Abstract
In Escherichia coli, the IclR protein regulates both the aceBAK operon and its own synthesis. Database homology searches have identified many IclR-like proteins, now known as the IclR family, which can be identified by a conserved C-terminal region. We have cloned and purified one of these proteins, which we have named GclR (glyoxylate carboligase repressor). Although purification is straightforward, both the IclR and GclR proteins are difficult to manipulate, requiring high salt (up to 0.6 M KCl) for solubility. With the advent of nanospray ionization, we could transfer the proteins into much higher concentrations of volatile buffer than had been practical with ordinary electrospray. In 0.5 M ammonium bicarbonate buffer, both proteins were stable as tetramers, with a small amount of dimer. In a separate experiment, we found that IclR protein selected from a random pool a sequence which matched exactly that of the presumed binding region of the GclR protein, although IclR does not regulate the gcl gene. We designed a 29 bp synthetic DNA to which IclR and GclR bind, and with which we were able to form noncovalent DNA-protein complexes for further mass spectrometry analysis. These complexes were far more stable than the proteins alone, and we have evidence of a stoichiometry which has not been described previously with (protein monomer : dsDNA) = (4 : 1).
Keywords: Protein-DNA interaction, noncovalent, nanospray ionization, IcIR family protein, time-of-flight mass spectrometry
IclR protein (an acronym for isocitrate lyase regulator) prevents constitutive expression of some genes required for acetate utilization in Escherichia coli (Kornberg 1963,Kornberg 1966; Brice and Kornberg 1968; Maloy and Nunn 1982; Chung et al. 1988; Sunnarborg et al. 1990; Cortay et al. 1991). IclR is the founding member of a family of proven and hypothetical transcription regulators that are distributed widely among bacteria. All members of the IclR family have a C-terminal sequence motif of ∼180 amino acids, designated PF01614 in the Pfam catalogue of motifs (Bateman et al. 2000), and probably have a helix-turn-helix motif around residues 30–50 (Nègre et al. 1991) in a polypeptide of 240–280 residues. Thus far, ∼100 examples of uncharacterized open reading frames containing the IclR family motif have been found by sequence homology searches (H. Duckworth, unpubl.). Most are known only as a gene sequence with no information about function, but in a few cases a role in transcription has been shown by genetic or in vitro techniques. In Streptomyces griseus and Streptomyces coelicolor, IclR family member GlyR controls expression of the operon concerned with glycerol metabolism (Smith and Chater 1988a,Smith and Chater 1988b; Bolotin and Biro 1990). In the plant pathogen Pectobacterium (i.e., Erwinia), the IclR family members KdgR (Liu et al. 1999), Pir (Nomura et al. 1998, 1999), and RexZ (Thomson et al. 1999) regulate genes concerned with the breakdown of plant cell wall polysaccharides. Other IclR family members regulate genes concerned with the metabolism of aromatic acids, such as protocatechuate in Acinetobacter spp. (Gerischer et al. 1998; Kok et al. 1998), Rhodococcus opacus (Eulberg et al. 1998; Eulberg and Schlomann 1998), and Pseudomonas putida (Guo and Houghton 1999), and 3-(3-hydroxyphenyl) pyruvate in E. coli (Spence et al. 1996; Ferrandez et al. 1997). There are many other cases in which the gene specifying an IclR family member is adjacent to a probable or demonstrated operon, but no regulatory role has yet been documented. Thus proteins of the IclR family have diverse regulatory roles in bacteria, and a proper understanding of their mode of action is of interest.
The most extensively characterized member of the family is IclR. Classical genetics experiments showed that IclR represses transcription of a cluster of genes coding for malate synthase A (aceB), isocitrate lyase (aceA), and isocitrate dehydrogenase phosphatase/kinase (aceK) by binding to an operator-like sequence 5′ to this aceBAK operon (Vanderwinkel and De Vlieghere 1968; Maloy and Nunn 1982; LaPorte and Chung 1985). Mutants of the iclR gene show constitutive expression of this gene cluster now known as the glyoxylate pathway (Brice and Kornberg 1968; Sunnarborg et al. 1990). IclR protein binds specifically to two DNA sequences. One target sequence is the aceBAK operator (Cortay et al. 1991; Nègre et al. 1991, 1992; Donald et al. 1996). The second sequence is in the promoter region of the iclR structural gene, thus allowing IclR to regulate its own synthesis (Gui et al. 1996). The two target sequences are variants of a palindrome of 15 bp (Pan et al. 1996; Hosfield 1996), which fits well with a helix-turn-helix interaction. However, in DNase footprinting experiments, 28–34 bp of DNA are protected (Cortay et al. 1991; Donald et al. 1996; Gui et al. 1996), which suggests that either more than a dimer is involved in the interaction or that the DNA is bent.
The study described here arose out of the discovery of a third potential target sequence for IclR protein in the E. coli genome, which is adjacent to the probable promoter for gcl, the glycolate carboligase gene. This region was implicated previously in expression of gcl (Chang et al. 1993). Cusa et al. (1999) reported their analysis of the transcriptional units in the gcl region and showed that deletion of a gene, which they call allR, results in constitutive expression of three groups of genes concerned with metabolism of glyoxylate and allantoin, one of which is gcl.
To understand the role of this third site better, we describe here the cloning, purification, and characterization of GclR (glycolate carboligase regulator), whose sequence indicates that it is a hitherto unrecognized member of the IclR family of transcription regulators. We have also used electrospray and nanospray ionization time-of-flight mass spectrometry to investigate the conformation and interactions of the IclR and GclR proteins, and present further evidence for a new kind of DNA-protein noncovalent complex (Donald et al. 2000).
Results
Identification of DNA sequences preferred by full-length IclR protein
We have shown previously that IclR protein is a target for protease ompT and that the resulting truncated protein (IclR-8) has a lower binding affinity for ace operon DNA than does the full-length protein (KD IclR = 0.36 ± 0.04 nM; KD IclR-8 = 14 ± 0.6 nM) (Donald et al. 1996). It appears that earlier studies of DNA binding by IclR protein in other laboratories had probably used the truncated form, so it seemed desirable to check the key findings with full-length protein.
To define the base sequence that is best able to bind to IclR protein in the absence of other constraints, we used full-length IclR to select from a mixture of random sequences. Of the 32 clones obtained, 21 contained a palindromic consensus site TGGAAANNNTTTCCA. Of the other 11 clones, seven contained one-half of this palindrome, and the remaining four showed no relationship to the other molecules and were presumably unselected sequences that had been carried through the enrichment procedure. This consensus sequence agrees with that reported by Pan et al. (1996), who as already noted, were probably using truncated IclR. Pan et al. (1996) have shown that some of the departures from the consensus sequence seen in the natural operators are necessary to achieve a working promoter within the same stretch of DNA.
We also used the EMSA method, with full-length IclR protein, to measure DNA binding to each of the two known natural operators, and also to six clones from the amplification/selection experiment: three containing the full palindrome, two containing a half-site only, and one of the apparently random sequences that had survived the selection procedure. Table 1 shows the binding data. The three full palindrome clones and the two natural operators bound IclR in much the same way, with approximately the same affinities and with marked cooperativity. Because the natural operators bound in much the same way as the consensus sequences, at least under in vitro conditions, it is clear that some features of the ideal palindrome defined in the selection experiments can be dispensed with without compromising recognition of the DNA by IclR protein. Binding to the two sequences containing half sites was weaker and noncooperative (Table 1) as might be expected because only one subunit of IclR should bind tightly to such a DNA sequence. The binding to the one apparently random DNA sample was negligible (not shown).
Table 1.
