, M., Mancheño, J.M., Laynez, J., Lacadena, J., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1995. Thermal unfolding of the cytotoxin α-sarcin. Phospholipid binding induces destabilization of the protein structure. Biochim. Biophys. Acta 1252 126–134. [DOI] [PubMed] [Google Scholar]
- Gavilanes, J.G., Vázquez, D., Soriano, F., and Méndez, E. 1983. Chemical and spectroscopic evidence on the homology of three antitumor proteins: α-Sarcin, mitogillin and restrictocin. J. Protein. Chem. 2 251–261. [Google Scholar]
- Inagaki, F., Kawano, Y., Shimada, I., Takahashi, K., and Miyazawa, T. 1981. Nuclear magnetic resonance study on the microenvironments of histidine residues of ribonuclease T1 and carboxymethylated ribonuclease T1. J. Biochem. 89 1185–1195. [PubMed] [Google Scholar]
- Kao, R. and Davies, J. 1995. Fungal ribotoxins: A family of naturally engineered targeted toxins? Biochem. Cell Biol. 73 1151–1159. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Molecular dissection of mitogillin reveals that the fungal ribotoxins are a family of naturally engineered ribonucleases. J. Biol. Chem. 274 12576–12582. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Single amino acid substitutions affecting the specificity of the fungal ribotoxin mitogillin. FEBS Lett. 466 87–90. [DOI] [PubMed] [Google Scholar]
- Kao, R., Shea, J.E., Davies, J., and Holden, D.W. 1998. Probing the active site of mitogillin, a fungal ribotoxin. Mol. Microbiol. 29 1019–1027. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wütrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A., Roberts, J.D., and Zakour, R.A. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154 367–382. [DOI] [PubMed] [Google Scholar]
- Lacadena, J., Martínez del Pozo, A., Barbero, J.L., Mancheño, J.M., Gasset, M., Oñaderra, M., López-Otín, C., Ortega, S., García, J.L., and Gavilanes, J.G. 1994. Overproduction and purification of biologically active native fungal α-sarcin in Escherichia coli. Gene 142 147–151. [DOI] [PubMed] [Google Scholar]
- Lacadena, J., Mancheño, J.M., Martínez-Ruiz, A., Martínez del Pozo, A., Gasset, M., Oñaderra, M., and Gavilanes, J.G. 1995. Substitution of histidine-137 by glutamine abolishes the catalytic activity of the ribosome-inactivating protein α-sarcin. Biochem. J. 309 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadena, J., Martínez del Pozo, A., Lacadena, V., Martínez-Ruiz, A., Mancheño, J.M., Oñaderra, M., and Gavilanes, J.G. 1998. The cytotoxin α-sarcin behaves as a cyclizing ribonuclease. FEBS Lett. 424 46–48. [DOI] [PubMed] [Google Scholar]
- Lacadena, J., Martínez del Pozo, A., Martínez-Ruiz, A., Pérez-Cañadillas, J.M., Bruix, M., Mancheño, J.M., Oñaderra, M., and Gavilanes, J.G. 1999. Role of histidine-50, glutamic acid-96 and histidine-137 in the ribonucleolytic mechanism of the ribotoxin α-sarcin. Proteins 37 474–484. [DOI] [PubMed] [Google Scholar]
- Mancheño, J.M., Gasset, M., Lacadena, J., Ramón, F., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1994a. Kinetic study of the aggregation and lipid-mixing produced by α-sarcin on phosphatidylglycerol and phosphatidylserine vesicles: Stopped-flow light-scattering and fluorescence energy transfer measurements. Biophys. J. 67 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancheño, J.M., Gasset, M., Oñaderra, M., Gavilanes, J.G., and D'Alessio, G. 1994b. Bovine seminal ribonuclease destabilizes negatively charged membranes. Biochem. Biophys. Res. Commun. 199 119–124. [DOI] [PubMed] [Google Scholar]
- Mancheño, J.M., Gasset, M., Lacadena, J., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1995a. Predictive study of the conformation of the cytotoxic protein α-sarcin: A structural model to explain α-sarcin-membrane interaction. J. Theor. Biol. 172 259–267. [DOI] [PubMed] [Google Scholar]
- Mancheño, J.M., Gasset, M., Albar, J.P., Lacadena, J., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1995b. Membrane interaction of a β-structure-forming synthetic peptide comprising the 116–139th sequence of the cytotoxic protein α-sarcin. Biophys. J. 68 2387–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancheño, J.M., Martínez del Pozo, A., Albar, J.P., Oñaderra, M., and Gavilanes, J.G. 1998. A peptide of nine amino acid residues from α-sarcin cytotoxin is a membrane-perturbing structure. J. Peptide Res. 51 142–148. [DOI] [PubMed] [Google Scholar]
- Martínez del Pozo, A., Gasset, M., Oñaderra, M., and Gavilanes, J.G. 1988. Conformational study of the antitumor protein α-sarcin. Biochim. Biophys. Acta 953 280–288. [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz, A., Kao, R., Davies, J., and Martínez del Pozo, A. 1999a. Ribotoxins are a more widespread group of proteins within the filamentous fungi than previously believed. Toxicon 37 1549–1563. [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz, A., Martínez del Pozo, A., Lacadena, J., Oñaderra, M., and Gavilanes, J.G. 1999b. Hirsutellin A displays significant homology to microbial extracellular ribonucleases. J. Invertebr. Pathol. 74 96–97. [DOI] [PubMed] [Google Scholar]
