Abstract
Homobifunctional chemical cross-linking reagents are important tools for functional and structural characterization of proteins. Accurate measures of the lengths of these molecules currently are not available, despite their widespread use. Stochastic dynamics calculations now provide quantitative measures of the lengths, and length dispersions, of 32 widely used molecular rulers. Significant differences from published data have been found.
Supplemental material: See www.proteinscience.org
Keywords: Protein cross-linking, cross-linking reagents, molecular rulers, stochastic dynamics
Bifunctional chemical cross-linking reagents and the intramolecular and intermolecular cross-linking of proteins and protein complexes have been instrumental in the structural and functional characterization of proteins. The importance of these investigations is reflected in the large and increasing arsenal of commercially available cross-linking reagents (Pierce Chemicals 1999). In the arena of structural studies, the cross-linking reagents are envisaged as molecular rulers that provide information on distances between the cross-linked amino acid residues that are pertinent to both the tertiary and quaternary arrangements of proteins (Fasold et al. 1971; Peters and Richards 1977). Such distance determinations are especially valuable in membrane proteins and other proteins that cannot be crystallized and frequently lead to important advances in mapping the protein topography (Swaney 1986; Kwaw et al. 2000). For crystallized proteins (Fasold et al. 1971) and lower resolution structures refined by computational methods (Holmes et al. 1990; Lorenz et al. 1993), the cross-linking results have provided valuable constraints for the modeling of such structures.
The most reliable information is derived from zero-length reagents that induce a direct covalent link between cross-linked sites. Disulfide and carbodiimide cross-linking reactions are probably the most popular in this category (Kunkle et al. 1986). In contrast, most bifunctional reagents introduce a bridge between the cross-linked residues and contain flexible bonds; these do not provide a rigid yardstick of interresidue distances. Nevertheless, the distances between cross-linked sites frequently are represented by a single mean value provided by the reagent manufacturer (Sun and Kaback 1997; Nagy et al. 2000). One result of using such mean cross-linking span values is the assignment of incorrect distances to the linked pairs of residues. In studies designed to probe macromolecular flexibility by using homologous series of bifunctional reagents, the dynamic features of the protein may be misrepresented if the flexibility of the reagents is not accounted for (Kliche et al. 1999). Despite the many uses of these substances, an accurate description of the cross-linking span of the bifunctional reagents is not currently available. In this study, we have used stochastic molecular dynamics techniques to provide a more realistic quantification of the lengths of 32 popular homobifunctional reagents. The data provide more accurate information on the structure and dynamics of proteins probed with these reagents. The dynamics of both the protein and the cross-linking reagent will have an influence on the rates of cross-linking. Our work deals with the most critical feature of how easy it is for the reagent to span various distances. Even then there will be some variation in the ability of various reagents to form cross-links with proteins containing diverse functional group orientation. These issues of orientation are not addressed in our considerations, and we only focus on the dynamic range of the reagents.
Computational methodology
Stochastic dynamics simulations are able to efficiently and completely sample the conformational space of a molecule without being limited to the local minimum in the region of the starting structure. The method simulates the effect of solvent on the conformation of small molecules by including the effects of random collisions with solvent and by introducing a frictional drag component (Leach 1996).
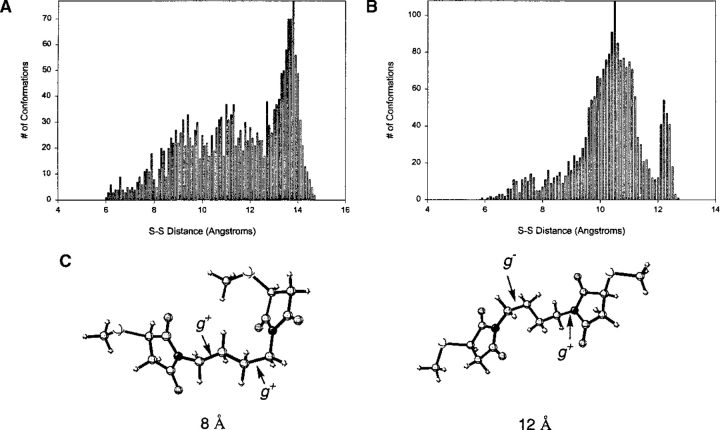
Each cross-linking reagent was fully minimized using the AMBER* force field (Mohamadi et al. 1990) as implemented in the Macromodel program. All simulations were performed at 298K. After a 50-psec equilibration period, each structure then was subjected to stochastic dynamics for 2.5 nsec using a generalized Born/solvent accessible (GB/SA) water solvation model (Still et al. 1990). The time step used was 1.5 femtosecond (fs). Every picosecond, a sample structure was taken, and the pertinent cross-linking span was measured. To ensure that there was adequate sampling, we plotted the ensemble of distances from each simulation as a function of time (Fig. 1B ▶). The frequency of occurrence for each distance during the simulation also was plotted, providing distribution statistics (Fig. 1A ▶). Statistics were collected at several time intervals to ensure that sufficient time was allowed to achieve equilibrium. The plots for oPDM are shown in Figure 1 ▶ as a representative example. Data of this type for all the cross-linking reagents studied here are given at www.proteinscience.org.
Fig. 1.
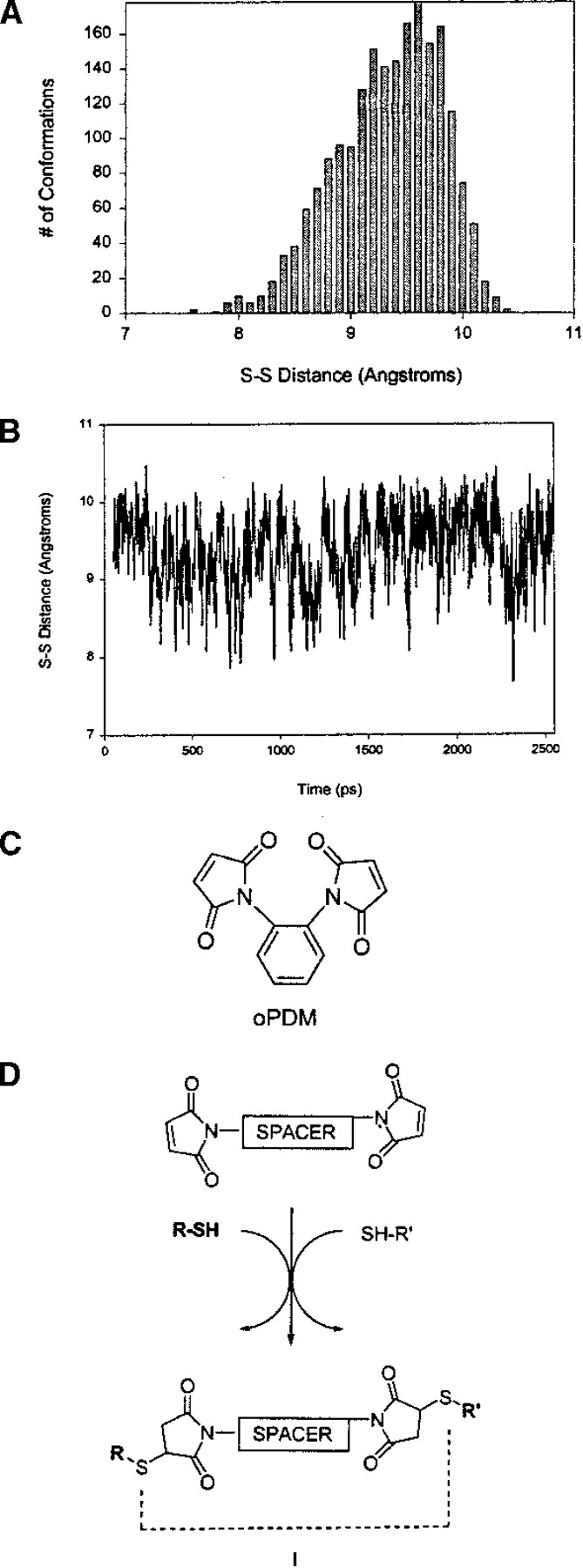
N,N′-1,2-phenylenedimaleimide (oPDM). (A) Frequency vs. cross-linking span for oPDM; (B) cross-linking span vs. time for oPDM; (C) structure of oPDM; (D) reaction of oPDM and other bis-maleimide cross-linking reagents with sulfhydryl groups. l is the S-S distance and the cross-linking span of the reagent. (oPDM) N,N′-1,2-phenylenedimaleimide.
In several of the cases studied, the effect of attaching one end of the cross-linker to a protein was estimated by conjugating the reagent to a molecule of pentaiodobenzene thiolate. This heavy, but sterically unhindering, group increases the time taken to achieve equilibrium. With sufficient calculation times, however, the conjugated and unconjugated molecules obtained the same distribution statistics. Because the conformational changes in the reagents occur on the nanosecond time scale, and most cross-linking reactions occur on the order of seconds, attachment of a heavy group free of steric encumbrance should not change the effective length of these molecular rulers.
Results and Discussion
All of the cross-linkers in the study are reactive with either amines or sulfhydryl groups. They can be divided into five different series based on the type of reagent and their reactivity. The first two series are sulfhydryl-reactive molecules. This first group reacts by alkylation chemistry, whereas the second cross-links via disulfide exchange. The remaining categories represent amine-reactive chemicals. They can react with either the α-amines at the N terminus or the ɛ-amines of lysine residues to give either stable amide or amidine linkages.
