Abstract
L-β-(Thieno[3,2-b]pyrrolyl)alanine and L-β-(thieno[2,3-b]pyrrolyl)alanine are mutually isosteric and pharmaceutically active amino acids that mimic tryptophan with the benzene ring in the indole moiety replaced by thiophene. Sulfur as a heteroatom causes physicochemical changes in these tryptophan surrogates that bring about completely new properties not found in the indole moiety. These synthetic amino acids were incorporated into recombinant proteins in response to the Trp UGG codons by fermentation in a Trp-auxotrophic Escherichia coli host strain using the selective pressure incorporation method. Related protein mutants expectedly retain the secondary structure of the native proteins but show significantly changed optical and thermodynamic properties. In this way, new spectral windows, fluorescence, polarity, thermodynamics, or pharmacological properties are inserted into proteins. Such an engineering approach by translational integration of synthetic amino acids with a priori defined properties, as shown in this study, proved to be a novel and useful tool for protein rational design.
Keywords: UV absorbance, fluorescence, bioincorporation, protein engineering, thia-surrogates of tryptophan
The immense functional diversity of naturally occurring proteins is based on the simple concept of varying the combination of the 20 canonical amino acids in polypeptide chains. In this context, the possibility of artificially expanding this diversity by in vivo protein synthesis is a challenging and attractive goal because newly incorporated noncanonical amino acids could deliver novel properties into existing protein structures. An a priori design of proteins with desired properties such as altered spectral windows or pharmacological activities should be feasible if some of the coded (i.e., canonical) amino acid chromophores are exchangeable with noncanonical ones in template-directed protein synthesis.
For the in vivo translational incorporation of a variety of noncanonical amino acids, an efficient method was developed that is based on the use of auxotrophic Escherichia coli host strains submitted to strong selective pressure (selective pressure incorporation; SPI) (Budisa et al. 1995, 1998a, 1999; Ross et al. 1997). It enables the expansion of the scope of protein biosynthesis, primarily because the main principle of this basically in vivo methodology is based on codon reassignments at the translational level under stringent fermentation conditions. Tryptophan represents an attractive target for such replacements because it is the main source of the absorption and fluorescence of proteins. Moreover, occurrence of Trp is rare as it represents only about 1% of all residues of globular proteins (McCaul and Ludescher 1999), and thus it may well provide a quasi-site-specific intrinsic probe for studying protein structure, dynamics, and function. Apart from its role as a basic building block in ribosome-mediated protein synthesis, Trp plays an essential role in the metabolism of living cells, as it is involved in the biosynthesis of hormones such as serotonin or melatonin in animals, and of indole alkaloids in plants (Phillips et al. 1995). Thus, noncanonical amino acids that mimic Trp could be of great interest as potential antagonists, drugs, or antibiotics, and their incorporation into proteins could be a promising approach for production of therapeutic agents (Budisa et al. 1998b; Minks et al. 2000b).
Tryptophan residues have already been replaced in proteins by various noncanonical aza-, fluoro-, and hydroxy-tryptophan analogs (Soumillion et al. 1995; Ross et al. 1997; Minks et al. 1999). In the present study, an additional expansion of the amino acid repertoire for in vivo protein synthesis was achieved by replacing Trp in model proteins with two isosteric sulfur-containing surrogates of Trp, L-β(thieno[3,2-b]pyrrolyl)alanine ([3,2]Tpa) and L-β-(thieno[2,3-b]pyrrolyl)alanine ([2,3]Tpa), in response to Trp UGG codon-containing DNA templates (plasmids) using the SPI method (Fig. 1 ▶).
Fig. 1.