- Miller, S.P. and Bodley, J.W. 1988. The ribosomes of Aspergillus giganteus are sensitive to the cytotoxic action of α-sarcin. FEBS Lett. 229 388–390. [DOI] [PubMed] [Google Scholar]
- Nayak, S.K. and Batra, J.K. 1997. A single amino acid substitution in ribonucleolytic toxin restrictocin abolishes its specific substrate recognition activity. Biochemistry 36 13693–13699. [DOI] [PubMed] [Google Scholar]
- Noguchi, S., Satow, Y., Uchida, T., Sasaki, C., and Matsuzaki, T. 1995. Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 Å resolution. Biochemistry 34 15583–15591. [DOI] [PubMed] [Google Scholar]
- Olson, B.H. and Goerner, G.L.. 1965. α-Sarcin, a new antitumor agent. I. Isolation, purification, chemical composition, and the identity of a new amino acid. Applied Microbiol. 13 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, B.H., Jennings, J.C., Roga, V., Junek, A.J., and Schuurmans, D.M. 1965. α-Sarcin, a new antitumor agent. II. Fermentation and antitumor spectrum. Applied Microbiol. 13 322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñaderra, M., Mancheño, J.M., Gasset, M., Lacadena, J., Schiavo, G., Martínez del Pozo, A., and Gavilanes, J.G. 1993. Translocation of α-sarcin across the lipid bilayer of asolectin vesicles. Biochem. J. 295 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñaderra, M., Mancheño, J.M., Lacadena, J., De los Ríos, V., Martínez del Pozo, A., and Gavilanes, J.G. 1998. Oligomerization of the cytotoxin α-sarcin associated to phospholipid membranes. Mol. Membr. Biol. 15 141–144. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Heinemann, U., Hahn, U., and Saenger, W. 1991. Ribonuclease T1: Structure, function and stability. Angew. Chem. Int. Ed. Engl. 30 343–360. [Google Scholar]
- Pérez-Cañadillas, J.M., Campos-Olivas, R., Lacadena, J., Martínez del Pozo, A., Gavilanes, J.G., Santoro, J., Rico, M., and Bruix, M. 1998. Characterization of pKa values and titration shifts in the cytotoxic ribonuclease α-sarcin by NMR. Relationship between electrostatic interactions, structure and catalytic function. Biochemistry 3715865–15876. [DOI] [PubMed] [Google Scholar]
- Pérez-Cañadillas, J.M., Santoro, J., Campos-Olivas, R., Lacadena, J., Martínez del Pozo, A., Gavilanes, J.G., Rico, M., and Bruix, M. 2000. The highly refined solution structure of the cytotoxic ribonuclease α-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity. J. Mol. Biol. 299 1061–1073. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schindler, D.G. and Davies, J.E. 1977. Specific cleavage of ribosomal RNA caused by α-sarcin. Nucleic Acids Res. 4 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert, J., Hallehg, K., Wyns, L., and Stansens, P. 1990. Histidine-40 of ribonuclease T1 acts as base catalyst when the true catalyst base, glutamic acid-58, is replaced by alanine. Biochemistry 29 9064–9072. [DOI] [PubMed] [Google Scholar]
- Struck, D.K., Hoekstra, D., and Pagano, R.E.1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry 20 4093–4099. [DOI] [PubMed] [Google Scholar]
- Szewczak, A.A. and Moore, P.B. 1995. The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 247 81–98. [DOI] [PubMed] [Google Scholar]
- Turnay, J., Olmo, N., Jiménez, J., Lizarbe, M.A., and Gavilanes, J.G. 1993. Kinetic study of the cytotoxic effect of α-sarcin, a ribosome inactivating protein from A. giganteus, on tumor cell lines: Protein biosynthesis inhibition and cell binding. Mol. Cell. Biochem. 122 39–47. [DOI] [PubMed] [Google Scholar]
- Utsugi, T., Schroit, A.J., Connor, J., Bucana, C., and Fidler, I.J. 1991. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 51 3062–3066. [PubMed] [Google Scholar]
- Wirth, J., Martínez del Pozo, A., Mancheño, J.M., Martínez-Ruiz, A., Lacadena, J., Oñaderra, M., and Gavilanes, J.G. 1997. Sequence determination and molecular characterization of gigantin, a cytotoxic protein produced by the mould Aspergillus giganteus IFO 5818. Arch. Biochem. Biophys. 343 188–193. [DOI] [PubMed] [Google Scholar]
- Wool, I.G., Glück, A., and Endo, Y. 1992. Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem. Sci. 17 266–269. [DOI] [PubMed] [Google Scholar]
- Yang, X.J. and Moffat, K. 1996. Insights into specificity of cleavage and mechanism of cell entry from the crystal structure of the highly specific Aspergillus ribotoxin, restrictocin. Structure 4 837–852. [DOI] [PubMed] [Google Scholar]
- Yasukawa, T., Kanei-Ishii, C., Mackaura, T., Fujimoto, J., Yamamoto, T., and Ishii, S. 1995. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 270 25328–25331. [DOI] [PubMed] [Google Scholar]