The statistics for each reagent are compiled in Tables 1–5. Every distance (in angstroms) obtained from the simulation was used to compute the statistics. In each table, the average distance between S or N atoms attached to the linker molecule, the standard deviation of this value, the mode—or most probable distance, median distance, and the range of all distances obtained in a simulation are given and compared with the commonly cited distances. The cited distances for mPDM, oPDM, pPDM, BM, and NDM were obtained from force field energy minimizations as described by Nitao and Reisler (1998), whereas the distances for the C-6 through C-9 disulfides were determined by Faulstich (Kliche et al. 1999). Cross-linking spans for all the remaining molecules studied were obtained from the Pierce Chemicals Double Agents Cross-Linking Reagents Selection Guide published in 1999 (Pierce Chemicals 1999).
Table 1.
Series 1: SH reactive Alkylation-based reagents
| Cross-linking reagent | Average S-S distance | Standard deviation | Mode | Median S-S distance | Other major modes | Range of S-S distances | Cited S-S distancea |
| mPDM | 10.65 | 0.55 | 10.5 | 10.67 | — | 8.84–11.87 | 9.6–11.5 |
| oPDM | 9.39 | 0.47 | 9.6 | 9.44 | — | 7.67–10.47 | 5.2–7.8 |
| pPDM | 11.13 | 0.52 | 11.4 | 11.19 | — | 9.20–12.29 | 12.1–12.4 |
| BM | 14.53 | 1.51 | 15.0 | 14.80 | — | 9.40–17.34 | 14.9–15.4 |
| NDM | 12.31 | 0.47 | 12.3 | 12.35 | — | 10.33–13.52 | 12.4–12.9 |
| BMOE | 8.18 | 0.75 | 7.8 | 8.09 | — | 6.27–10.52 | 8.0 |
| BMB | 10.44 | 2.04 | 12.0 | 11.00 | 8.0 | 4.52–14.14 | 10.9 |
| BMDB | 11.99 | 0.52 | 12.0 | 12.06 | — | 9.48–13.18 | 10.2 |
| BMH | 10.16 | 2.41 | 11.5 | 10.55 | — | 3.47–15.64 | 16.1 |
| DTME | 12.43 | 1.60 | 13.6 | 12.61 | — | 6.68–16.12 | 13.3 |
| BM[PEO]3 | 8.83 | 1.81 | 9.0 | 8.83 | — | 3.51–14.26 | 14.7 |
| BM[PEO]4 | 9.51 | 2.34 | 9.2 | 9.39 | — | 3.51–16.56 | 17.8 |
(mPDM) N,N′1,3-phenylenedimaleimide; (oPDM) N,N′-1,2-phenylenedimaleimide; (pPDM) N,N′-1,4-phenylenedimaleimide; (BM) N,N′-(methylene-4-1-phenylene)bismaleimide; (NDM) naphthalene-1,5-dimaleimide; (BMOE) bismaleimidoethane; (BMB) 1,4-bismaleimidobutane; (BMDB) 1,4-bismaleimidyl-2,3-dihydroxybutane; (BMH) 1,6-bismaleimidohexane; (DTME) dithio-bis-maleimdoethane; (BM[PEO]3) 1,8-bis-maleimidotriethyleneglycol; (BM[PEO]4) 1,11-bis-maleimidotetraethyleneglycol.
a For mPDM, oPDM, pPDM, BM, and NDM, see Nitao and Reisler (1998). All others, see Pierce Chemicals (1999).
Table 2.
Series 2: Sh-reactive disulfide exchange reagents
| Cross-linking reagent | Average S-S distance | Standard deviation | Mode | Median S-S distance | Other major modes | Range of S-S distances | Cited S-S distance |
| C-3 disulfide | 6.87 | 1.35 | 7.8 | 7.3 | 4.3, 5.9 | 3.90–8.90 | 8.6 |
| C-4 disulfide | 8.07 | 1.42 | 9.7 | 8.2 | 8.2 | 4.10–10.30 | 9.9 |
| C-6 disulfide | 10.37 | 1.24 | 10.4 | 10.50 | 12.3 | 5.95–12.70 | 12.5 |
| C-7 disulfide | 11.61 | 1.40 | 12.5 | 12.04 | — | 6.89–14.10 | 13.8 |
| C-8 disulfide | 11.50 | 2.09 | 13.8 | 11.75 | — | 6.08–14.71 | 15.1 |
| C-9 disulfide | 12.24 | 2.02 | 12.2 | 12.37 | — | 6.73–16.16 | 16.4 |
| DPBDP | 14.65 | 1.44 | 15.7 | 14.88 | — | 9.29–18.20 | 19.9 |
The activated disulfides are the C-3, C-4, and C-6 through C-9 disulfides and 1,4-di-(3′-[2-pyridyldithio]-propionamido) butane (DPDPB).
a See reference Pierce Chemicals (1999).
Table 3.
Series 3: NH3 reactive imidoester class of cross-linking reagents
| Cross-linking reagent | Average N-N distance | Standard deviation | Mode | Median N-N distance | Other major modes | Range of N-N distances | Cited N-N distancea |
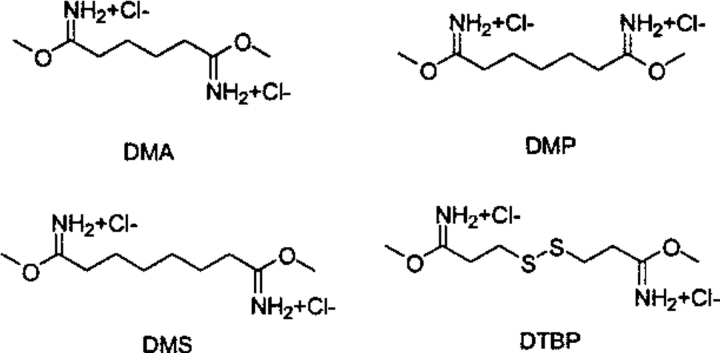
| DMA | 7.40 | 0.58 | 7.4 | 7.47 | — | 4.03–8.99 | 8.6 |
| DMP | 8.36 | 0.90 | 8.5 | 8.47 | — | 4.46–10.29 | 9.2 |
| DMS | 9.39 | 1.03 | 9.5 | 9.59 | — | 6.82–11.60 | 11.0 |
| DTBP | 8.41 | 1.05 | 8.3 | 8.54 | — | 3.93–11.07 | 11.9 |
(DMA) dimethyl adipimidate; (DMP) dimethyl pimelimidiate; (DMS) dimethyl suberimidate; (DTBP) dimethyl 3,3′-dithiobis-propionimidate.
a See reference Pierce Chemicals (1999).
Table 4.
Series 4: NH3 reactive N-hydroxysuccinimide esters
| Cross-linking reagent | Average N-N distance | Standard deviation | Mode | Median N-N distance | Other major modes | Range of N-N distances | Cited N-N distancea |
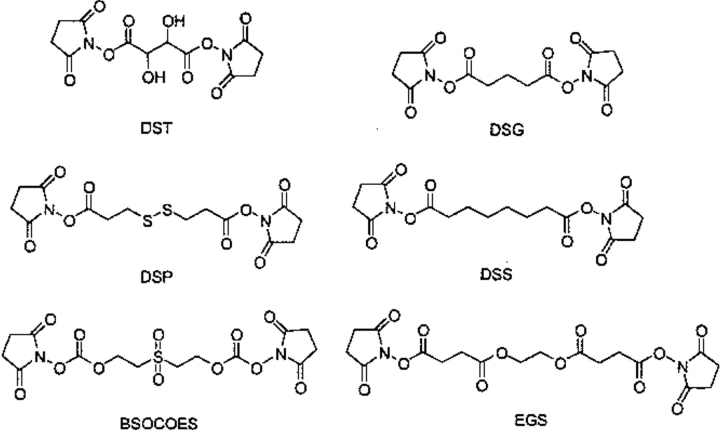
| DSG | 6.22 | 0.68 | 6.4 | 6.35 | — | 3.12–7.49 | 7.7 |
| DSP | 8.04 | 1.08 | 8.1 | 8.11 | — | 4.63–10.65 | 12.0 |
| DSS | 8.88 | 1.20 | 9.2 | 9.16 | — | 5.58–11.42 | 11.4 |
| DST | 4.06 | 0.62 | 3.6 | 4.03 | — | 2.45–5.84 | 6.4 |
| EGS | 9.11 | 2.31 | 9.8 | 9.40 | — | 3.23–14.79 | 16.1 |
| BSOCOES | 10.62 | 0.97 | 10.7 | 10.72 | 11.6 | 6.81–12.43 | 13.0 |
(DSG) disuccinimidyl glutarate; (DSP) dithiobis(succinimidylpropionate); (DSS) disuccinimidyl suberate; (DST) disuccimimidyl tartarate; (EGS) ethyleneglycol bis-(succinimidylsuccinate); (BSOCOES) bis(2-[succinimidooxycarbonyloxy]ethyl)sulfone.
a See reference Pierce Chemicals (1999).
Table 5.
Series 5 miscellaneous rigid cross-linking reagents
| Cross-linking reagent | Average distance | Standard deviation | Mode | Median distance | Other major modes | Range of distances | Cited distancea |
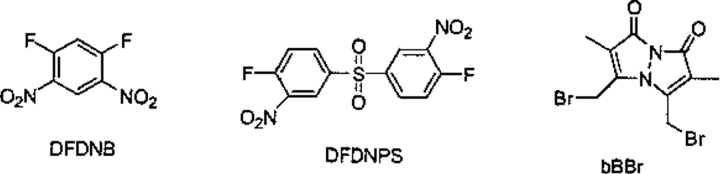
| DFDNB | 4.91 | 0.07 | 4.9 | 4.91 | — | 4.73–5.11 | 3.0 |
| DFDNPS | 9.74 | 0.33 | 9.7 | 9.75 | — | 8.28–10.64 | 3.0 |
| bBBr | 4.88 | 0.57 | 5.0 | 4.99 | 3.6 | 3.17–6.61 |
(DFDNB) 1,5-difluro-2,4-dinitrobenzene; (DFDNPS) 4,4′-difluoro3,3′-dinitrodiphenylsulfone; (bBBr) dibromobimane.
a See reference Pierce Chemicals (1999).