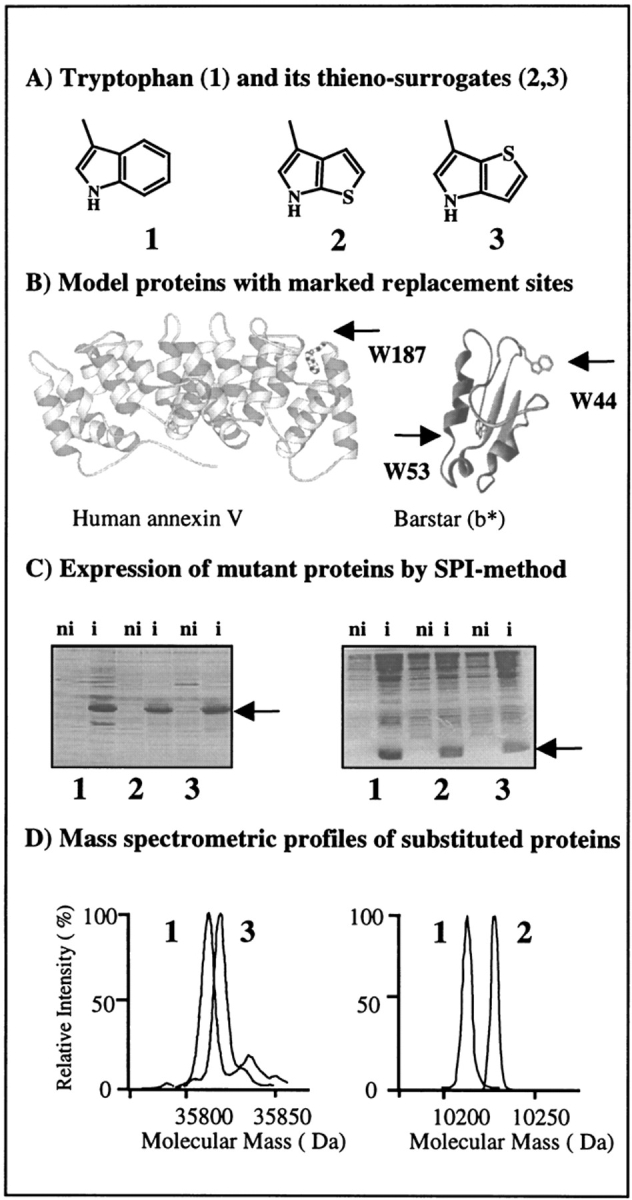
Flow chart for in vivo incorporation of tryptophan and β-(thienopyrrolyl)alanines by the SPI method into model proteins. (A) Structural representations of side chains of the canonical amino acid tryptophan (1) and its noncanonical thia-containing surrogates [2,3]Tpa (2) and [3,2]Tpa (3). (B) Ribbon plots of barstar (right) based on its PDB coordinates (Martin et al. 1999) with labeled locations of two tryptophan residues, W44 and W53, and side view of the AxV structure (left) with W187 buried in the hydrophobic pocket at the convex side of the molecule (Huber et al. 1990). Figures were prepared with MOLSCRIPT (Kraulis 1991). (C) Analysis of the expression profiles in cell lysates of E. coli ATCC49980 with overexpressed AxV (left) and b* (right) in the defined minimal medium with Trp (1), [2,3]Tpa (2), and [3,2]Tpa (3). (ni) Noninduced cells; (i) cells induced for protein expression. Arrows indicate position of overexpressed substituted proteins. (D) Analytical proof for substitutions by mass analyses of native and substituted proteins. Two different electrospray mass-spectrometric measurements were superimposed at the same mass-scale: (left) native and [3,2]Tpa-AxV; (right) wild-type and [2,3]Tpa-b*. Experimental conditions are described in Materials and Methods.
Results
Human annexin V and barstar as model proteins
A suitable model protein for replacement experiments should have a known three-dimensional structure and well established biochemical, biophysical, genetic, and kinetic properties. In addition, controllable, robust, and efficient plasmid-directed expression should be available in a suitable auxotrophic host strain. Barstar, a small bacterial ribonuclease inhibitor, is an excellent model protein for incorporation studies because it meets all these criteria: Both high-resolution crystal and solution structures are known (Martin et al. 1999), and it undergoes completely reversible unfolding on either thermal or chemical denaturation. Thus, it is frequently used as a model protein for protein folding, stability, and dynamic studies (Nath and Udgaonkar 1997). Barstar contains three Trp residues at positions 38, 44, and 53. In the present study, the replacement experiments were performed using the pseudo-wild-type mutant C40A/C82A/P27A/W38F (b*), which is expressed in the form of inclusion bodies and is refolded into the native form during the purification procedure (Golbik et al. 1999). This mutant (b*) contains only two Trp residues: W44, which is partially buried, and W53, which is buried completely in the protein hydrophobic core (Fig. 1 ▶). The latter Trp residue is essential for the protein conformational integrity as it cannot be replaced by any of 20 canonical amino acids by routine DNA mutagenesis (Nath and Udgaonkar 1997).