Parameters for binding of full-length IclR protein to selected oligonucleotides and naturally-occuring DNA sequences
| DNA | Selected sequence | [IclR] needed for 50% binding, nM | Hill number |
| Natural sequences | |||
| aceBAK promoter | AAATGGAAATTGTTTTTGATT | 0.36 ± 0.04a | 1.7 ± 0.2a |
| iclR promoter | AAATGAAAATGATTTCCACGA | 0.9 ± 0.1 | 1.6 ± 0.2 |
| gcl promoter | AGTTGGAAAAATTTTCCAATA | 0.5 ± 0.1 | 1.2 ± 0.1 |
| IclR-selected sequences, full palindromes | |||
| Isolate 8a | AATTGGAAACCGTTTCCAAAG | 1.8 ± 0.2 | 1.4 ± 0.2 |
| Isolate 25 | CAATGGAAATTCTTTCCATTC | 0.5 ± 0.1 | 1.4 ± 0.2 |
| Isolate 26 | AAATGGAAATCCTTTCCAATT | 1.9 ± 0.2 | 1.4 ± 0.2 |
| IclR-selected sequences, half palindromes | |||
| Isolate 7 | TGGTGGAAACTAGGTATGTGC | 6.7 ± 0.3 | 0.7 ± 0.1 |
| Isolate 23 | GGGTGGAAATTGTTATTCAGC | 10.3 ± 0.4 | 0.7 ± 0.1 |
| Synthetic DNA | |||
| 29 bp consensus | AAATGGAAATGATTCCACTA | 0.07 ± 0.03 | 0.9 ± 0.1 |
Bases conforming to the core consensus sequence are underlined. In isolate 8a the random region had expanded to 29 bases during the selection process; in the other isolates, this region had remained at 24 bases.
a Data for aceBAK promotor from Donald et al. (1996).
We then made a 29 bp ideal IclR target sequence, designed to contain our consensus palindrome and all other bases that seem to be preferred among the sequences selected by IclR. In the standard EMSA experiment, the binding of IclR to this DNA (Table 1) was indeed better than to any of the other sequences studied so far, with a KD of 0.07 ± 0.03 nM. This DNA was chosen for the mass spectrometry experiments because it was of high purity, and the differences in binding constants were well below the concentration required for characterization by mass spectrometry.
Discovery of a third potential IclR target in the E. coli genome and evaluation of its significance in IclR regulation
A search of the full E. coli genome revealed three good matches to the IclR consensus sequence. Two were the already known operator sequences in the promoter regions of the aceBAK and iclR genes. The third, a new discovery, was the only perfect match and was located at bp 39–53 of Section 47 of the 400-section E. coli genome sequence (GenBank Accession No. AE000157). This location overlaps predicted promoters (bp 20–49 or 29–55) for gcl, the structural gene for glyoxylate carboligase, and a deletion of this region leads to constitutive expression of the carboligase (Chang et al. 1993; Cusa et al. 1999). We therefore investigated whether IclR protein binds to this sequence and whether it has a role in the expression of gcl. First, a 153-bp piece of E. coli genomic DNA containing the putative IclR target was amplified, cloned, and labeled, and its binding to IclR was measured by the EMSA method. We obtained good binding to full-length IclR protein, with an [IclR]50% value of 0.5 ± 0.1 nM and a Hill number of 1.2 ± 0.1.
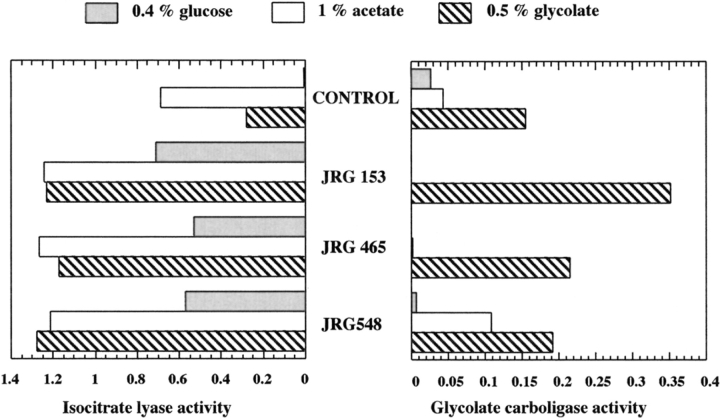
To see whether this binding affinity was associated with regulation of gcl by IclR protein, we assayed the activity of glyoxylate carboligase in iclR− strains. When grown on glucose minimal medium, these strains are constitutive for isocitrate lyase (because of the known defect in regulation of the aceBAK operon), and, if the mechanism of gcl regulation were similar, we would expect a similar elevation of glyoxylate carboligase activity (the product of the gcl gene). This is not the case (Fig. 1 ▶). The control strain, DL41, had a low amount of isocitrate lyase activity when cells were grown with glucose as the carbon source and a considerable increase in enzyme activity when acetate or glycolate were used. These are typical wild-type results. The three different iclR− derivatives of DL41 tested were constitutive for isocitrate lyase, with 40 to 500 times as much enzyme activity in glucose-grown cells as the iclR+ strain had. In contrast, the activity of glyoxylate carboligase increased only when cells were grown on glycolate (or glyoxylate [data not shown]), a known feature of this enzyme (Gupta and Vennesland 1964). Thus, although IclR protein binds well to the proposed target sequence flanking the gcl gene, the presence or absence of a functional iclR gene in E. coli cells appears to be irrelevant to gcl gene expression in vivo. This result indicates that the mode of action of IclR is very complex.
Fig. 1.
Activity of isocitrate lyase and glycolate carboligase in a control and three different IclR− strains in response to different carbon sources. Isocitrate lyase activity is expressed as μmoles of glyoxylate phenylhydrazone/min/mg protein. Glycolate carboligase activity is expressed as μmoles CO2 /min/mg protein.
Characterization of GclR, a new member of the IclR protein family, and its role in regulation of the gcl gene
Because bacterial regulatory proteins are often closely linked to the genes they control, we searched for a gene for a potential IclR homologue in the sections of the E. coli genome near gcl. A likely candidate was found as an open reading frame, designated ybbU, at bp 9996–10811 of Segment 46. The stop codon of this open reading frame is only 54 bp from the new IclR target sequence and 90 bp from the initiation codon of the gcl gene. The inferred amino acid sequence of the putative ybbU-produced protein is 42% identical to that of IclR itself. We will refer to the protein product as GclR because deletion of the gene results in constitutive expression of glycolate carboligase (Chang et al. 1993), although these investigators did not establish what genes were present in the deleted region. Cusa et al. (1999) refer to the ybbU gene as allR, but they did not characterize the protein product, although their deletions confirm the earlier results of Chang et al. (1993).
To clone the ybbU region into pET15b, the second amino acid was changed from threonine to alanine. Assuming that the N-terminal methionine would be removed, the expected mass of the recombinant GclR protein was 29,109 daltons. Purification was achieved by slight modifications to the procedure we use for IclR, producing a protein with a measured mass of 29,104 ± 2 daltons, indicating that the polypeptide was intact. In some preparations, however, in which β-mercaptoethanol was used in the column buffers, we found a second polypeptide of mass 29,181 daltons. This indicated the possibility that the protein might be covalently modified under some conditions.
Identifying the covalent modification in preparations of GclR
To understand the origin of the anomalous protein with mass 29,181 daltons, we prepared a tryptic digest of this protein and analyzed the mixture of fragments by ESI Qq TOF MS/MS. All tryptic peptides predicted from the inferred GclR sequence could be identified at isotopic resolution (data not shown), with the exception of T15 (aa 140–147, 834.40 daltons), T19 (aa 200–207, 924.47 daltons) and T20 (aa 208–240, 3315.65 daltons). Attention was then focused on two doubly charged ions at m/z = 455.71 (910.42 daltons) and m/z = 921.49 (1841.98 daltons), whose masses fit T15 + 76 and T15 + T16 + 76, respectively. The doubly charged ion at m/z = 455.71 was then subjected to partial sequencing by MS/MS, from which it was possible to identify the modification as a 76.01 dalton addition to Cys141 (data not shown). This mass increase is consistent with the formation of a mixed disulfide between Cys141 and β-mercaptoethanol. Replacement of β-mercaptoethanol by DTT eliminated the modification. Thus, the extra peak is an artifact of preparation, but its formation indicates that Cys141 is at a reactive, surface position in native GclR protein. Packman and Berry (1995) describe in some detail the identification of one β-mercaptoethanol-modified cysteine on the surface of an aldolase.