Most of the distributions obtained were basically gaussian in nature, although some were skewed slightly toward longer lengths. This is not surprising given that the lowest energy conformation of the alkyl bridged reagents is an extended conformation. There is only one fully extended conformation, however, and very little energy is required to rotate one or more bonds into gauche conformations. The number of populated conformations with eclipsed bonds is much lower than gauche conformations because of the relatively high energy of eclipsed conformations. Therefore, the mode of the distribution represents an ensemble of conformations all with the same cross-linking span. In a normal distribution, 97.7% of the data should fall within three standard deviations of the mean. In almost all of the cases studied, this is the case. Therefore, the average distance, coupled with the standard deviation, provides an accurate sense of the distribution data. For mPDM, for example, our recommended ruler length is 10.6 ± 0.5 Å.
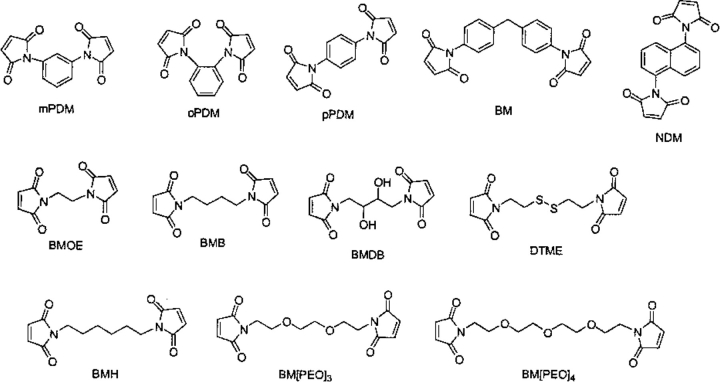
The first series (Fig. 2 ▶) consists of maleimide-capped reagents that cross-link by addition of the sulfhydryl group of a cysteine residue to the double bond in the maleimide (Fig. 1D ▶). In this series, the resultant sulfur to sulfur distance was monitored in the derivatives formed by the addition of methanethiol to the double bond of each maleimide. This group includes N,N′1,3-phenylenedimaleimide (mPDM; for some representative examples, see Chang and Flaks 1972; Moroney et al. 1982; Nadeau et al. 1997; Nitao and Reisler 1998), N,N′-1,2-phenylenedimaleimide (oPDM; for some representative examples, see Chang and Flaks 1972; Oda and Funatsu 1979; Wells et al. 1980; Masuho et al. 1982; Moroney et al. 1982; Nadeau et al. 1997; Nitao and Reisler 1998; Yu et al. 1998; Wang and Kaback 1999), N,N′-1,4-phenylenedimaleimide (pPDM; for some representative examples, see Ohara et al. 1982; Valenzuela et al. 1984; Chaussepied et al. 1988; King et al. 1991; Hesterkamp et al. 1993; Bubis et al. 1995; Nadeau et al. 1997; Polosukhina and Highsmith 1997; Nitao and Reisler 1998; Wang and Kaback 1999), N,N′-(methylene-4-1-phenylene)bismaleimide(BM; for some representative examples, see Coggins 1996; Nitao and Reisler 1998), naphthalene-1,5-dimaleimide (NDM; for some representative examples, see Wells et al. 1980; Miller et al. 1982; Moroney et al. 1982; Perkins et al. 1984; Nitao and Reisler 1998), bismaleimidoethane (BMOE; e.g., see Cheronis et al. 1992; Tanaka et al. 1996), 1,4-bismaleimidobutane (BMB; e.g., see Cheronis et al. 1992; Schwarzer et al. 1997; Franke et al. 1999), 1,4-bis-maleimidyl-2,3-dihydroxybutane (BMDB; e.g., see Bauer and Hagen 1997; Schwarzer et al. 1997), 1,6-bismaleimidohexane (BMH; e.g., see Goldberg et al. 1991; King et al. 1991; Ishiguro et al. 1992; Greve and McClelland 1994; Hansen and Barklis 1995; McDermott et al. 1996; Stults 1997; Margolin et al. 1998; Franke and Pingoud 1999; Wang and Kaback 1999; Archambault 2000; Rappsilber et al. 2000), dithio-bis-maleimdoethane (DTME); e.g., see Moroney et al. 1982; Kato et al. 1986), 1,8-bismaleimidotriethyleneglycol (BM[PEO]3; e.g., see Alexander and Speranza 1989; Kossmehl et al. 1995), and 1,11-bis-maleimidotetraethyleneglycol (BM[PEO]4).
Fig. 2.
The bis-maleimide cross-linking reagents. (mPDM) N,N′1,3-phenylenedimaleimide; (oPDM) N,N′-1,2-phenylenedimaleimide; (pPDM) N,N′-1,4-phenylenedimaleimide; (BM) N,N′-(methylene-4-1-phenylene)bismaleimide; (NDM) naphthalene-1,5-dimaleimide; (BMOE) bismaleimidoethane; (BMB) 1,4-bismaleimidobutane; (BMDB) 1,4-bis-maleimidyl-2,3-dihydroxybutane; (DTME) dithio-bis-maleimdoethane; (BMH) 1,6-bismaleimidohexane; (BM[PEO]3) 1,8-bismaleimidotriethyleneglycol; (BM[PEO]4) 1,11-bis-maleimidotetraethyleneglycol.
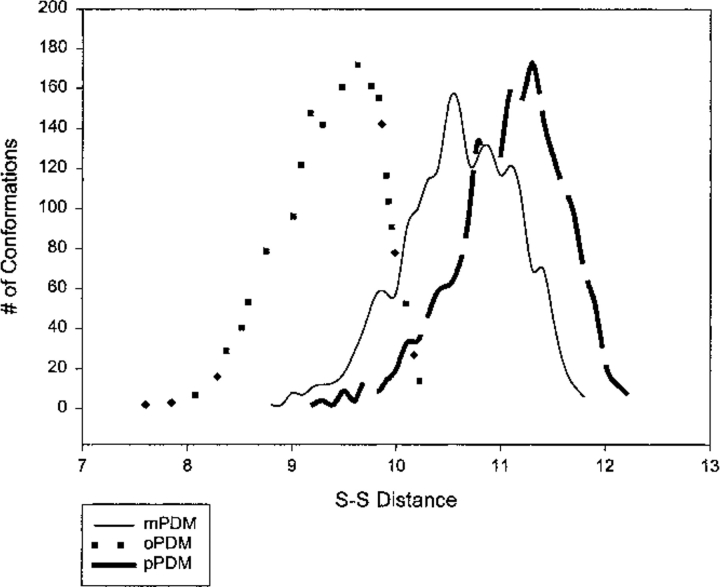
The bis-maleimide reagents oPDM, mPDM, pPDM, and BM are perhaps among the most frequently used molecular rulers of intersulfide distances in proteins. They owe their popularity to their high reactivity and specificity and the perceived lack of length overlap among them (Perkins et al. 1984; Nitao and Reisler 1998; Kwaw et al. 2000). Our results (Table 1) show an overlap in the range of S-S distances that can be bridged by these reagents, particularly mPDM and pPDM (Fig. 3 ▶). The range of distances in the tables is derived from all distances with a contribution of one of more conformers to the distribution. The values obtained for mPDM are quite close to those cited from literature whereas the width of the distribution found here for pPDM is very different from cited, and the cited oPDM values (5.2–7.8 Å) are entirely different from those found here (7.67–10.47 Å). Although in some cases the overlap regions may represent only a small fraction of the overall conformations of the reagents, the very existence of such overlap must be taken into account in the analysis of protein cross-linking kinetics by these reagents. The various rates of cross-linking may be as much influenced by protein dynamics as by that of the reagent making it more difficult to assess the conformational fluctuations of the protein.
Fig. 3.
Frequency plots showing overlap of mPDM, oPDM, and pPDM spans. (mPDM) N,N′1,3-phenylenedimaleimide; (oPDM) N,N′-1,2-phenylenedimaleimide; (pPDM) N,N′-1,4-phenylenedimaleimide.
BM[PEO]3 and BM[PEO]4 prefer gauche conformations in solution and, as such, provide two of the most striking examples of large differences between the cited sulfur–sulfur distance and the most common distance from dynamics. In triethylene glycol, the distance cited for the spacer arm length is 14.7 Å, whereas we obtain a distance of 8.8 Å. An analysis of this conformation, obtained from the dynamics simulation, shows that the CO bonds adopt gauche conformations, thus bringing the two reactive ends of the molecule closer together. This gauche preference is well known in such molecules (Eliel 1965) and makes an even larger difference in the tetraethylene glycol case. Our calculations show that the most populated distance is ∼9.5 Å. Literature values commonly cited are almost twice this length (17.8 Å).