A second model protein that meets the above-mentioned conditions is recombinant human annexin V (AxV), which is expressed in soluble form under the control of the T5 promoter/polymerase system. AxV binds in a calcium-dependent manner to acidic phospholipid membrane head groups (Huber et al. 1990; Berendes et al. 1993; Liemann and Huber 1997). AxV contains only one, but an essential, Trp residue (W187) that in the crystalline state can have two molecular conformations: one in which Trp187 is buried in the hydrophobic niche of the domain III (Fig. 1 ▶) and another in which Trp187 is completely exposed to the bulk solvent (Berendes et al. 1993; Concha et al. 1993). In previous studies, we have shown that this residue can be replaced with fluoro-substituted Trp analogs without affecting the protein structure in solution and in crystals, although the thermodynamic properties were altered drastically (Minks et al. 1999).
In the present work, W187 of AxV as well as the two Trp residues of b* were replaced by [3,2]Tpa and [2,3]Tpa, respectively, using the Trp-auxotrophic E. coli strain ATCC 49980 and a T5-based expression system in the context of the SPI methodology as outlined in Figure 1 ▶.
Amino acid toxicity and optimal fermentation conditions
Met-auxotrophic E. coli strains grow in the presence of selenomethionine (SeMet) as a sole source for Met in minimal media because of its moderate toxicity for the cells (Budisa et al. 1995). However, this was not the case with the thia-surrogates of Trp, as shown clearly in Figure 2 ▶. Like fluorinated Trp analogs that did not support cellular growth of E. coli ATCC 49980 in our earlier experiments (Minks et al. 1999) and behaved as competitive inhibitors of their native Trp counterpart, the growth of E. coli strain ATCC 49980 is also strongly inhibited from the beginning of the fermentation in defined minimal medium that contains 0.015 mM (3.1 μg/mL) of each thia-amino acid as the sole source of Trp. To our surprise, when the β-(thienopyrrolyl)alanines were supplied together with Trp in a ratio of 1 : 1 in fermentation experiments (Fig. 2 ▶), this mixture allowed cells to grow to the stationary phase; that is, the growth was obviously supported even after the Trp supply was exhausted. These findings showed clearly that in a mixture with Trp as a supply for cellular growth, the thieno compounds are not only well tolerated by the auxotrophic cells, but are also used to some extent as substrates for cellular growth. Thus, the toxicity of these substances must be rather moderate when compared with fluoro-Trp analogs, but still relatively strong compared with SeMet. Moreover, in all fermentation experiments, both [3,2]Tpa and [2,3]Tpa as free amino acids proved to be stable, and despite exposure to aerobic conditions and light for days, a degradation of this amino acid was not detected.
Fig. 2.
The growth curves at 30°C of the transformed Trp-auxotrophic E. coli strain ATCC 49980 in NMM with different concentrations of L-Trp and its thia-analogs. The cells were cotransformed with two plasmids: ampicillin-resistant pQIA-30b* with the gene for pseudo-wild-type barstar and kanamycin-resistant pREP4 containing a repressor gene lacIq (Qiagen). Six measured cultures with NMM were supplied with the following amounts of native substrate and β-(thienopyrrolyl)alanines: (1) 0.03 mM L-Trp; (2) 0.015 mM L-Trp; (3) 0.015 mM [2,3]Tpa; (4) 0.015 mM [3,2]Tpa; (5) 0.015 mM [2,3]Tpa + 0.015 mM L-Trp; (6) 0.015 mM [3,2]Tpa + 0.015 mM L-Trp. Note that the mixtures of L-Trp with [2,3]Tpa and [3,2]Tpa, respectively (5, 6), allow cells to reach stationary phase but the overall growth rate is lowered. The growth was followed by measuring the changes in the OD600 by UV spectrometry.