Mass spectrometric analysis of the native proteins
No mass spectrum could be obtained for full-length IclR unless the protein was prepared in waterbugs (Orr et al. 1995) and the denatured protein solubilized with acetic acid. The protein came out of solution rapidly, but the closed configuration of the waterbugs allowed for a quantitative recovery. Attempts to exchange the full-length protein into 0.5 M NH4HCO3, 0.1 mM DTT buffer by Centricon filtration caused the protein to precipitate before buffer exchange was complete. We did have limited success with the truncated version, IclR-8, some of which remained in solution during the buffer exchange. Considerable material was lost, and the protein that remained in solution gave good spectra for < 24 h. This was in marked contrast to the DNA-protein complexes (see below), which were stable for at least 3 d and could tolerate much lower ionic strength. This behavior is reminiscent of our experience with TrpR protein, which is partially denatured in ESI compatible buffers, whereas its complex with DNA is stable (Potier et al. 1998).
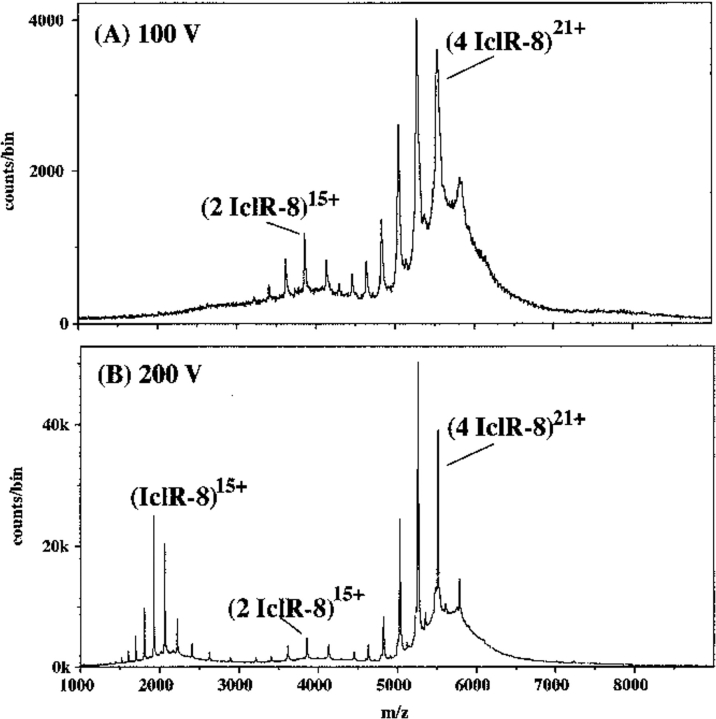
Nanospray ionization of the IclR-8 protein from 0.5 M NH4HCO3, 0.1 mM DTT showed two major ion envelopes at 100 V declustering voltage (Fig. 2A ▶), which can be identified as dimer and tetramer. The major ions at m/z = 5000–6000 represent the 20–24+ charge states of a species with mass 116,175 daltons, 817 daltons FWHM (expected mass for a tetramer of IclR-8 is 115,728 daltons). The second ion envelope represents the 14–16+ charge states of a species with mass 57,977 daltons, 144 daltons FWHM (expected mass for a dimer is 57,864 daltons). When the declustering voltage was increased to 200 V (Fig. 2B ▶), the amount of dimer decreased, and ions attributable to monomer began to appear. The peaks were now much sharper, and the measured masses closer to the expected values (tetramer 115,795 daltons, 168 daltons FWHM; dimer 57,907 daltons, 104 daltons FWHM; monomer 28,933 daltons, 24 daltons FWHM). At a declustering voltage of 300 V, the monomer ions became the major species in the spectrum and the tetramer became less distinct, whereas at 50 V declustering voltage, the spectrum was similar to that in Figure 2A ▶, except the peaks were broader and not as clearly defined (data not shown). Dilution of the buffer to 0.25 M produced a spectrum with approximately equal monomer, dimer, and tetramer ions. At 10 mM buffer, only monomer ions were present. This shows that IclR-8 protein is not stable in low salt in the absence of DNA, and we were fortunate to be able to obtain a spectrum with 0.5 M ammonium bicarbonate, which is near the working limit for this buffer in the nanospray apparatus.
Fig. 2.
Nanospray spectra of 11 μM IclR-8 protein (the truncated protein) in 0.5 M NH4HCO3, 1 mM DTT. (A) At 100 V declustering voltage, showing two ion envelopes, the 20–24+ ions of tetramer, and the 14–17+ ions of dimer. There is little evidence of fragmentation or of monomer ions. (B) At 200 V declustering voltage, showing three ion envelopes, with highly charged monomer ions produced at the expense of the others.
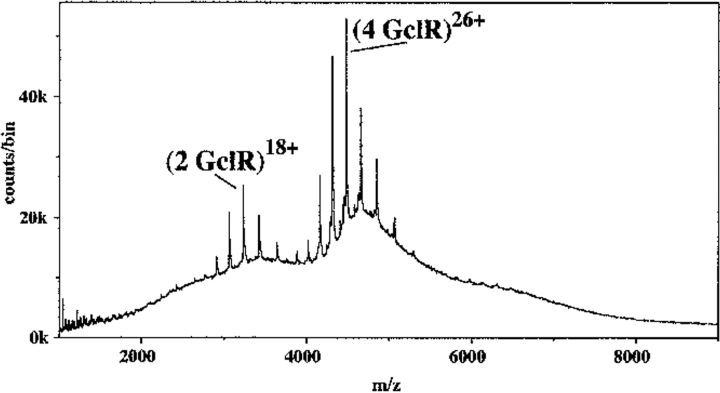
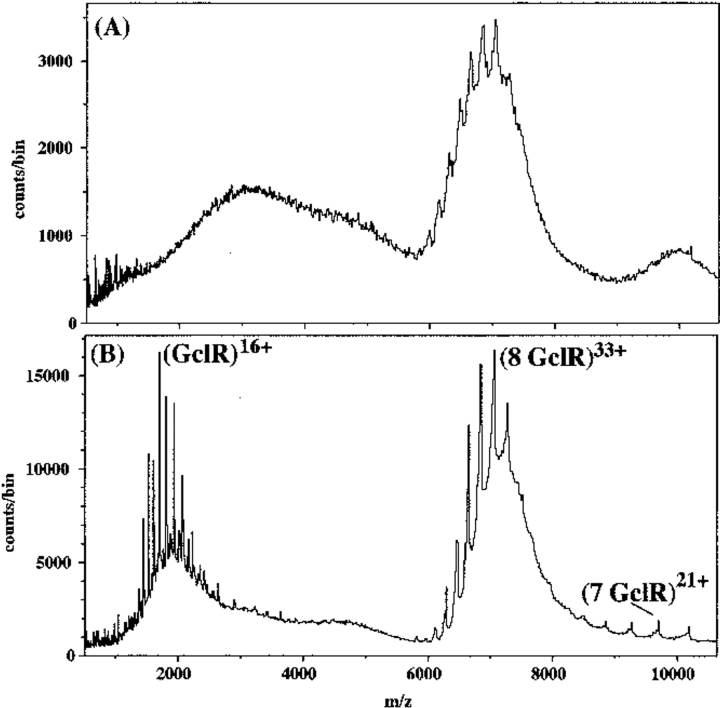
Native GclR protein was much easier to characterize by nanospray ionization TOF MS than the IclR-8 protein, and it had the same kind of ion distribution. In Figure 3 ▶, the major ions at 4000–5000 m/z represent the 23–28+ ions of a species with mass 116,521 daltons, 172 daltons FWHM, (expected mass for tetramer = 116,436 daltons). The second ion envelope is the 16–20+ ions of a species with mass 58,253 daltons, 87 daltons FWHM, (expected mass for dimer = 58,218 daltons). As with the IclR-8 protein, an increase in the declustering voltage led to the appearance of ions from monomer at the expense of dimer ions. Unlike the case with IclR-8 protein, a decrease in the buffer concentration below 0.3 M (Fig. 4 ▶) produced a new species at 7000 m/z, which can best be explained as an octamer (observed mass 233,022 daltons [expected mass = 232,872 daltons]). The ions of the octamer could be resolved only by increasing the declustering voltage, which caused some fragmentation as shown by the appearance of ions from heptamer at higher m/z and ions from monomer which have very high charge states. When the buffer concentration was < 0.1 M, the octamer was replaced by an unresolvable large aggregate (data not shown), which may mean that the octamer is some sort of intermediate.
Fig. 3.
Nanospray spectrum for 13 μM GclR protein in 0.5 M NH4HCO3, 1 mM DTT. At 120V declustering voltage, showing two ion envelopes, the 23–28+ ions of tetramer, and the 16–20+ ions of dimer. As with the IclR protein under gentle ionization conditions, there is little evidence of either monomer formation or fragmentation.