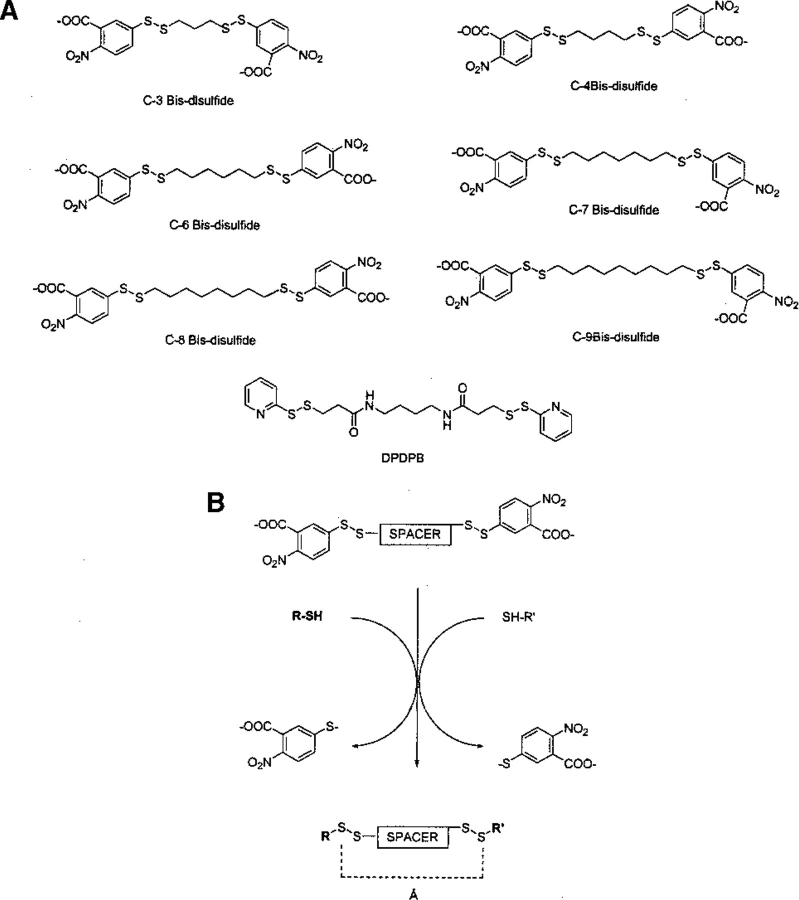
The second series (Fig. 4A ▶) consists of sulfhydryl-reactive reagents capped by aryl disulfides. These activated disulfides are the C-3, C-4, and C-6 through C-9 disulfides (Kliche et al. 1999), and 1,4-di-(3′-[2-pyridyldithio]-propionamido) butane (DPDPB) (e.g., see Russell-Jones et al. 1994; Zecherle et al. 1992; Lai 1997; Middleton et al. 1997; Wolff et al. 1998, 1999). The distance between the outermost sulfurs was monitored, because these reagents cross-link through disulfide exchange reactions (Fig. 4B ▶). In addition, the cross-linker can be cleaved through reducing reagents like dithiothreitol (DTT; Kumar et al. 1991; Hovinen et al. 1993) or other disulfide reductants.
Fig. 4.
Disulfide exchange cross-linking reagents. (A) Structures of the arene-disulfide exchange reagents. (B) Reaction of bis-disulfide cross-linking reagents with sulfhydryl groups. Å is the cross-linking span of the reagent.
The arene disulfide reagents of series 2 also show striking differences between the cited values and those obtained from this work (Table 2). The implications are significant. For example, Faulstich and coworkers (Kliche et al. 1999) tried to determine the distance between SH1 and SH2 in rabbit skeletal myosin subfragment 1 by monitoring the cross-linking kinetics by using a series of these arene disulfide reagents. The reagent with the fastest kinetics was believed to be the one with the optimal fit. From these kinetic experiments, the authors concluded that the distance between SH1 and SH2 must be >15 Å. The dynamic nature of the cross-linking reagents, however, was not taken into account, nor was the fact that the most frequently observed conformations had distances much shorter than those cited. In addition, there is substantial overlap in the lengths that are easily obtainable in this series of reagents, adding another layer of difficulty. The observed kinetics probably result from both the conformational fluctuations of the cross-linking reagent and the dynamic processes of the protein. When highly flexible cross-links are used, concrete conclusions about protein structures are difficult.
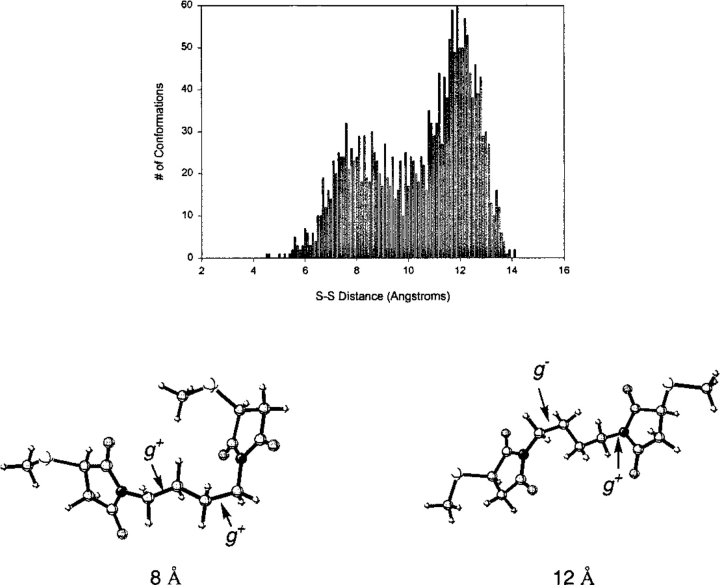
The distributions of BMB, C-6 disulfide, C-7 disulfide, and C-8 disulfide appear to be a composite of two overlapping Gaussian functions. In all cases, one of the modes is much higher than the other, and this mode is what is reported in the tabulated data. All other statistics take into account the full distribution of data. The existence of the second mode can be understood by examining the conformations of the reagent that compose the lesser mode. For example, in BMB (Fig. 5 ▶) the mode at 12 Å represents the case in which there are two gauche N-C-C-C conformations, one g+ and the other g−. The minor mode at 8 Å, however, is the case in which both of these are g+, and the terminal groups are closer together. Some of the sparsely populated distances between two modes represent cases in which there may be one gauche and one or two anticonformations. Similar situations exist for the C-7 and C-8 arene disulfide (Fig. 6A ▶) molecules, except that there is a relatively narrow distribution overlapping with a very broad one. This is not surprising, because as the molecule becomes more flexible, there are many more possible conformations that can be populated without a large energy penalty. The mode in both of these cases represents conformations that have two gauche carbon–carbon bonds.
Fig. 5.
Frequency plot for 1,4-bismaleimidobutane (BMB). The structures shown represent conformations of BMB with cross-linking spans corresponding to the two modes. Both conformations have two gauche NCCC dihedral angles.
Fig. 6.
C-6 and C-8 disulfides. (A) Frequency plot for C-8 arene disulfide; (B) frequency plot for C-6 arene disulfide; (C) two major conformations of the C-6 disulfide reagent and the corresponding S-S distances.
The C-6 arene bis-disulfide (Figure 6B ▶) shows some of the features of the longer arene disulfides but has a notable narrow distribution with a mode at 12.2 Å. The mode with the longer distance represents a molecule in which both of the C-S-S-C dihedral angles are ∼90°. The most common length, on the other hand, results from one of the C-C bonds being in a gauche conformation in addition to the two C-S-S-C dihedral angles being ∼90°. An example of one of these conformations is shown in Figure 6C ▶.
Series 3, shown in Figure 7 ▶ and Table 3, represents the imidoester class of cross-linking reagents. This class of reagents shows considerable selectivity for primary amines and has very little cross-reactivity with other possible nucleophiles in the protein. The imidoester reacts with an amine to form an amidine linkage that carries a positive charge at physiological pH (Kiehm and Ji 1977; Wilbur 1992). The N(amidine) to N(amidine) distance—resulting from displacement of methoxide by methyl amine—was used to determine the cross-linking span. The reagents studied were dimethyl adipimidate (DMA; e.g., see Hartman and Wold 1967; Niehaus and Wold 1970; Wang and Kassell 1974; Yu and Carter 1976; Pennathur-Das et al. 1982; Wasylewska et al. 1987; Garlick et al. 1992; Erarslan and Ertan 1995; Rappsilber et al. 2000), dimethyl pimelimidiate (DMP; e.g., see Cohlberg et al. 1972; Davies and Kaplan 1972; Hitchcock 1975; Sinha and Brew 1981; Koga 1987; Bar-Peled and Raikhel 1996), dimethyl suberimidate (DMS; e.g., see Shoshan-Barmatz et al. 1995; Gotte et al. 1997; Vanhoutte and Malaisse 1997; Watty et al. 1997, 1998), and dimethyl 3,3′-dithiobis-propionimidate (DTBP; e.g., see Lloyd and Weitzman 1987; Stros and Kolibalova 1987; Shivdasani and Thomas 1988; Cornell 1989; Dubey et al. 1989; Erarslan and Ertan 1995).
Fig. 7.
The bis-imidoester cross-linking reagents. (DMA) dimethyl adipimidate; (DMP) dimethyl pimelimidiate; (DMS) dimethyl suberimidate; (DTBP) dimethyl 3,3′-dithiobis-propionimidate.