The E. coli ATCC 49980 strain shows typical growth curves in the minimal medium with 0.3 mM tryptophan. As shown in Figure 2 ▶, the optimal limiting concentration essential for successful incorporation of both β-(thienopyrrolyl)alanines is 0.015 mM Trp. This concentration leads to Trp depletion in the culture in the mid-logarithmic phase of growth (OD600: 0.7–1.0), a fact that is almost ideal for induction of protein expression. Thus, the cells were grown in the minimal medium (NMM) with 0.015 mM Trp as natural substrate (Minks et al. 1999) until its exhaustion, followed by simultaneous addition of the β-(thienopyrrolyl)alanines and target gene induction with IPTG. Under these conditions, optimal production of the labeled proteins was achieved in yields comparable with those of the wild-type protein (10–30 mg/L in the case of AxV and 50–100 mg/L in the case of b*).
Analytical characterization and spectroscopic properties of the protein mutants
In routine bioexpression protocols based on the SPI method, an almost quantitative incorporation of both β-(thienopyrrolyl)alanines in AxV and b* was readily achieved (Fig. 1 ▶). To assess incorporation of these amino acids, the fluorescence profiles of the protein mutants were recorded, because replacement of the benzene ring of the indole moiety with thiophene provides an efficient fluorescence quenching, as shown in Figure 3 ▶. Thus, the absence of the characteristic Trp emission fluorescence profile was used as a qualitative analytical criterion to monitor successful labeling. Conversely, for quantitative analysis mass spectrometry was applied, because the molecular mass difference between Trp and both thia-variants is sufficiently large (6 daltons) to be determined experimentally. AxV (expected mass: 35,809.2 daltons; experimental value: 35,808 ± 2.0 daltons) contains only one Trp residue that on replacement with the β-(thienopyrrolyl)alanines leads to a slight increase in the protein molecular mass (expected mass: 35,815.2 daltons; experimental value: 35,814 ± 1.0 daltons for [3,2]Tpa-AxV and 35,815 ± 2.0 daltons for [2,3]Tpa-AxV). The barstar mutant b* (expected mass: 10,214 daltons; experimental value: 10,214 ± 3.2 daltons) contains two Trp residues; thus, the mass differences on substitution are much more pronounced (expected mass: 10,226.2 daltons; experimental value: 35,226 ± 2.0 daltons for both protein variants) (Fig. 1 ▶). These data confirm also that thia-analogs once incorporated into proteins are stable. This is especially pronounced in the case of b*, which is refolded under air-oxygen before its purification.