Fig. 4.
Nanospray spectra of 13 μM GclR protein in 0.1 M NH4HCO3, 1 mM DTT. (A) At 150V declustering voltage, one ion envelope, which is not well resolved. (B) At 250 V declustering voltage, the envelope resolves into the 32–37+ ions of an octamer, accompanied by the appearance of highly charged monomer ions and some evidence of heptamer. There is little evidence of ions from the tetramer or dimer.
DNA binding by GclR protein
The ability of the purified GclR protein to bind to known IclR target DNA sequences was studied by the EMSA method under the same conditions used for IclR protein. GclR protein binds the gcl promoter fragment with [GclR]50% = 39 ± 4 nM, and the aceBAK promoter with [GclR]50% = 76 ± 17 nM. The equivalent values for IclR binding are 0.5 ± 0.1 nM and 0.36 ± 0.04 nM, respectively, which indicates that the binding conditions for the GclR protein have not been optimized. The GclR binding was not affected by the addition of 1 mM glycolate or glyoxylate to all the buffers in the system.
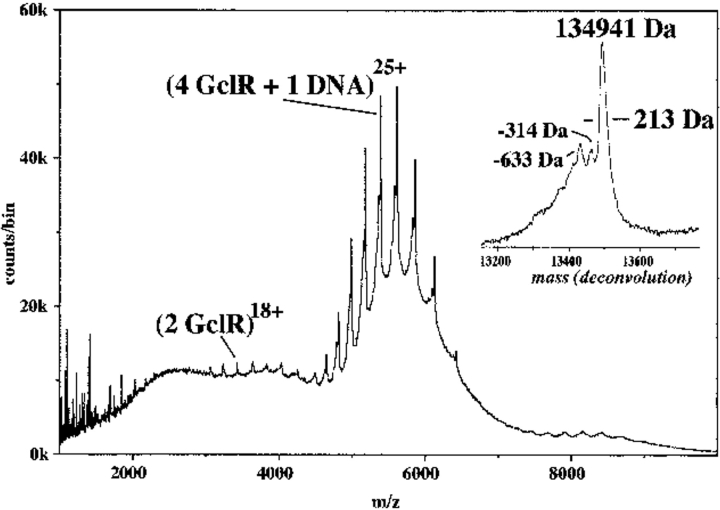
GclR binding to IclR target DNA could also be shown by nanospray ionization TOF MS, using the 29 bp IclR consensus oligonucleotide. There was one major ion envelope (Fig. 5 ▶) with the 22–28+ ions of a species with measured mass 134,941 daltons (expected for a GclR tetramer plus one dsDNA [double-stranded DNA] = 134,844 daltons). Close inspection of the deconvoluted spectrum (Fig. 5 ▶, inset) shows two minor species: 134,627 daltons and 134,308 daltons, 314 and 633 daltons smaller, respectively, than the major ion. The simplest explanation for these extra peaks is that there are failure sequences in the "top" strand of the consensus DNA. The complex with mass 134,627, then, would lack the 5′ adenine nucleotide (313 daltons), and the complex with mass 134,308 would lack this and the adenine nucleotide next to it. By adjusting the spray conditions, the excess uncomplexed DNA could be observed as ssDNA (single-stranded DNA) using a declustering voltage of 110 V but as dsDNA at 80 V. There were no ions that could be assigned to a complex made up of GclR dimer plus one dsDNA or GclR tetramer plus two dsDNA.
Fig. 5.
Nanospray spectrum of 13 μM GclR protein with 6 μM consensus DNA in 10 mM NH4HCO3, 1 mM DTT. This spectrum was acquired continuously over 40 min. Only one major ion series is found, with the 22–28+ of a complex of mass 134,939 daltons, which can be explained as a tetrameric protein with one dsDNA. This complex was stable at 160 V (compare to the spectrum of the protein alone in Fig. 3 ▶). Addition of excess DNA made the peaks broader (increase in the FWHM value from 213 daltons), but did not produce any different ions. The deconvolution of the major ion series (inset) shows three species, the most abundant representing a complete dsDNA and four protein subunits. The minor species probably represent failure synthesis of the DNA. The ions at m/z 3000–4000, are the 14–20+ ions from protein dimer (see also Fig. 3 ▶).
DNA binding by IclR-8 protein
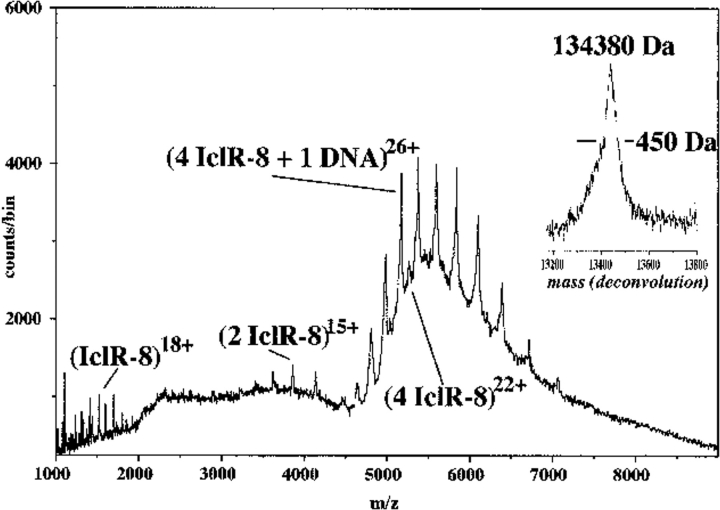
The IclR-8-DNA complex was analyzed initially by electrospray ionization from 5 mM NH4HCO3, but spectral quality improved by using 10 mM NH4HCO3 and ionization by nanoelectrospray. However, resolution was never as good as for the GclR-DNA complex. The complex formed between IclR-8 protein and the 29-bp consensus oligomer gave one major ion envelope (Fig. 6 ▶). This contained the 20–28+ ions from a species with measured mass of 134,380 daltons, corresponding to tetrameric IclR-8 protein plus one ds DNA fragment (expected mass 134,136 daltons). Between the 24+ and 27+ ions of the complex there were ions that matched the 21–23+ ions from protein tetramer (see also Fig. 2A ▶). There was also a small amount of dimer (14–16+ ions) and some monomer. There were no ions that matched either a dimer plus one dsDNA or a tetramer plus two dsDNA, even when twice as much DNA was used. Analysis of the same sample after 24 h showed more of the dimer protein and less of the DNA-protein complex. A similar experiment with IclR-8 protein and the opTB DNA (data not shown) also had one major ion envelope giving a mass for the complex 133,640 daltons (expected for one DNA plus one IclR-8 tetramer, 133,518 daltons). This DNA sample was not as pure, and this severely restricted the resolution that could be obtained. As with the GclR-DNA complex, there were no ions to match any other complex.
Fig. 6.
Nanospray spectrum of 11 μM IclR-8 protein plus 5 μM consensus DNA in 10 mM NH4HCO3, 1 mM DTT, at 140 V declustering voltage. This spectrum was acquired over 40 min (compare to identical conditions used for the complex in Fig. 5 ▶). The major ion species represent a tetramer plus one dsDNA. The deconvolution (inset) gives mass 134,380 daltons at poor resolution (450 daltons FWHM). There is also evidence of uncomplexed protein tetramer, dimer, and monomer.
Evidence that full-length IclR protein binds one operator DNA
IclR protein contains a good match to the helix-turn-helix DNA binding motif (Nègre et al. 1991), and was expected to interact with its quasi-palindromic DNA target as a dimer. We showed earlier that both full-length and truncated versions of IclR protein are tetrameric at the relatively high concentrations used for molecular weight determination by gel filtration (Donald et al. 1996), measurements that agree with the mass spectrometry data reported herein. However, gel shift assays at lower concentrations of protein and DNA allowed us to test the possibility that IclR could bind two operators simultaneously, as has been shown for the lactose repressor (Lewis et al. 1996). To test this, we adopted a procedure used to identify such complexes (Krämer et al. 1987) by incubating IclR protein with pairs of 32P-labeled operator DNAs of different lengths, and looking for complexes of slower mobility than those formed when only one such DNA was used. The operator DNAs used were restriction fragments of 333-bp (XmnI-MluI fragment from pCHicl), 227-bp (EcoRI-HindIII fragment from pTZop), and 170-bp (BamHI fragment from pTZop), used in the three possible pairwise combinations. No new bands were seen (data not shown). The lack of a complex of novel mobility indicated that IclR binds only one DNA fragment under standard gel shift conditions, which agrees with the MS measurements. The unusually large amount of DNA protected from DNase by IclR (Cortay et al. 1991; Donald et al. 1996; Gui et al. 1996) may also be explained if a tetramer, rather than a dimer, forms the complex.