A fourth class (Fig. 8 ▶; Table 4) represents the amine-reactive N-hydroxysuccinimide esters. The esters react with amine functionality on the protein to form stable amide bonds and release two molecules of N-hydroxysuccinimide (NHS). The esters tend to react with the α-amines at the N terminus and the ɛ-amines of lysine residues. It is also possible for these reagents to react with sulfhydryl and hydroxyl groups, but this does not lead to stable adduct formation. The cross-linking spans of disuccinimidyl glutarate (DSG; e.g., see Eisman and Schnaare 1996; Horiguchi et al. 1997; Pan et al. 1998; Holwerda 1999; Xu et al. 1999; Korsgren et al. 2000), dithiobis(succinimidylpropionate) (DSP; e.g., see Walleczek et al. 1989; Meunier et al. 1991; Poruchynsky and Atkinson 1991; Jansson et al. 1996; Stout and Kirley 1996; Tanaka et al. 1996; Carl et al. 1998), disuccinimidyl suberate (DSS; e.g., see Chain and Malkin 1991; Pliszka 1993; Persson and Ezban 1994; Loester et al. 1995; Stout and Kirley 1996; Carl et al. 1998; Leonard et al. 1998), disuccimimidyl tartarate (DST; e.g., see Bragg and Hou 1986; Farries et al. 1988; Mita et al. 1989; Miyazaki et al. 1990; Plouet and Moukadiri 1990; Amiranoff and Lorinet-Laburthe 1991; Chen et al. 1992; Maman et al. 1994; Horiguchi et al. 1997), ethyleneglycol bis-(succinimidylsuccinate) (EGS; e.g., see Huey and Hugli 1985; Bladon et al. 1989; Caplow and Shanks 1990; Geisler et al. 1992), and bis(2-[succinimidooxycarbonyloxy]ethyl)sulfone (BSOCOES; e.g., see Smith et al. 1986; Svoboda et al. 1988a, 1988b; Schoffelmeer et al. 1989; Fujioka et al. 1990) were studied. In this series, methyl amine was again added to both ends, and the N(amide) to N(amide) bond distance was monitored.
Fig. 8.
The bis-N-hydroxysuccinimide ester cross-linking reagents. (DST) disuccimimidyl tartarate; (DSG) disuccinimidyl glutarate; (DSP) dithiobis(succinimidylpropionate); (DSS) disuccinimidyl suberate; (BSOCOES) bis(2-[succinimidooxycarbonyloxy]ethyl)sulfone; (EGS) ethyleneglycol bis-(succinimidylsuccinate).
The cited distances for all of the molecules studied in this series are outside the range of distance found through our calculations. Minimization of the fully extended conformations of these molecules with AMBER and measurement of the maximum achievable distances reveal that the longer distances cited could be obtained only by introducing considerable angle strain into the linker.
The final group (Fig. 9 ▶; Table 5) consists of three short and nearly rigid cross-linking reagents: 1,5-difluro-2,4-dinitrobenzene (DFDNB; e.g., see Mayeux et al. 1991; Herzig et al. 1995; Krupenko et al. 1995; Shoshan-Barmatz et al. 1995; Herzig et al. 1996), 4,4′-difluoro3,3′-dinitrodiphenylsulfone (DFDNPS; e.g., see Givol 1969; Modesto and Pesce 1971; Hsia et al. 1984), and dibromobimane (bBBr; e.g., see Kim and Raines 1995; Bhattacharjee and Rosen 1996; Wu et al. 1996; Loo and Clarke 1997, 1999; Konno et al. 2000). The first two are amine reactive through nucleophilic aromatic substitution to give aryl amines. Reactions with other nucleophiles (such as sulfhydryls, imidazoles, and phenolates) are possible, but these are reversible. In these two cases, the resultant aryl amine nitrogen to aryl amine nitrogen was measured for each conformation. The various cross-linking spans result mainly from molecular vibrations because the reagents are basically rigid. Of greatest interest is that both reagents have cross-linking spans significantly greater than the 3 Å cited in the literature. Nevertheless, DFDNB and bBBr are the reagents of choice among the large group of thiol cross-linking reagents (Tables 1 and 2) for bridging cysteine residues over relatively short distances (∼5 Å).
Fig. 9.
The small rigid cross-linking reagents. (DFDNB) 1,5-difluro-2,4dinitrobenzene; (DFDNPS) 4,4′-difluoro3,3′-dinitrodiphenylsulfone; (bBBr) dibromobimane.
Dibromobimane reacts mainly with thiol groups in an SN2 manner to yield thioether linkages. As before, the cross-linking span was determined from the sulfur to sulfur distance resulting from nucleophilic displacement of the bromides by methyl thiolate. The various conformers in this case result from rotations about the resultant sulfur–carbon bonds.
Conclusions
The most striking result of this work is that the distances cited for many commonly used protein cross-linkers are highly improbable. In other cases, our calculations show that the linker can achieve a broader range of end-to-end distances than normally has been recognized in the literature.
In most cases, the cross-linking distances cited previously in the literature were obtained by measuring the distances between the two reactive groups in a fully extended conformation (Pierce Chemical Company, pers. comm.)— even in polyethylene glycol molecules BM[PEO]3 and BM[PEO]4 that are known to prefer gauche conformations; these cited values assume that there is only one significantly populated conformation of the molecule in solution. In fact, even in mostly rigid molecules there are several different conformations (with differing cross-linking spans) that are easily accessible in solution. In only two of the 32 cases studied was the average distance obtained from the dynamics simulation within 0.5 Å of the cited distance. In all other cases, the most populated states had lengths that were considerably different from the fully extended conformation. In addition, the literature value was only minimally populated.
The distances obtained from these dynamics simulations give more realistic lengths for these molecular rulers. We recommend the use of the statistical average plus or minus the standard deviation as a new measure of these lengths. The new distances should be a useful tool for studies of protein structure and function and should provide guidelines for the appropriate choice of cross-linking reagents.
Acknowledgments
We are grateful to the National Institutes of Health (GM 61402 and AR 22031), the National Science Foundation (Grant MCB 9904599), and to the UCLA Graduate Division for a Dissertation Year Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/
References
- Alexander, D.C. and Speranza, G.P. 1989. Novel bismaleimide derivatives. Eur. Pat. Appl. 16 pp.
- Amiranoff, B. and Lorinet-Laburthe, M. 1991. A clonal rat pancreatic Δ cell line (Rin14B) expresses a high number of galanin receptors negatively coupled to a pertussis-toxin-sensitive cAMP-production pathway. Eur. J. Biochem. 195 459–463. [DOI] [PubMed] [Google Scholar]
- Archambault, J. 2000. The oligomerization domain of the E1 helicase of papillomavirus and its role in viral replication and therapeutic use. PCT Int. Appl. 101 pp.
- Bar-Peled, M. and Raikhel, N.V. 1996. A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 241 140–142. [DOI] [PubMed] [Google Scholar]
- Bauer, P.J. and Hagen, V. 1997. Dimaleimido-substituted dihydroxy alkanes which can be used as crosslinking reagents and process for their preparation. PCT Int. Appl. 12 pp.
- Bhattacharjee, H. and Rosen, B.P. 1996. Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J. Biol. Chem. 271 24465–24470. [DOI] [PubMed] [Google Scholar]
- Bladon, C.M., Mitchell, R., and Ogier, S.A. 1989. Preparation and use of biotinylated ligands for LHRH receptor purification. Tetrahedron Lett. 30 1401–1404. [Google Scholar]
- Bragg, P.D. and Hou, C. 1986. Chemical crosslinking of α subunits in the F1 adenosine triphosphatase of Escherichia coli. Arch. Biochem. Biophys. 244 361–372. [DOI] [PubMed] [Google Scholar]
- Bubis, J., Ortiz, J.O., Moeller, C., and Millan, E.J. 1995. Identification and characterization of transducin functional cysteines, lysines, and acidic residues by group-specific labeling and chemical crosslinking. Methods Protein Struct. Anal. 227–250.
- Caplow, M. and Shanks, J. 1990. Mechanism of the microtubule GTPase reaction. J. Biol. Chem. 265 8935–8941. [PubMed] [Google Scholar]
- Carl, S.A.L., Smith, T.M., and Kirley, T.L. 1998. Crosslinking induces homodimer formation and inhibits enzymic activity of chicken stomach ecto-apyrase. Biochem. Mol. Biol. Int. 44 463–470. [DOI] [PubMed] [Google Scholar]
- Chain, R.K. and Malkin, R. 1991. The chloroplast cytochrome b6f complex can exist in monomeric and dimeric states. Photosynth. Res. 28 59–68. [DOI] [PubMed] [Google Scholar]
- Chang, F.N. and Flaks, J.G. 1972. Specific crosslinking of two proteins from the Escherichia coli 30S ribosomal subunit. J. Mol. Biol. 68 177–180. [DOI] [PubMed] [Google Scholar]
- Chaussepied, P., Morales, M.F., and Kassab, R. 1988. The myosin (SH2-50-kilodalton fragment) cross-link: Location and consequences. Biochemistry 27 1778–1785. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Couvineau, A., Laburthe, M., and Amiranoff, B. 1992. Solubilization and molecular characterization of active galanin receptors from rat brain. Biochemistry 31 2415–2422. [DOI] [PubMed] [Google Scholar]
- Cheronis, J.C., Whalley, E.T., Nguyen, K.T., Eubanks, S.R., Allen, L.G., Duggan, M.J., Loy, S.D., Bonham, K.A., and Blodgett, J.K. 1992. A new class of bradykinin antagonists: Synthesis and in vitro activity of bissuccinimidoalkane peptide dimers. J. Med. Chem. 35 1563–1572. [DOI] [PubMed] [Google Scholar]
- Coggins, J.R. 1996. Crosslinking reagents for proteins. Proteins Labfax 307–314.