Fig. 3.
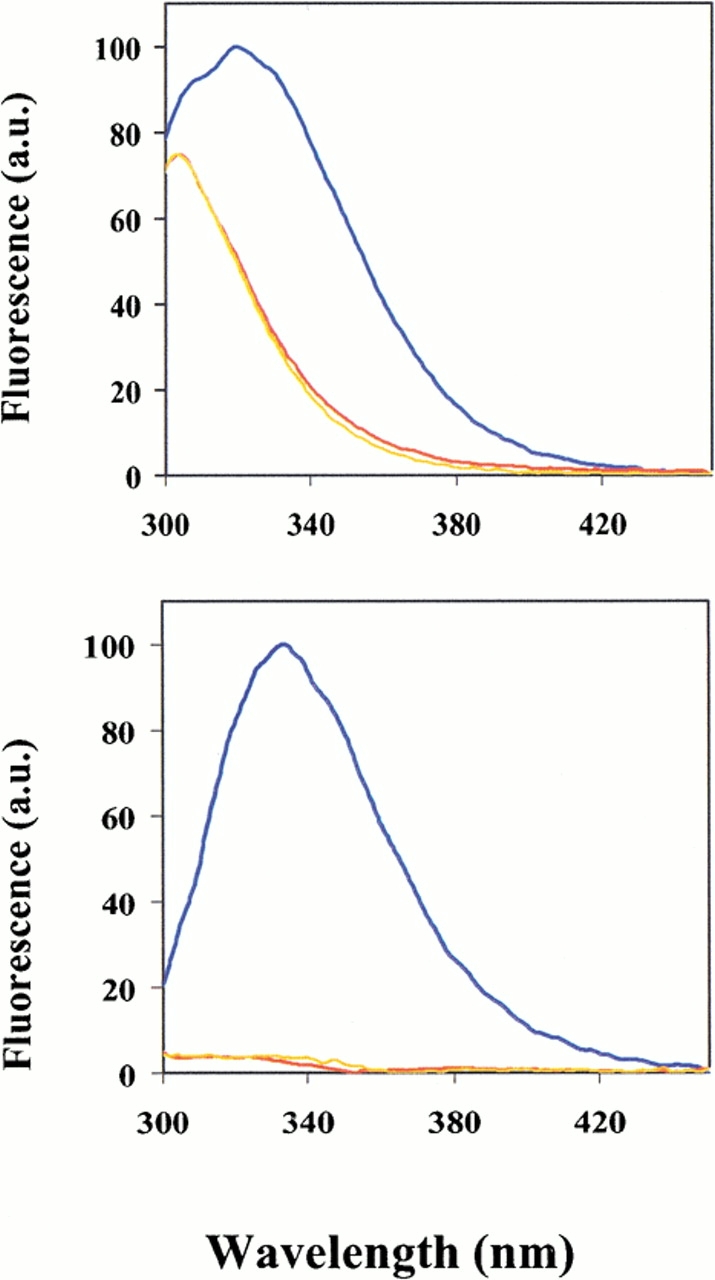
Fluorescence emission spectra of native and mutant proteins of AxV (top) and b* (bottom) excited at 280 nm. (Blue) wild-type proteins; (yellow) [2,3]Tpa proteins; (red) [3,2]Tpa proteins. The emission profile of AxV revealed tyrosine contribution (with λmax at ∼307 nm) to the overall protein fluorescence profile, whereas in the case of b* the fluorescence of substituted proteins is almost completely quenched when compared with the native one. Experiments were performed at 20°C as described in Materials and Methods.
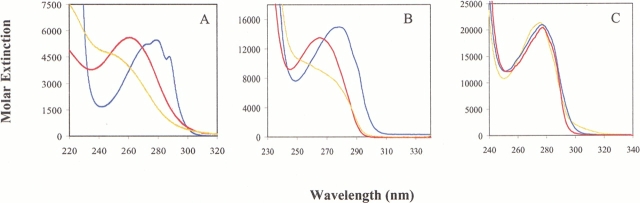
As expected from the UV spectra of the two β-(thienopyrrolyl)alanines (Table 1, Fig. 4A ▶), the optical properties of the protein mutants are affected by their incorporation at the Trp positions. The UV spectra of the AxV mutants are not changed dramatically because of dominant contribution of the Tyr residues to the overall absorbance (Fig. 4C ▶). Conversely, in the case of the barstar mutants the UV spectral properties of the β-(thienopyrrolyl)alanines are reflected fully in the spectra of the mutant proteins (Fig. 4B ▶), with a blue shift of the absorption maximum (Table 1). Such blue shift originates from the hydrophobic environment of the partially (position 38) and fully buried (position 53) aromatic β-substituted alanines.
Table 1.
Molar extinction coefficients (ɛM) and absorption maxima (λmax) of L-Trp, [3,2]Tpa, and [2,3]Tpa as free amino acids in buffered solution and of Annexin V and b* related mutants produced by Trp replacements
| Free amino acida | Annexin Vb | b* (C40A/C82A/P27A/W38F)a | ||||
| Residue | ɛMc | λmaxd | ɛM | λmax | ɛM | λmax |
| L-Trp | 5579 ± 257 | 280 | 21,500 ± 931 | 277 | 14,920 ± 531 | 280 |
| [3,2]Tpa | 5632 ± 373 | 260 | 21,500 ± 791 | 277 | 13,060 ± 197 | 261 |
| [2,3]Tpa | 4897 ± 169 | 241 | 22,800 ± 287 | 277 | 10,712 ± 331 | 250 |
Experimental conditions are described in Materials and Methods, and related spectra are presented in Fig. 4 ▶.
d Expressed in nm. cExpressed in M−1 cm−1. aIn 50 mM Na-dihydrogenphosphate (pH 8.0). bIn 10 mM TrisCl (pH 8.0).
Fig. 4.