Discussion
We have shown previously that DNA-protein complexes can be studied by electrospray ionization mass spectrometry, and that the results are consistent with those from other methods. Using samples prepared in 5 mM NH4OAc, we found that the tryptophan repressor protein (TrpR) binds only specific DNA with the correct spacing of the recognition sequence, and in the expected stoichiometry of 2 protein subunits : 1 dsDNA (Potier et al. 1998). These results are consistent with the findings of both NMR (Ramesh et al. 1994; Zhang et al. 1994) and X-ray crystallography (Otwinowski et al. 1988; Lawson and Carey 1993). A similar experiment with the lactose repressor protein (LacI) and the 20-bp dsDNA used for crystallography analysis (Lewis et al. 1996), also produced the expected complex with stoichiometry of 4 protein subunits : 2 dsDNA. In addition, the LacI + DNA clearly showed evidence for 2 protein subunits : 1 dsDNA and 4 protein subunits : 1 dsDNA (Donald et al. 1998). Although these protein/DNA complexes were very stable, the proteins by themselves were not, and we were only able to resolve monomers of both proteins at that time.
With the introduction of nanospray ionization, we have shown that it is now possible to analyze proteins directly in buffer concentrations up to 0.5 M. Using this technique, we have shown that both IclR-8 and GclR proteins behave as tetramers when the buffer concentration is high enough. The small amount of dimer shown in the mass spectrometer may indicate a dimer-tetramer equilibrium, and it may be possible to study this by nanospray in the future. The IclR-8 workup was still difficult, given the instability of this protein in solution, but with electrospray it had been impossible. Unlike the acid-denatured proteins, which give sharp ion peaks, high resolution, and reasonable error measurements, the nondenatured proteins produce broader ion peaks with poorer resolution. The masses of the complexes seen are consistently somewhat larger (100–500 daltons) than what is calculated. This is always found for macromolecules studied by nanospray or ESI TOF MS under nondenaturing conditions, perhaps because of the presence of trapped water molecules; it contrasts with the experience of acid-denatured molecules, whose measured masses agree exactly with the calculated values.
As with TrpR and LacI, the IclR-8 and GclR complexes with DNA are very stable and tolerate much lower concentrations of buffer than the proteins do themselves. However, the stoichiometry of DNA binding, 4 protein subunits : 1 dsDNA, was unexpected. The gel-shift experiments with different sizes of DNA confirm that only one dsDNA interacts with the protein, but do not give any information on the conformation of the protein in the complex. Previous attempts to measure the mass of the complex of IclR and the opTB DNA (Donald et al. 1996) showed only that it was much larger than a simple dimer plus one dsDNA. Our previous experiments with TrpR and LacI and their specific DNA have shown that mass spectrometry analysis gives the same stoichiometry as other methods. Given our experience with these well-known systems, we are confident that our mass spectrometry results with the IclR-8 and GclR proteins are valid.
If these proteins indeed contain helix-turn-helix motifs, why do we not see the expected stoichiometry of 2 protein subunits : 1 dsDNA? If the proteins have no functional reason to bind two target DNA sequences simultaneously (and there is no evidence for this), the second pair of helix-turn-helix motifs may simply be unavailable for DNA binding for steric reasons. This differs from the LacI system in which, it has been proposed, binding to two of three available operators in the lac operon region could allow tighter regulatory control by a DNA looping mechanism (Lewis et al. 1996).
Our discovery that IclR and GclR proteins have the same DNA sequence specificity raises broader questions about the mechanism of action of IclR family proteins. We have shown that IclR binds well to a sequence at or near the probable gcl promoter, yet bacteria lacking a functional IclR protein show no elevation in levels of the gcl product, glyoxylate carboligase. The recent work of Cusa et al. (1999) on the gcl region indicates that the GclR protein (product of the gene they call allR) does repress transcription of the gcl gene, and it is natural to assume that this process involves GclR binding to the IclR/GclR target we have identified. Finally, although GclR binds reasonably well to IclR target operators as if it were a repressor itself, more than three decades of research on regulation of the glyoxylate cycle operon in E. coli have never revealed a regulatory element for that operon that maps anywhere near the structural gene for GclR. In summary, we believe that although IclR and GclR bind to the same DNA sequences, they do not regulate the same genes. The resolution of this paradox must be that one or both of the two proteins also interact with some other entity, perhaps another regulatory protein yet to be defined.
Materials and methods
Preparation of DNA
Oligonucleotides were synthesized by phosphoroamidite chemistry at 0.2 μM scale, isolated from the column support, and desalted using standard procedures (Applied Biosystems Bulletin #30). For selection of the consensus binding site, we designed a 60-mer with a central tract of 24 random bases, flanked by the sequence 5′-CCGAAGCGGAATTCAATT-3′ on the 5′ side of the tract and 5′-GCATTGGATCCTGCTCGC-3′ on the 3′ side. This oligonucleotide was further purified by electrophoresis through a 12% polyacrylamide gel, followed by Sep-Pak purification. The second strand was synthesized using Klenow extension of a specific primer (Maniatis et al. 1982).
The synthetic promoter region opTB was made with two 29-mers, 5′-GATCCAAAATGGAAATTGTTTTTGATTTG-3′ and 5′-GATCCAAATCAAAAACAATTTCCATTTTG-3′. Equimolar amounts of the two oligonucleotides were mixed in 50 mM ammonium bicarbonate buffer, heated to 80°C, and cooled slowly to give a 25-bp fragment with BamHI sticky ends, and a calculated mass of 17,790.6 daltons. We previously used this dsDNA in an attempt to measure the mass of DNA-IclR complex by a gel electrophoresis method (Donald et al. 1996).
The two HPLC purified 30-mers used to form the 29-bp consensus binding site, 5′-AACTAAAATGGAAATGATTTCCAC TATACA-3′ and 5′TTGTATAGTGGAAATCATTTCCATTT TAGT-3′, were purchased from the Midland Certified Reagent Company. Duplex DNA was formed the same way as for opTB (see above) and had a calculated mass of 18,408.0 daltons. For gel shift assays, the upper strand was first end-labeled (Maniatis et al. 1982), then annealed to the second strand.
For most gel-shift experiments, we used the natural promoter/operator region from the aceBAK operon, obtained as a 227-bp EcoRI-HindIII restriction fragment from plasmid pTZop (Donald et al. 1996). In one experiment we also used a smaller BamHI fragment of 170 bp from the same plasmid. The promoter/operator region of the iclR gene was the 333-bp XmnI-MluI restriction fragment from pCHicl (Donald et al. 1996). For gel-shift experiments with the gcl promoter region, we amplified the relevant fragment by PCR, using E. coli genomic DNA as template with two oligonucleotide primers, 5′-GGCGTTGGGACTGAATTCA CATCCAT-3′ and 5′-GGCGGTAGTGAAAGCTTCTTTCTCC A-3′. These correspond to nucleotides 12–37 and 194–169, respectively, of the gcl gene (Chang et al. 1993), but introduce an EcoRI and a HindIII restriction site, respectively. The amplified product was digested with the two enzymes and cloned into phage M13mp18 to give product M13gcl. We used the EcoRI-HindIII insert from M13gcl (153 bp) for EMSA experiments. DNA was purified and labeled using standard methods (Maniatis et al. 1982).
Preparation and characterization of full-length and truncated IclR proteins
Expression and purification of full-length wild-type IclR protein was performed as described previously, using the IclR expression plasmid pKKICL8 and the host cell line LD103, which is ompT− (Donald et al. 1996). To produce truncated protein (IclR-8), the host cell line JM103 was transformed with pKKICL8, and the cells were lysed in the presence of EDTA to allow cleavage of IclR by OmpT protease (Sugimura and Nishihara 1988; Donald et al. 1996). Purification of the protein was essentially the same as for the full-length protein except that HEPES buffer at pH 7.6 was substituted for Tris-Cl at pH 7.8. This allowed preparation of IclR-8 protein without the HU protein contaminant (Donald et al. 1996). Assessment of the OmpT cleavage was measured by mass spectrometry on a denatured sample.