- Cohlberg J.A., Pigiet, Jr., V.P, and Schachman, H.K. 1972. Structure and arrangement of the regulatory subunits in aspartate transcarbamylase. Biochemistry 11 3396–3411. [DOI] [PubMed] [Google Scholar]
- Cornell, R. 1989. Chemical cross-linking reveals a dimeric structure for CTP : phosphocholine cytidylyltransferase. J. Biol. Chem. 264 9077–9082. [PubMed] [Google Scholar]
- Davies, G.E. and Kaplan, J.G. 1972. Use of a diimidoester crosslinking reagent to examine the subunit structure of rabbit muscle pyruvate kinase. Can. J. Biochem. 50 416–422. [DOI] [PubMed] [Google Scholar]
- Dubey, A.K., Bisaria, V.S., Mukhopadhyay, S.N., and Ghose, T.K. 1989. Stabilization of restriction endonuclease Bam Hi by cross-linking reagents. Biotechnol. Bioeng. 33 1311–1316. [DOI] [PubMed] [Google Scholar]
- Eisman, J.M. and Schnaare, R.L. 1996. The formation of a crosslinked carboxyhemoglobin membrane at an organic-aqueous interface. Artif. Cells Blood Substit. Immobil. Biotechnol. 24 185–196. [DOI] [PubMed] [Google Scholar]
- Eliel, E.L. 1965. Conformational analysis. Interscience Publishers, New York.
- Erarslan, A. and Ertan, H. 1995. Thermostabilization of penicillin G acylase obtained from a mutant of Escherichia coli ATCC 11105 by bisimidoesters as homobifunctional crosslinking agents. Enzyme Microb. Technol. 17 629–635. [Google Scholar]
- Farries, T.C., Lachmann, P.J., and Harrison, R.A. 1988. Analysis of the interaction between properdin and factor B, components of the alternative-pathway C3 convertase of complement. Biochem. J. 253 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasold, H., Klappenberger, J., Meyer, C., and Remold, H. 1971. Bifunctional reagents for the cross-linking of proteins. Angew. Chem. Int. Ed. Engl. 10 795–801. [DOI] [PubMed] [Google Scholar]
- Franke, I. and Pingoud, A. 1999. Synthesis and biochemical characterization of obligatory dimers of the sugar non-specific nuclease from Serratia marcescens using specifically designed bismaleimidoalkanes as SH-specific crosslinking reagents. J. Protein Chem. 18 137–146. [DOI] [PubMed] [Google Scholar]
- Franke, I., Pingoud, A., Wende, W., and Meiss, G. 1999. Immobilized nuclease variants from Serratia marcescens which do not dissociate into subunits. PCT Int. Appl. 21 pp.
- Fujioka, T., Inoue, F., Kuriyama, M., Miura, Y., and Higashijima, T. 1990. Identification of opioid-binding materials of rat brain. Chem. Pharm. Bull. 38 168–171. [DOI] [PubMed] [Google Scholar]
- Garlick, R.L., Martin, Jr., J.P., and Lyle, S.B. 1992. Imidoester cross-linked hemoglobin compositions. PCT Int. Appl. 22 pp.
- Geisler, N., Schuenemann, J., and Weber, K. 1992. Chemical cross-linking indicates a staggered and antiparallel protofilament of desmin intermediate filaments and characterizes one higher-level complex between protofilaments. Eur. J. Biochem. 206 841–852. [DOI] [PubMed] [Google Scholar]
- Givol, D. 1969. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by a bifunctional reagent. FEBS Lett. 5 153–156. [DOI] [PubMed] [Google Scholar]
- Goldberg, M., Knudsen, K.L., Platt, D., Kohen, F., Bayer, E.A., and Wilchek, M. 1991. Specific interchain crosslinking of antibodies using bismaleimides. Repression of ligand leakage in immunoaffinity chromatography. Bioconjug. Chem. 2 275–280. [DOI] [PubMed] [Google Scholar]
- Gotte, G., Testolin, L., Costanzo, C., Sorrentino, S., Armato, U., and Libonati, M. 1997. Cross-linked trimers of bovine ribonuclease A: Activity on double-stranded RNA and antitumor action. FEBS Lett. 415 308–312. [DOI] [PubMed] [Google Scholar]
- Greve, J.M. and McClelland, A. 1994. Multimeric forms of human rhinovirus receptor protein ICAM and their use in the treatment of infection. PCT Int. Appl. 69 pp.
- Hansen, M.S.T. and Barklis, E. 1995. Structural interactions between retroviral Gag proteins examined by cysteine crosslinking. J. Virol. 69 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, F.C. and Wold, F. 1967. Cross-linking of bovine pancreatic ribonuclease A with dimethyl adipimidate. Biochemistry 6 2439–2448. [DOI] [PubMed] [Google Scholar]
- Herzig, M.C.S. and Leeb-Lundberg, L.M.F. 1995. The agonist binding site on the bovine bradykinin B2 receptor is adjacent to a sulfhydryl and is differentiated from the antagonist binding site by chemical crosslinking. J. Biol. Chem. 270 20591–20598. [DOI] [PubMed] [Google Scholar]
- Herzig, M.C.S., Nash, N.R., Connolly, M., Kyle, D.J., and Leeb-Lundberg, L.M.F. 1996. The N terminus of bradykinin when bound to the human bradykinin B2 receptor is adjacent to extracellular Cys20 and Cys277 in the receptor. J. Biol. Chem. 271 29746–29751. [DOI] [PubMed] [Google Scholar]
- Hesterkamp, T., Weeds, A.G., and Mannherz, H.G. 1993. The actin momomers in the ternary gelsolin: 2 actin complex are in an antiparallel orientation. Eur. J. Biochem. 218 507–513. [DOI] [PubMed] [Google Scholar]
- Hitchcock, S.E. 1975. Crosslinking of troponin with dimethylimido esters. Biochemistry 14 5162–5167. [DOI] [PubMed] [Google Scholar]
- Holmes, K.C., Popp, D., Gebhard, D., and Kabsch, W. 1990. Atomic model of the actin filament. Nature 347 44–49. [DOI] [PubMed] [Google Scholar]
- Holwerda, B. 1999. Activity in monomers of human cytomegalovirus protease. Biochem. Biophys. Res. Commun. 259 370–373. [DOI] [PubMed] [Google Scholar]
- Horiguchi, T., Miwa, Y., and Shigesada, K. 1997. The quaternary geometry of transcription termination factor ρ: Assignment by chemical crosslinking. J. Mol. Biol. 269 514–528. [DOI] [PubMed] [Google Scholar]
- Hovinen, J., Guzaev, A., Azhayev, A., and Lonnberg, H. 1993. Synthesis of 3′-functionalized oligonucleotides on a single solid support. Tetrahedron Lett. 34 8169–8172. [Google Scholar]
- Hsia, J.C., Wong, L.T., Tan, C.T., Er, S.S., Kharouba, S., Balaskas, E., Tinker, D.O., and Feldhoff, R.C. 1984. Bovine serum albumin: Characterization of a fatty acid binding site on the N-terminal peptic fragment using a new spin-label. Biochemistry 23 5930–5932. [DOI] [PubMed] [Google Scholar]
- Huey, R. and Hugli, T.E. 1985. Characterization of a C5a receptor on human polymorphonuclear leukocytes (PMN). J. Immunol. 135 2063–2068. [PubMed] [Google Scholar]
- Ishiguro, M., Matori, Y., Tanabe, S., Kawase, Y., Sekine, I., and Sakakibara, R. 1992. Biochemical studies on oral toxicity of ricin. V. The role of lectin activity in the intestinal absorption of ricin. Chem. Pharm. Bull. 40 1216–1220. [DOI] [PubMed] [Google Scholar]
- Jansson, S., Andersen, B., and Scheller, H.V. 1996. Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 112 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, Y., Umemoto, N., Saito, M., and Hara, T. 1986. Preparation of cytotoxic complexes containing pharmaceuticals, albumins, and antibodies to target tissues. Jpn. Kokai Tokkyo Koho 18 pp.
- Kiehm, D. and Ji, T.H. 1977. Photochemical crosslinking of cell membranes. A test for natural and random collisional crosslinks by millisecond crosslinking. J. Biol. Chem. 252 8524–8531. [PubMed] [Google Scholar]
- Kim, J.S. and Raines, R.T. 1995. Dibromobimane as a fluorescent crosslinking reagent. Anal. Biochem. 225 174–176. [DOI] [PubMed] [Google Scholar]
- King, D.J, Mountain, A., Owens, R.J., and Yarranton, G.T. 1991. Multivalent antigen-binding proteins and their production by recombinant DNA technology. PCT Int. Appl. 55 pp.
- Kliche, W., Pfannstiel, J., Tiepold, M., Stoeva, S., and Faulstich, H. 1999. Thiol-specific cross-linkers of variable length reveal a similar separation of SH1 and SH2 in myosin subfragment 1 in the presence and absence of MgADP. Biochemisty 38 10307–10317. [DOI] [PubMed] [Google Scholar]
- Koga, R. 1987. Affinity of chemically cross-linked immunoglobulin to antigen. Seirigaku Kenkyusho Gijutsuka Hokoku 2 5–7. [Google Scholar]
- Konno, K., Ue, K., Khoroshev, M., Martinez, H., Ray, B., and Morales, M.F. 2000. Consequences of placing an intramolecular crosslink in myosin S1. Proc. Natl. Acad. Sci. 97 1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsgren, P., Ahlberg, P., and Baltzer, L. 2000. Covalent control of polypeptide folding. Induction of helix-loop-helix motifs by bridging. Perkin 2 643–647. [Google Scholar]
- Kossmehl, G., Nagel, H.I., and Pahl, A. 1995. Crosslinking reactions on polyamides by bis- and tris(maleimide)s. Angew. Makromol. Chem. 227 139–157. [Google Scholar]
- Krupenko, S.A., Kolesnik, O.I., Krupenk, N.I., and Strelchyonok, O.A. 1995. Organization of the transcortin-binding domain on placental plasma membranes. Biochim. Biophys. Acta 1235 387–394. [DOI] [PubMed] [Google Scholar]
- Kumar, P., Bose, N.K., and Gupta, K.C. 1991. A versatile solid phase method for the synthesis of oligonucleotide-3′-phosphates. Tetrahedron Lett. 32 967–970. [Google Scholar]
- Kunkle, G.R., Mehrebian, M., and Matinson, G.H. 1986. Contact-site cross-linking reagents. Mol. Cell. Biochem. 234 826–836. [DOI] [PubMed] [Google Scholar]
- Kwaw, I., Sun, J., and Kaback, H.R. 2000. Thiol cross-linking of cytoplasmic loops in the lactose permease of Escherichia coli. Biochemistry 39 3134–3140. [DOI] [PubMed] [Google Scholar]
- Lai, C.S. 1997. Antibodies directed against dithiocarbamates. PCT Int. Appl. 31 pp.