UV spectra of free amino acid Trp and its thia-surrogates and of native proteins and related mutants (A), b* (B), and AxV (C) at 20°C. (Blue) Trp and parent proteins; (yellow) [2,3]Tpa and [2,3]Tpa proteins; (red) [3,2]Tpa and [3,2]Tpa proteins. Note that the overall spectral shape and intensity of AxV are not significantly affected by substitutions due to the Tyr dominance in the AxV structure (13 Tyr and only 1 Trp). Native and substituted annexins have an absorption maximum at 277 nm. On the other hand, in b*, with only three Tyr and two Trp residues, replacements indeed caused significant blue shift in the absorption maximum and changes in overall spectral shape and intensity. Molar extinctions are calculated for the whole absorption range and are expressed as M−1cm−1. Absorption maximum values with related ɛM values are presented in Table 1. Experimental procedures and calculations are as described in Materials and Methods.
Conformational analysis of the protein mutants in solution
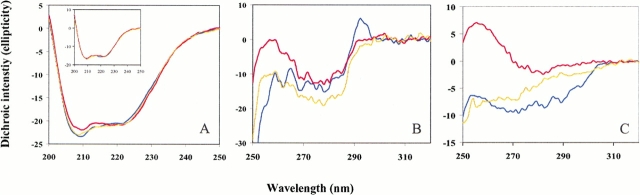
The far-UV CD spectra of AxV and its mutants show the typical pattern of α-helical proteins with the two characteristic minima at 222 nm and at 208 nm of similar intensity (Fig. 5A ▶, inset). The signal ratio between these two minima ([θ]R)222/([θ]R)208 is ∼0.96 for the wild-type protein and the [2,3]Tpa mutant and reduced only slightly for [3,2]Tpa-AxV (0.95). Similarly, the parent b* mutant and its [2,3]Tpa-b* analog (Fig. 5A ▶) show almost identical spectra and signal ratios: ([θ]R)222/([θ]R)208 of ∼0.90, whereas for the [3,2]Tpa-b* protein this ratio is increased significantly (0.97).
Fig. 5.
Far-UV CD profiles of the parent and substituted b* (A) and AxV (inset) protein forms at 20°C determined as described in Materials and Methods. The mean residual ellipticity ([θ]R) is expressed in deg cm2/dmol. Near-UV CD spectra for native and mutant proteins at 20°C: (B) AxV; (C) b*. The mean molar ellipticity ([θ]M) is expressed in deg cm2/dmol. (Blue) Spectra of wild-type protein forms; (yellow) [2,3]Tpa mutants; (red) [3,2]Tpa protein mutants. Note that the differences in the spectral shapes in the far-UV CD spectrum of [3,2]Tpa-b* might not be entirely related to changes in secondary structure. Namely, strong alternations in near-UV CD of this mutant, as is obvious from panel C, might influence the shape and intensities of far-UV CD spectra, as well.
The near-UV CD spectrum of wild-type AxV shows the contribution of the Phe residues with the sharp fine structure (255–270 nm) of the Tyr residues, with the maximum centered between 275 and 282 nm, and of the Trp residue with its maximum around 290 and 305 nm (Minks et al. 1999). Replacement of the Trp residues in both proteins by the two β-(thienopyrrolyl)alanines leads expectedly to significant spectral alterations in the aromatic region that arise mainly from the intrinsic dichroic properties of the Trp variants. A comparative analysis of the near-UV CD spectra of Trp and the β-(thienopyrrolyl)alanines as free amino acids revealed that the spectra of Trp and [2,3]Tpa are more or less similar whereas the spectrum of [3,2]Tpa shows a strong positive band between 280 and 250 nm (data not shown). This is fully reflected in the spectra of both mutant proteins and especially for b*, in which the contribution of Trp residues to the overall protein spectrum is dominant (Fig. 5 ▶).
In the near-UV, the CD spectra of [2,3]Tpa-containing proteins are similar to those of the native proteins (Fig. 5B,C ▶), with only small differences in the dichroic intensity of the positive bands. This is stronger for wild-type AxV than for [2,3]Tpa-AxV (Fig. 5B ▶), but weaker for the parent b* protein than for the [2,3]Tpa mutant (Fig. 5C ▶).