Cloning, preparation and characterization of GclR protein
Two primers, 5′-GTTAACGTAGCCATGGCGGAAGTTAG-3′ and 5′-GTCAAAAAAACCGGATCCGCAGAGC-3′, were designed to produce an NcoI site at the 5′ end, and a BamHI site at the 3′ end of the E. coli genomic open reading frame ybbU (in genomic segment 46/400, GenBank Accession No. AE000156). After PCR from E. coli genomic DNA, the product was ligated into the SrfI site of PCRScript (Stratagene). Integrity of the clone was confirmed by sequencing with the T3, T7, and two internal primers. The NcoI-BamHI fragment was then subcloned into pET15B (Novagen) to create plasmid pETgclR. For expression of protein, this plasmid was transformed into strain BL21(DE3)pLysS. Liquid cultures were grown to midlog phase, then protein expression was induced with 1 mM IPTG. After 3 h, cells were harvested by centrifugation, suspended in 3× volume of 20 mM Tris-Cl, 1 mM EDTA, 1 mM DTT, and 150 mM KCl at pH 7.8 and lysed in a French pressure cell. The cellular debris was pelleted by a further centrifugation at 18,000 g for 1 h, and the clear supernatant loaded directly onto a Bio-Rex70 (Biorad) column equilibrated with the same buffer. Proteins were eluted with a KCl gradient to 0.5 M and the fractions were then analyzed by SDS-PAGE, absorbance at 280 nm, and conductivity. The GclR protein normally eluted at 0.3 M KCl. Fractions were pooled, concentrated by vacuum dialysis and passed through a G100 (Pharmacia) column equilibrated in 20 mM Tris-Cl, 1 mM EDTA, 300 mM KCl, and 1 mM DTT at pH 7.8. Fractions were analyzed as before, pooled, and concentrated.
Concentrations of pure DNA-binding proteins were determined from the absorbance of their solutions at 280 nm. For IclR-8, a molar extinction coefficient of 15,040 M−1cm−1 was determined previously (Donald et al. 1996). For GclR, the predicted amino acid sequence includes two tryptophans and six tyrosines. We used the Edelhoch (1967) method to measure the tryptophan and tyrosine concentrations in a solution of GclR whose A280 was known. From these measurements, the molar extinction coefficient of GclR was determined to be 21,600 M−1cm−1.
Selection of DNA sequences binding preferentially to IclR protein
The approach of Thiesen and Bach (1990) was followed, in which IclR protein was used to select DNA sequences from a pool of random sequences; the selected molecules were amplified by PCR, and the selection and amplification were repeated until the tightly-bound sequences were highly enriched. Freshly diluted IclR protein and the 60-bp DNA fragment were mixed in EMSA buffer (20 mM Tris-Cl at pH 7.8, 50 mM KCl, 8.3 mM MgCl2, 0.6 mM EDTA, 0.4 mM DTT, 42 μg/mL BSA, and 5% glycerol). After 30 min at room temperature, the IclR-DNA complex was collected on nitrocellulose (Thiesen and Bach 1990). The selected DNA was eluted with 0.5 M KCl and then PCR was used to amplify the selected sequences using primers corresponding to the 5′-flanking sequence and to the reverse complement of the 3′-flanking sequence in the 60-mer. Reaction mixtures were heated to 80°C before addition of enzyme, followed by amplification for 10 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. These selected and amplified sequences were then subjected to five more rounds of selection and amplification using the same conditions. The concentration of IclR used was 175 nM, 100 nM, and 75 nM in the first three selection steps, and 22 nM in the remaining selection steps. The first two steps were performed with 1 μg DNA with 0.2 μg used in the latter steps. To monitor success in enriching IclR-specific DNA, samples of selected DNA were removed at each cycle, amplified by PCR using 32P-dATP in place of unlabelled dATP, and used in gel shift experiments to determine the concentration of IclR protein needed for 50% binding. The unselected starting DNA bound only weakly to IclR even at a protein concentration of 5.6 μM, whereas the product of six cycles of selection/amplification had an [IclR]50% value of about 2 nM. This final selected product was digested with EcoRI and BamHI and ligated into M13mp18 cut with the same enzymes. Clones were sequenced by dideoxy chain termination (Sanger et al. 1977).
DNA binding measurements
Gel shift (EMSA) experiments and analysis of the data were performed as in Donald et al. (1996).
Glyoxylate carboligase in IclR strains
Isogenic strains lacking functional IclR protein were constructed from strains known to be constitutive for the aceA gene product, isocitrate lyase. Three iclR−, metA+ strains and one iclr+, metA− strain were obtained from the E. coli Genetics Stock Center (CGSC) at Yale University. P1 lysates of each of the IclR− strains, JRG 153 (CGSC#4457 [Herbert and Guest 1968]), JRG 465 (CGSC#4456 [Herbert and Guest 1969]), and JRG 548 (CGSC#5859 [Creaghan and Guest 1972]) were prepared (Miller 1972), transduced into DL41 (CGSC#7177 [Hendrickson et al. 1990]) and plated onto minimal M9 glucose plates (Miller 1972), selecting for metA+. Because the metA and iclR loci are tightly linked, most metA+ isolates from this experiment were expected to be constitutive for isocitrate lyase. To confirm this, all transductants were assayed for both isocitrate lyase and glycolate carboligase. Isolates showing high constitutive levels of isocitrate lyase were grown in minimal M9 medium with alternative carbon sources: 0.4% glucose, 0.4% glycolate, 0.5% glyoxylate, or 1% acetate. For strain DL41, methionine was also added at a concentration of 12.5 μg/mL. Cells were grown with shaking overnight at 37°C, harvested by centrifugation, washed with 5 mL of 50 mM K-PO4 at pH 7.0, pelleted, and stored at −20°C. Pellets were thawed in 1 mL of 50 mM K-PO4 at pH 7.0, 0.1 mM TPP, 2 mM MgCl2, sonicated (Artek Sonic dismembrator, set at 40) for three, 30 sec bursts, and the debris removed by centrifugation at 24,000 g for 15 min. Glyoxylate carboligase assays were performed by manometry as described by Gupta and Vennesland (1964), except that the total volume was 3.2 mL and the incubations were performed in air for 10 min, conditions were similar to those used by Chang et al. (1993). Activity was calculated as μmoles of CO2 released per min per mg protein. Isocitrate lyase was assayed by the method of Dixon and Kornberg (1959) in which activity is calculated as μmoles of glyoxylate phenylhydrazone per min per mg protein. The Lowry et al. (1951) method was used to measure protein. All assays were performed at least in duplicate.
Mass spectrometry
All measurements of intact proteins and complexes were performed on a time-of-flight mass spectrometer (TOF3) constructed at the University of Manitoba (Verenchikov et al. 1994; Krutchinsky et al. 1998) using the singly and doubly charged ions of substance P for calibration. Data were acquired and analyzed using TOFMA, an in-house software program. Analysis of tryptic fragments of GclR protein and MS/MS sequencing of individual peaks were performed on an ESI Qq TOF tandem instrument (Shevchenko et al. 1997). The technology is reviewed in Chernushevich et al. (1999).
For routine monitoring of all protein preparations, 2–3 nmole (in ∼100 μL) of pure protein was dialyzed against at least three changes of 1 L of nanopure water using waterbugs (Orr et al. 1995). All the protein was recovered by adding 2–5 μL of acetic acid directly to the sample. This denatured protein was diluted to about 20 μM monomer, mixed with an equal volume of methanol, and characterized by ESI TOF MS.
For characterization of the native proteins, 2–3 nmole of pure protein was diluted into 2 mL of cold, filtered, 0.5 M NH4HCO3, 0.1 mM DTT in a Centricon 10 (Amicon) ultrafiltration unit. The volume was reduced to 100 μL by centrifuging at 6000 g for 1 h at 4°C. The sample was then diluted with fresh buffer to 2 mL, and the centrifugation was repeated eight times. The recovery volume was adjusted to give a final concentration of 10–15 μM monomer. The protein was recovered with a GELoader tip (Eppendorf) and loaded directly into a nanospray capillary (New Objective Picotip).