- Leach, A. 1996. Molecular modelling: Principles and applications. Addison Wesley Longman Limited, Essex, UK.
- Leonard, A.S., Davare, M.A., Horne, M.C., Garner, C.C., and Hell, J.W. 1998. SAP97 is associated with the α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J. Biol. Chem. 273 19518–19524. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.J. and Weitzman, P.D.J. 1987. Modification of the regulatory properties of Acinetobacter citrate synthase by cross-linking. Biochem. Soc. Trans. 15 840. [Google Scholar]
- Loester, K., Baum, O., Hofmann, W., and Reutter, W. 1995. Characterization of molecular aggregates of α1β1-integrin and other rat liver membrane proteins by combination of size-exclusion chromatography and chemical crosslinking. J. Chromatogr. A 711 187–199. [DOI] [PubMed] [Google Scholar]
- Loo, T.W. and Clarke, D.M. 1999. Identification of residues in the drug-binding domain of human P-glycoprotein. Analysis of transmembrane segment 11 by cysteine-scanning mutagenesis and inhibition by dibromobimane. J. Biol. Chem. 274 35388–35392. [DOI] [PubMed] [Google Scholar]
- Loo, T.W. and Clarke, D.M. 1997. Identification of residues in the drug-binding site of human P-glycoprotein using a thiol-reactive substrate. J. Biol. Chem. 272 31945–31948. [DOI] [PubMed] [Google Scholar]
- Lorenz, M., Popp, D., and Holmes, K.C. 1993. Refinement of the F-actin model against x-ray fiber diffraction data by the use of a directed mutation algorithm. J. Mol. Biol. 234 826–836. [DOI] [PubMed] [Google Scholar]
- Maman, J.D., Yager, T.D., and Allan, J. 1994. Self-association of the globular domain of histone H5. Biochemistry 33 1300–1310. [DOI] [PubMed] [Google Scholar]
- Margolin, A.L., Persichetti, R.A., St. Clair, N.L., Khalaf, N.K., and Shenoy, B.C. 1998. Controlled dissolution crosslinked protein crystals. PCT Int. Appl. 153 pp.
- Masuho, Y., Umemoto, N., Hara, T., and Ohtomo, N. 1981. Cytotoxicity of a hybrid prepared by coupling diphtheria toxin A-chain with immunoglobulin Fab′ with N,N′-o-phenylenedimaleimide. Biochem. Biophys. Res. Commun. 102 561–567. [DOI] [PubMed] [Google Scholar]
- Masuho, Y., Kishida, K., Saito, M., Umemoto, N., and Hara, T. 1982. Importance of the antigen-binding valency and the nature of the crosslinking bond in ricin A-chain conjugates with antibody. J. Biochem. (Tokyo) 91 1583–1591. [DOI] [PubMed] [Google Scholar]
- Mayeux, P., Lacombe, C., Casadevall, N., Chretien, S., Dusanter, I., and Gisselbrecht, S. 1991. Structure of the murine erythropoietin receptor complex. Characterization of the erythropoietin cross-linked proteins. J. Biol. Chem. 266 23380–23385. [PubMed] [Google Scholar]
- McDermott, J., Farrell, L., Ross, R., and Barklis, E. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J. Virol. 70 5106–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier, A.C., Voisin, C.P., Van Camp, G., Cenatiempo, Y., and Muller, J.M. 1991. Molecular characterization and peptide specificity of two vasoactive intestinal peptide (VIP) binding sites in the chicken pineal. Neuropeptides 19 1–8. [DOI] [PubMed] [Google Scholar]
- Middleton, S.A., Johnson, D., McMahon, F.J., Mulkahy, L.S., and Jolliffe, L.K. 1997. Purification of human erythropoietin-binding protein from microbial fermentations and its uses in drug discovery. PCT Int. Appl. 64 pp.
- Miller, L., Coppedge, J., and Reisler, E. 1982. The reactive SH1 and SH2 cysteines in myosin subfragment 1 are crosslinked at similar rates with reagents of different length. Biochem. Biophys. Res. Commun. 106 117–122. [DOI] [PubMed] [Google Scholar]
- Mita, S., Tominaga, A., Hitoshi, Y., Sakamoto, K., Honjo, T., Akagi, M., Kikuchi, Y., Yamaguchi, N., and Takatsu, K. 1989. Characterization of high-affinity receptors for interleukin 5 on interleukin 5-dependent cell lines. Proc. Natl. Acad. Sci. 86 2311–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, H., Kondoh, M., Watanabe, H., Masuda, Y., Murakami, K., Takahashi, M., Yanagisawa, M., Kimura, S., Goto, K., and Masaki, T. 1990. Affinity labeling of endothelin receptor and characterization of solubilized endothelin-endothelin-receptor complex. Eur. J. Biochem. 187 125–129. [DOI] [PubMed] [Google Scholar]
- Modesto, R.R. and Pesce, A.J. 1971. Reaction of 4,4′-difluoro-3,3′-dinitrodiphenyl sulfone with γ-globulin and horseradish peroxidase. Biochim. Biophys. Acta 229 384–395. [DOI] [PubMed] [Google Scholar]
- Mohamadi, F., Richards, N.G., Guida, W.C., Liskamp, R., Lipton, M., Canfield, C., Chang, G., Hendrickson, T., and Still, W.C. 1990. MacroModel—An integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 11 440–467. [Google Scholar]
- Moroney, J.V., Warncke, K., and McCarty, R.E. 1982. The distance between thiol groups in the g subunit of coupling factor 1 influences the proton permeability of thylakoid membranes. J. Bioenerg. Biomembr. 14 347–359. [DOI] [PubMed] [Google Scholar]
- Nadeau, O.W., Sacks, D.B., and Carlson, G.M. 1997. Differential affinity crosslinking of phosphorylase kinase conformers by the geometric isomers of phenylenedimaleimide. J. Biol. Chem. 272 26196–26201. [DOI] [PubMed] [Google Scholar]
- Nagy, J.K., Lau, F.W., Bowie, J.U., and Saunders, C.R. 2000. Mapping the oligomeric interface of diacylglycerol kinase by engineered thiol cross-linking: Homologous sites in the transmembrane domain. Biochemistry 39 4154–4164. [DOI] [PubMed] [Google Scholar]
- Niehaus, Jr., W.G. and Wold, F. 1970. Cross-linking of erythrocyte membranes with dimethyl adipimidate. Biochim. Biophys. Acta 196 170–175. [DOI] [PubMed] [Google Scholar]
- Nitao, L.K. and Reisler, E. 1998. Probing the conformational states of the SH1-SH2 helix in myosin: A crosslinking approach. Biochemistry 37 16704–16710. [DOI] [PubMed] [Google Scholar]
- Oda, T. and Funatsu, G. 1979. Biochemical studies on ricin. Part XXIV. Cross-linking of the two constituent polypeptide chains of ricin D with N,N′-o-phenylenedimaleimide. Agric. Biol. Chem. 43 547–554. [Google Scholar]
- Ohara, O., Takahashi, S., Ooi, T., and Fujiyoshi, Y. 1982. Cross-linking study on skeletal muscle actin: Properties of suberimidate-treated actin. J. Biochem. (Tokyo) 91 1999–2012. [DOI] [PubMed] [Google Scholar]
- Pan, W., Ko, Y.H., and Pedersen, P.L. 1998 Subunit of rat liver mitochondrial ATP synthase: Molecular description and novel insights into the nature of its association with the F1-moiety. Biochemistry 37 6911–6923. [DOI] [PubMed]
- Pennathur-Das, R., Heath, R.H., Mentzer, W.C., and Lubin, B.H. 1982. Modification of hemoglobin S with dimethyl adipimidate. Contribution of individual reacted subunits to changes in properties. Biochim. Biophys. Acta 704 389–397. [DOI] [PubMed] [Google Scholar]
- Perkins, W.J., Wells, J.A., and Yount, R.G. 1984. Characterization of the properties of ethenoadenosine nucleotides bound or trapped at the active site of myosin subfragment 1. Biochemistry 23 3994–4002. [DOI] [PubMed] [Google Scholar]
- Persson, E. and Ezban, M. 1994. Characterization of chemically crosslinked human factor VIIIa. Biochem. Biophys. Res. Commun. 200 233–238. [DOI] [PubMed] [Google Scholar]
- Peters, K. and Richards, F.M. 1977. Chemical cross-linking: Reagents and problems in studies of membrane structure. Annu. Rev. Biochem. 46 523–551. [DOI] [PubMed] [Google Scholar]
- Pierce Chemicals. 1999. Double agents cross-linking reagents selection guide. Pierce Chemicals, Rockford, IL.