Conversely, replacing the Trp residues in both proteins with [3,2]Tpa results in drastic changes of the near-UV CD spectra (Fig. 5B,C ▶). In the near-UV CD spectrum of [3,2]Tpa-AxV, these changes dominate the spectral region between 250 and 268 nm, whereas between 270 and 290 nm the intensities are only slightly decreased compared with the spectrum of the wild-type protein (Fig. 5B ▶). A similar, but more pronounced, effect is observed for [3,2]Tpa-b* (Fig. 5C ▶), in which the changes could originate from a strong local alteration of the spatial array of neighboring aromatic residues or from the intrinsic spectral nature of the [3,2]Tpa as chromophore as well as from a combination of both effects. However, in the absence of spectral information from model peptides, any attempt to explain the observed effects in a more reliable and detailed manner would be too speculative.
A comparison of the far-UV CD spectra of all AxV proteins shows that the overall dichroic patterns are nearly identical (Fig. 5A ▶), whereas for [3,2]Tpa-b* the spectrum is slightly different around 208 nm when compared with native protein variant. This could be indicative of small changes in the secondary structure of this mutant. However, the drastic alterations detected in the aromatic region can affect the far-UV CD spectrum without changes in secondary structure, as was already observed in other systems (Woody and Dunker 1996).
Thermal denaturation
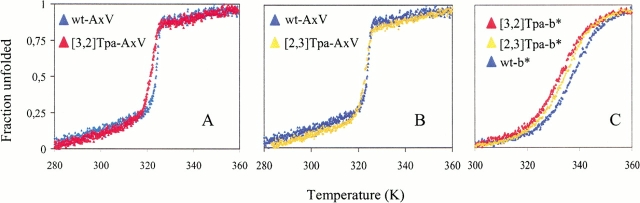
Thermal unfolding of both model proteins is known to occur as a two-state transition from native to denatured state (Golbik et al. 1999; Minks et al. 1999). Compared with the parent proteins, all mutants containing [3,2]Tpa or [2,3]Tpa are characterized by lower thermal stabilities (Fig. 6 ▶); the related Tm values are listed in Table 2. Thereby, the proteins substituted with [3,2]Tpa not only show the lowest Tm values, but the cooperativity of their unfolding process is also affected as indicated by changes in the steepness of the transition profiles (Fig. 6A,C ▶) and consequently by the van't Hoff enthalpies (ΔHm; Table 2). This effect is more pronounced in the case of the barstar mutant, in which replacement of the structurally critical and buried W53 with [3,2]Tpa possibly provokes local perturbations of the hydrophobic environment. Incorporation of [2,3]Tpa residues into the model proteins leads in both cases to folding cooperativities that are even enhanced by almost 15% if compared with the parent proteins (Table 2). It has been observed previously that isosteric replacements of Met and Trp are accompanied by rather large differences in ΔHm values despite the moderate shifts of the Tm values (Budisa et al. 1998a; Minks et al. 1999).
Fig. 6.
Thermal denaturing of wild-type and mutant protein forms of AxV (A and B) and barstar (C). Fractions of denatured proteins are calculated from CD data monitored at 222 nm as described in Materials and Methods, and derived thermodynamic parameters are presented in Table 2. Note that differences in the unfolding transition profiles are lower among wild-type and [2,3]Tpa protein melting curves, whereas these differences are higher between wild-type and [3,2]Tpa protein forms.
Table 2.
Thermodynamic parameters of the denaturation of the wild-type and substituted protein variants of AxV and b*
| Protein variant | Tm value (K) | ΔTma | ΔHm (kJ/mol) |
| wt-AxV | 323.76 ± 0.11 | — | 703.92 ± 18.8 |
| [2,3]Tpa-AxV | 323.70 ± 0.03 | −0.06 | 817.32 ± 17.7 |
| [3,2]Tpa-AxV | 322.69 ± 0.03 | −1.07 | 677.06 ± 14.1 |
| wt-b* | 337.28 ± 0.20 | — | 154.14 ± 9.51 |
| [2,3]Tpa-b* | 335.57 ± 0.23 | −1.71 | 181.90 ± 14.7 |
| [3,2]Tpa-b* | 332.45 ± 0.18 | −4.83 | 153.63 ± 9.45 |
Experimental conditions and calculation methods for all unfolding profiles are described in Materials and Methods, and related curves are presented in Fig. 6 ▶. aΔTm is the difference between Tm of the native and substituted proteins.