To form the DNA-protein complexes, we started with the protein at ∼1 mg/mL in the high salt buffer used for protein purification (1 mM EDTA, 1 mM DTT, and either 20 mM Tris-Cl at pH 7.8 and 0.3 M KCl for GclR, or 20 mM HEPES at pH 7.6 and 0.4 M KCl for IclR-8). The actual volume varied with experiment, but was in the range of 2–3 nmole protein in 50–70 μL. To this was added 1–5 μL of the annealed dsDNA, either the 29-bp consensus binding site or the 25-bp opTB natural operator sequence, and the mixture was left for 30 min at room temperature. In all experiments, the amount of DNA added was calculated to be slightly in excess of an exact mixture. Initially, we used 2 nmole protein (monomer) : 1 nmole DNA, because we expected a 2 : 1 stoichiometry. Later experiments were performed with less DNA, which improved resolution but did not change the ion distribution. We have shown that an excess of DNA can affect the quality of the spectra and can cause adducts on the major complexes (Potier et al. 1998). The complex was then exchanged into 10 mM NH4HCO3, 0.1 mM DTT buffer. The samples were prepared initially using the waterbug procedure (Orr et al. 1995), which had been successful with TrpR-DNA complexes (Potier et al. 1998), but spectral quality improved when the samples were washed repeatedly in Microcon 50 (Amicon) ultrafiltration units. Up to 10 washes were required.
Initially the protein-DNA complexes were diluted to 5 mM buffer and analyzed by ordinary electrospray ionization. However, better quality spectra were obtained with 10 mM buffer and nanoelectrospray ionization as described by Wilm and Mann (1996). All spectra of native proteins and complexes were acquired with SF6 as the carrier gas, because it improves resolution for large biomolecules (Krutchinsky et al. 2000) and a minimum back pressure of air to maintain liquid at the orifice. The nozzle voltage was set at 1 KeV, and the collisional voltage (Vc) adjusted to give either the best resolution of ions of interest or to give some information on the stability of the complex.
Samples destined for tryptic digest were first dialyzed in waterbugs (Orr et al. 1995) into 0.1 M NH4HCO3. TPCK-trypsin was added at a ratio of 1 : 100 mg target protein and incubated for 12–24 h at 37°C. Samples were lyophilized, taken up in 1% acetic acid, 50% methanol, and sprayed directly into the mass analyzer. Spectra were analyzed as for the complete proteins. These data yielded isotopic resolution, so charge states could be assigned directly.
Acknowledgments
We thank Andrew Krutchinsky for assembling the apparatus for nanospray ionization and the members of the TOF laboratory for their technical expertise, especially Sasha Loboda, Vic Spicer, and Jim McNabb. We are grateful to the Department of Microbiology, University of Manitoba for use of their facilities, to Jack Switala who synthesized the oligonucleotides, and to Dr. H. Halvorson who loaned us his manometry equipment. Lorraine Lamaga provided excellent technical assistance. This work was supported by Research Grants to H.W.D., W.E., and K.G.S. from the Natural Sciences and Engineering Research Council, Canada.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
PCR, polymerase chain reaction
EMSA, electrophoretic mobility shift assay
TPP, thiamine pyrophosphate (cocarboxylase)
DTT, dithiothreitol
ESI, electrospray ionization
TOF MS, time-of-flight mass spectrometry
SDS-PAGE, sodium dodecyl sulphate polyacrylamide gel electrophoresis
MS/MS, tandem mass spectrometry (for sequencing)
FWHM, full width half maximum (error measurement).
Article and publication are at http://www.proteinscience.org/cgi/doi/10. 1101/
References
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L.L. 2000. The Pfam protein families database. Nucleic Acids Res. 28 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin, A. and Biro, S. 1990. Nucleotide sequence of the putative regulatory gene and major promoter region of the Streptomyces griseus glycerol operon. Gene 87 151–152. [DOI] [PubMed] [Google Scholar]
- Brice, C.D. and Kornberg, H.L. 1968. The role of acetate in isocitrate lyase induction. J. Bacteriol. 96 2185–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.Y., Wang, A.Y., and Cronan, Jr., J.E. 1993. Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase. J. Biol. Chem. 268 3911–3919. [PubMed] [Google Scholar]
- Chernushevich, I.V., Ens, W., and Standing, K.G. 1999. Orthogonal-injection time-of-flight mass spectrometry for analysis of biomolecules. Anal. Chem. 71 452A–461A. [DOI] [PubMed] [Google Scholar]
- Chung, T., Klumpp, D.J., and LaPorte, D.C. 1988. Glyoxylate bypass operon of Escherichia coli: Cloning and determination of the functional map. J. Bacteriol. 170 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortay, J.C., Nègre, D., Galinier, A., Duclos, B., Perrière, G., and Cozzone, A.J. 1991. Regulation of the acetate operon in Escherichia coli: Purification and functional characterization of the IclR repressor. EMBO J. 10 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creaghan, I.T. and Guest, J.R. 1972. Amber mutants of the α-ketoglutarate dehydrogenase gene of Escherichia coli K12. J. Gen. Microbiol. 71 207–220. [DOI] [PubMed] [Google Scholar]
- Cusa, E., Obradors, N., Baldoma, L., Badia, J., and Aguilar, J. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181 7479–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, G. and Kornberg, H.L. 1959. Assay methods for key enzymes of the glyoxylate cycle. Biochem. J. 72 3P. [Google Scholar]
- Donald, L.J., Chernushevich, I.V., Zhou, J., Verentchikov, A., Poppe-Schriemer, N., Hosfield, D.J., Westmore, J.B., Ens, W., Duckworth, H.W., and Standing, K.G. 1996. Preparation and properties of pure, full-length IclR protein of Escherichia coli. Use of time-of-flight mass spectrometry to investigate the problems encountered. Protein Sci. 5 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, L.J., Loboda, A.V., Potier, N., Swint-Kruse, L., Ens, W., Matthews, K.S., Duckworth, H.W., and Standing, K.G. 1998. ESI/TOF measurements of a noncovalent complex between lactose repressor protein (LacI) and double-stranded DNA containing its specific operator sequence. In Proc. 46th ASMS Conf. Mass Spectrom. Allied Topics. Orlando, FL, p. 379.
- Donald, L.J., Duckworth, H.W., Ens, W., and Standing, K.G. 2000. Characterization of the IclR protein and its complex with two different ds DNA fragments. In Proc. 48th ASMS Conf. Mass Spectrom. Allied Topics. Long Beach, CA.
- Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6 1948–1954. [DOI] [PubMed] [Google Scholar]
- Eulberg, D. and Schlomann, M. 1998. The putative regulator of catechol catabolism in Rhodococcus opacus 1CP — An IclR-type, not a LysR-type transcriptional regulator. Antonie Van Leeuwenhoek 74 71–82. [DOI] [PubMed] [Google Scholar]
- Eulberg, D., Lakner, S., Golovleva, L.A., and Schlomann, M. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: Evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandez, A., Garcia, J.L., and Diaz, E. 1997. Genetic characterization and expression in heterologous hosts of the 3-3-(hydroxyphenyl)-propionate catabolic pathway of Escherichia coli K-12. J. Bacteriol. 170 2573–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerischer, U., Segura, A., and Ornston, L.N. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, L., Sunnarborg, A., Pan, B., and LaPorte, D.C. 1996. Autoregulation of iclR, the gene encoding the repressor of the glyoxylate bypass operon. J. Bacteriol. 178 321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z. and Houghton, J.E. 1999. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the -35 and the -10 promoter elements. Mol. Microbiol. 32 253–263. [DOI] [PubMed] [Google Scholar]
- Gupta, N.K. and Vennesland, B. 1964. Glyoxylate carboligase of Escherichia coli: A flavoprotein. J. Biol. Chem. 239 3787–3789. [PubMed] [Google Scholar]
- Hendrickson, W.A., Horton, J.R., and LeMaster, D.M. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): A vehicle for direct determination of three-dimensional structure. EMBO J. 9 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, A.A. and Guest, J.R. 1968. Biochemical and genetic studies with lysine + methionine mutants of Escherichia coli: Lipoic acid and α-ketoglutarate dehydrogenase-less mutants. J. Gen. Microbiol. 53 363–381. [DOI] [PubMed] [Google Scholar]
- ———. 1969. Studies with α-ketoglutarate dehydrogenase mutants of Escherichia coli. Mol. Gen. Genetics 105 182–190. [DOI] [PubMed] [Google Scholar]
- Hosfield, D.J. 1996. An investigation into the DNA binding properies of the Escherichia coli repressor protein, IclR . MSc thesis, University of Manitoba, Canada.