- Pliszka, B. 1990. Influence of nucleotide on chemical crosslinking between alkali light chains and the heavy chain of myosin subfragment 1. Biochim. Biophys. Acta 1040 89–94. [DOI] [PubMed] [Google Scholar]
- Pliszka, B. 1993. Mapping of the region of the heavy chain of myosin subfragment 1 that can be crosslinked to the alkali light chains. Biochem. Mol. Biol. Int. 31 381–388. [PubMed] [Google Scholar]
- Plouet, J. and Moukadiri, H. 1990. Characterization of the receptor to vasculotropin on bovine adrenal cortex-derived capillary endothelial cells. J. Biol. Chem. 265 22071–22074. [PubMed] [Google Scholar]
- Polosukhina, K. and Highsmith, S. 1997. Kinetic investigation of the ligand dependence of rabbit skeletal muscle myosin subfragment 1 Cys-697 and Cys-707 reactivities. Biochemistry 36 11952–11958. [DOI] [PubMed] [Google Scholar]
- Poruchynsky, M.S. and Atkinson, P.H. 1991. Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J. Virol. 65 4720–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J., Siniossoglou, S., Hurt, E.C., and Mann, M. 2000. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem. 72 267–275. [DOI] [PubMed] [Google Scholar]
- Russell-Jones, G.J., Westwood, S.W., Gould, A.R., and McInerney, B.V. 1994. Amplification of the vitamin B12 uptake system using polymers. PCT Int. Appl. 36 pp.
- Schoffelmeer, A.N.M., Yao, Y.H., and Simon, E.J. 1989. Cross-linking of human 125I-β-endorphin to a μ-Δ opioid receptor complex in rat striatum. Eur. J. Pharmacol. 166 357–358. [DOI] [PubMed] [Google Scholar]
- Schwarzer, A., Kim, T.S.Y., Hagen, V., Molday, R.S., and Bauer, P.J. 1997. The Na/Ca-K exchanger of rod photoreceptor exists as dimer in the plasma membrane. Biochemistry 36 13667–13676. [DOI] [PubMed] [Google Scholar]
- Shivdasani, R.A. and Thomas, D.W. 1988. Molecular associations of IA antigens after T-B cell interactions. I. Identification of new molecular associations. J. Immunol. 141 1252–1260. [PubMed] [Google Scholar]
- Shoshan-Barmatz, V., Hadad-Halfon, N., and Ostersetzer, O. 1995. Crosslinking of the ryanodine receptor/Ca2+ release channel from skeletal muscle. Biochim. Biophys. Acta 1237 151–161. [DOI] [PubMed] [Google Scholar]
- Sinha, S.K. and Brew, K. 1981. A label selection procedure for determining the location of protein–protein interaction sites by cross-linking with bisimidoesters. Application to lactose synthase. J. Biol. Chem. 256 4193–4204. [PubMed] [Google Scholar]
- Smith, R.A., Kirstein, M., Fiers, W., and Baglioni, C. 1986. Species specificity of human and murine tumor necrosis factor. A comparative study of tumor necrosis factor receptors. J. Biol. Chem. 261 14871–14874. [PubMed] [Google Scholar]
- Still, W.C., Tempczyk, A., Hawley, R.C., and Hendrickson, T. 1990. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 121 6127–6129. [Google Scholar]
- Stout, J.G. and Kirley, T.L. 1996. Control of cell membrane ecto-ATPase by oligomerization state: intermolecular crosslinking modulates ATPase activity. Biochemistry 35 8289–8298. [DOI] [PubMed] [Google Scholar]
- Stros, M. and Kolibalova, A. 1987. Interaction of non-histone proteins HMG1 and HMG2 with core histones in nucleosomes and core particles revealed by chemical cross-linking. Eur. J. Biochem. 162 111–118. [DOI] [PubMed] [Google Scholar]
- Stults, N.L. 1997. Preparation of photoprotein conjugates and methods of use thereof. U.S. 21 pp. Cont.-in-part of U.S. 5,486,455.
- Sun, J. and Kaback, H.R. 1997. Proximity of periplasmic loops in the lactose permease of Escherichia coli determined by site-directed crosslinking. Biochemistry 36 11959–11965. [DOI] [PubMed] [Google Scholar]
- Svoboda, M., De Neef, P., Tastenoy, M., and Christophe, J. 1988a. Molecular characteristics and evidence for internalization of vasoactive intestinal peptide (VIP) receptors in the tumoral rat-pancreatic acinar cell line AR 4-2 J. Eur. J. Biochem. 176 707–713. [DOI] [PubMed] [Google Scholar]
- Svoboda, M., Poloczek, P., Winand, J., Robberecht, P., and Christophe, J. 1988b. Species differences in the molecular characteristics of vasoactive intestinal peptide receptors in the pancreas from rat and guinea pig. Eur. J. Biochem. 174 59–66. [DOI] [PubMed] [Google Scholar]
- Swaney, J.B. 1986. Use of cross-linking reagents to study lipoprotein structure. Methods Enzymol. 128 613–626. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Inoue, M., Sakamoto, J., and Sone, N. 1996. Intra- and inter-complex crosslinking of subunits in the quinol oxidase super-complex from thermophilic Bacillus PS3. J. Biochem. 119 482–486. [DOI] [PubMed] [Google Scholar]
- Valenzuela, D., Leon, O., and Schulman, L.H. 1984. Modification of specific lysine residues in E. coli methionyl-tRNA synthetase by crosslinking to E. coli formylmethionine tRNA. Biochem. Biophys. Res. Commun. 119 677–684. [DOI] [PubMed] [Google Scholar]
- Vanhoutte, C. and Malaisse, W.J. 1997. d-Glucose metabolism in dimethyl suberimidate-treated tumoral pancreatic islet cells. Biochem. Mol. Biol. Int. 41 1209–1216. [DOI] [PubMed] [Google Scholar]
- Walleczek, J., Martin, T., Redl, B., Stoeffler-Meilicke, M., and Stoeffler, G. 1989. Comparative cross-linking study on the 50S ribosomal subunit from Escherichia coli. Biochemistry 28 4099–4105. [DOI] [PubMed] [Google Scholar]
- Wang, Q. and Kaback, H.R. 1999. Helix packing in the lactose permease of Escherichia coli determined by site-directed thiol crosslinking: Helix I is close to helices V and XI. Biochemistry 38 3120–3126. [DOI] [PubMed] [Google Scholar]
- Wang, T.W. and Kassell, B. 1974. Preparation of a chemically cross-linked complex of the basic pancreatic trypsin inhibitor with trypsin. Biochemistry 13 698–702. [DOI] [PubMed] [Google Scholar]
- Wasylewska, E., Dulinska, J., Trubetskoi, V.S., Torchilin, V.P., and Ostrowski, W.S. 1987. Stabilization of human prostate acid phosphatase by cross-linking with diimidoesters. Acta Biochim. Pol. 34 145–156. [PubMed] [Google Scholar]
- Watty, A., Methfessel, C., and Hucho, F. 1997. Fixation of allosteric states of the nicotinic acetylcholine receptor by chemical crosslinking. Proc. Natl. Acad. Sci. 94 8202–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watty, A., Weise, C., Dreger, M., Franke, P., and Hucho, F. 1998. The accessible surface of the nicotinic acetylcholine receptor. Identification by chemical modification and crosslinking with 14C-dimethyl suberimidate. Eur. J. Biochem. 252 222–228. [DOI] [PubMed] [Google Scholar]
- Wells, J.A., Knoeber, C., Sheldon, M.C., Werber, M.M., and Yount, R.G. 1980. Cross-linking of myosin subfragment 1. Nucleotide-enhanced modification by a variety of bifunctional reagents. J. Biol. Chem. 255 11135–11140. [PubMed] [Google Scholar]
- Wilbur, D.S. 1992. Radiohalogenation of proteins: An overview of radionuclides, labeling methods and reagents for conjugate labeling. Bioconjug. Chem. 3 433–470. [DOI] [PubMed] [Google Scholar]
- Wolff, J.A., Hagstrom, J.E., Budker, V.G., Trubetskoy, V.S., Slattum, P.M., and Hanson, L.J. 1998. Method for making a compound for delivery to cells by forming a polymer in the presence of a template drug, especially nucleic acid. PCT Int. Appl. 79 pp.
- Wolff, J.A., Hagstrom, J.E., and Budker, V.G. 1999. Polymer formation in the presence of nucleic acid using template polymerization. PCT Int. Appl. 73 pp.
- Wu, J., Voss, J., Hubbell, J.W., and Kaback, H.R. 1996. Site-directed spin labeling and chemical crosslinking demonstrate that helix V is close to helixes VII and VIII in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. 93 10123–10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., Vasilyeva, E., and Forgac, M. 1999. Subunit interactions in the clathrin-coated vesicle vacuolar (H+)-ATPase complex. J. Biol. Chem. 274 28909–28915. [DOI] [PubMed] [Google Scholar]
- Yu, R. and Carter, J. 1976. Cross-linking phytochrome to its receptor in situ using imidoesters. J. Exp. Bot. 27 283–293. [Google Scholar]
- Yu, Y.Y., Wie, B.V.J., Koch, A.R., Moffett, D.F., and Davis, D.C. 1998. Preparation and characterization of bifunctional biopolymers for receptor-based liposomal immunosensing. Biotechnol. Prog. 14 310–317. [DOI] [PubMed] [Google Scholar]
- Zecherle, G.N., Oleinikov, A., and Traut, R.R. 1992. The proximity of the C-terminal domain of Escherichia coli ribosomal protein L7/L12 to L10 determined by cysteine site-directed mutagenesis and protein-protein cross-linking. J. Biol. Chem. 267 5889–5896. [PubMed] [Google Scholar]