- Kok, R.G., D'Argenio, D.A., and Ornston, L.N. 1998. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J. Bacteriol. 180 5058–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg, H.L. 1963. The role of acetate in isocitrate lyase induction. Biochim. Biophys. Acta 73 517–519. [DOI] [PubMed] [Google Scholar]
- ———. 1966. The role and control of the glyoxylate cycle. Biochem. J. 99 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer, H., Niemöller, M., Amouyal, M., Revet, B., von Wilcken-Bergmann, B., and Müller-Hill, B. 1987. Lac repressor forms loops with linear DNA carrying the suitably spaced lac operators. EMBO J. 6 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutchinsky, A.N., Chernushevich, I.V., Spicer, V.L., Ens, W., and Standing, K.G. 1998. A collisional damping interface for an electrospray ionization time-of-flight mass spectrometer. J. Am. Soc. Mass Spectrom. 9 569–579. [Google Scholar]
- Krutchinsky, A.N., Ayed, A., Donald, L.J., Ens, W., Duckworth, H.W., and Standing, K.G. 2000. Studies of noncovalent complexes in an electrospray ionization/time-of-flight mass spectrometer. Methods Molec. Biol. 146 239–249. [DOI] [PubMed] [Google Scholar]
- LaPorte, D.C. and Chung, T. 1985. A single gene codes for the kinase and phosphatase which regulate isocitrate dehydrogenase. J. Biol. Chem. 260 15291–15297. [PubMed] [Google Scholar]
- Lawson, C.L. and Carey, J. 1993. Tandem binding in crystals of a trp repressor/operator half-site complex. Nature 366 178–182. [DOI] [PubMed] [Google Scholar]
- Lewis, M., Chang, G., Horton, N.C., Kercher, M.A., Pace, H.C., Schumacher, M.A., Brennan, R.G., and Lu, P. 1996. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271 1247–1254. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Jiang, G., Cui, Y., Mukherjee, A., Ma, W.L., and Chatterjee, A.K. 1999. kdgREcc negatively regulates genes for pectinases, cellulase, protease, HarpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J. Bacteriol. 181 2411–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, O.H., Rosebrough, N.J., Farr, A.L., and Randall, R.J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193 265–275. [PubMed] [Google Scholar]
- Maloy, S.R. and Nunn, W.D. 1982. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J. Bacteriol. 149 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. 1982. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Miller, J.H. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Nègre, D., Cortay, J.C., Old, I.G., Galinier, A., Richaud, C., Saint Girons, I., and Cozzone, A.J. 1991. Overproduction and characterization of the iclR gene product of Escherichia coli K-12 and comparison with that of Salmonella typhimurium LT2. Gene 97 29–37. [DOI] [PubMed] [Google Scholar]
- Nègre, D., Cortay, J.C., Galinier, A., Sauve, P., and Cozzone, A.J. 1992. Specific interactions between the IclR repressor of the acetate operon of Escherichia coli and its operator. J. Mol. Biol. 228 23–29. [DOI] [PubMed] [Google Scholar]
- Nomura, K., Nasser, W., Kawagishi, H., and Tsuyumu, S. 1998. The pir gene of Erwinia chrysanthemi EC16 regulates hyperinduction of pectate lyase virulence genes in response to plant signals. Proc. Natl. Acad. Sci. 95 14034–14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K., Nasser, W., and Tsuyumu, S. 1999. Self-regulation of pir, a regulatory protein responsible for hyperinduction of pectate lyase in Erwinia chrysanthemi EC16. Mol. Plant Microbe Interact. 12 385–390. [DOI] [PubMed] [Google Scholar]
- Orr, A., Ivanova, V.S., and Bonner, W.M. 1995. `Waterbug` dialysis. Biotechniques 19 204–206. [PubMed] [Google Scholar]
- Otwinowski, Z., Schevitz, R.W., Zhang, R.G., Lawson, C.L., Joachimiak, A., Marmorstein, R.Q., Luisi, B.F., and Sigler, P.B. 1988. Crystal structure of trp repressor/operator complex at atomic resolution. Nature 335 321–329. [DOI] [PubMed] [Google Scholar]
- Packman, L.C. and Berry, A. 1995. A reactive, surface cysteine residue of the class-II fructose-1,6-biphosphate aldolase of Escherichia coli revealed by electrospray ionisation mass spectrometry. Eur. J. Biochem. 227 510–515. [DOI] [PubMed] [Google Scholar]
- Pan, B., Unnikrishnan, I., and LaPorte, D.C. 1996. The binding site of the IclR repressor protein overlaps the promoter of aceBAK. J. Bacteriol. 178 3982–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier, N., Donald, L.J., Chernushevich, I., Ayed, A., Ens, W., Arrowsmith, C.H., Standing, K.G., and Duckworth, H.W. 1998. Study of a noncovalent trp repressor: DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 7 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh, V., Frederick, R.O., Syed, S.H.E., Gibson, C.F., Yang, J.-C., and Roberts, G.C.K. 1994. The interactions of Escherichia coli trp repressor with tryptophan and with an operator nucleotide. Eur. J. Biochem. 225 601–608. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Chernushevich, I., Ens, W., Standing, K.G., Thomson, B., Wilm, M., and Mann, M. 1997. Rapid `de novo' peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun. Mass Spec. 11 1015–1024. [DOI] [PubMed] [Google Scholar]
- Smith, C.P. and Chater, K.F. 1988a. Cloning and transcription analysis of the entire glycerol utilization (gylABX) operon of Streptomyces coelicolor A3(2) and identification of a closely associated transcription unit. Mol. Gen. Genet. 211 129–137. [DOI] [PubMed] [Google Scholar]
- ———. 1988b. Structure and regulation of controlling sequences for the Streptomyces coelicolor glycerol operon. J. Mol. Biol. 204 569–580. [DOI] [PubMed] [Google Scholar]
- Spence, E.L., Kawamukai, M., Sanvoisin, J., Braven, H., and Bugg, T.D. 1996. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): Sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J. Bacteriol. 178 5249–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura, K. and Nishihara, T. 1988. Purification, characterization and primary structure of Escherichia coli protease VII with specificity for paired basic residues: Identity of protease VII with ompT. J. Bacteriol. 170 5625–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnarborg, A., Klumpp, D., Chung, T., and LaPorte, D.C. 1990. Regulation of the glyoxylate bypass operon: Cloning and characterization of IclR. J. Bacteriol. 172 2642–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen, H.J. and Bach, C. 1990. Target detection assay (TDA): A versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 18 3203–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, N.R., Nasser, W., McGowan, S., Sebaihia, M., and Salmond, G.P. 1999. Erwinia carotovora has two KdgR-like proteins belonging to the IclR family of transcriptional regulators: Identification and characterization of the RexZ activator and the KdgR repressor of pathogenesis. Microbiology 145 1531–1545. [DOI] [PubMed] [Google Scholar]
- Vanderwinkel, E. and DeVlieghere, M. 1968. Physiologie et génétique de l'isocitratase et des malate synthases chez Escherichia coli K12. Eur. J. Biochem. 5 81–90. [DOI] [PubMed] [Google Scholar]
- Verentchikov, A.N., Ens, W., and Standing, K.G. 1994. Reflecting time-of-flight mass spectrometer with an electrospray ion source and orthogonal extraction. Anal. Chem. 66 126–133. [DOI] [PubMed] [Google Scholar]
- Wilm, M. and Mann, M. 1996. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68 1–8. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Zhao, D., Revington, M., Lee, W., Jia, X., Arrowsmith, C., and Jardetzky, O. 1994. The solution structures of the trp repressor-operator complex. J. Mol. Biol. 238 592–614. [DOI] [PubMed] [Google Scholar]