The findings on thermal stabilities agree with the observed changes in the near-UV CD spectra of both proteins (Fig. 5 ▶). The [3,2]Tpa mutants that show significantly enhanced dichroism in the near-UV are characterized by markedly lower Tm values and cooperativities than the [2,3]Tpa mutants (Table 2). Conversely, the higher cooperativity in unfolding of the [2,3]Tpa mutants would indicate an enhanced order of the internal architecture.
Biological activities
Mutants of both proteins are biologically active in our qualitative activity tests. The barstar mutants inhibit the RNase activity of barnase in standard inhibition assays on RNA agar-containing plates (Golbik et al. 1999) to a similar extent as the parent variant. Similarly, the AxV mutants are able to bind to liposomes in the presence of higher Ca2+ concentrations (Berendes et al. 1993).
Discussion
Expression of proteins with β-(thienopyrrolyl)alanines by the SPI method
The main prerequisite for the successful use of the SPI method is the availability of expression hosts with a stable genotype (i.e., auxotrophism) that is also suitable for transformations with target plasmids. In addition, optimal plasmid-directed protein translation when fermented in minimal medium should be achieved in terms of a well controllable and stringent expression system. In this study, we could show not only that the E. coli strain ATCC 49980 can grow robustly in NMM, but also that it is sufficiently stable to afford efficient limitation of the cellular growth at the mid-logarithmic phase (OD600 at about 0.7–0.9). The T5 expression system was found to suppress basal protein expression completely before induction ("gene-leakage"), as shown in Figure 1 ▶. Thus, the availability of the proper bacterial host and expression system together with the experimentally optimized fermentation parameters were decisive to achieve high-level incorporation, as shown previously by replacements of Met (Budisa et al. 1995), Pro (Budisa et al. 1998b), Trp (Minks et al. 1999), Phe (Minks et al. 2000a), and Tyr residues (Minks et al. 2000b) using the SPI method.
In expression experiments, the toxicity of noncanonical amino acids may represent an additional problem. In fact, all amino acids outside the 20 canonical amino acids are generally not substrates for cellular growth. Especially those amino acid analogs that are very close to the native ones or that display reactive functionalities show the highest toxicity (Liu and Schultz 1999). Exceptions are SeMet (Cohen and Cowie 1957) and trifluoroleucine (Rennert and Anker 1963), which allow Met- and Leu-auxotrophic bacterial strains to be fully adapted and grow in their presence in minimal medium. Bacteria adapted in this way have acquired the ability to charge all cognate tRNAs with SeMet and trifluoroleucine in in vivo biosynthesis. However, in most cases the amino acid analogs are efficient inhibitors of cellular growth. Thus, to achieve high-level to almost quantitative substitutions while avoiding this toxicity problem in the context of the SPI method, a strong auxotrophism of the host cells, controlled amino acid supply in fermentation media, and a stringent expression system are necessary (Budisa et al. 1998b).
Chromophores with new spectral windows
Although Trp as the main chromophore in proteins has the unique advantage of an intrinsic probe, it is less suitable for investigating protein–protein or protein–nucleic acid interactions because the absorption spectra of nucleic acids overlap that of Trp and thus prevent the assignment of the spectral contribution of Trp residues to the total signal output. Similar difficulties arise when protein–protein interactions are investigated because many interacting proteins in multiprotein complexes contain Trp residues and, thus, their absorption and fluorescence signals are more or less indistinguishable (Ross et al. 1997).
A very common approach to study the functional role of Trp in proteins is to use site-directed mutagenesis. Thereby, in most of the cases Trp residues are mutated to Phe in an attempt to minimize structural perturbations by replacing one aromatic planar moiety with another. However, this strategy is limited because Trp residues are often essential for the structural integrity and functionality of proteins, as in the case of Trp187 in AxV and Trp53 in barstar (Fig. 1 ▶), and therefore cannot be replaced by any of the remaining 19 canonical amino acids. Even if such replacements are possible, local structural perturbations could alter the spectral contributions of the remaining chromophores, as the most similar canonical counterparts Phe or Tyr always bring relatively large alterations both in the size and charge.
By the use of noncanonical Trp analogs, much more subtle alterations are expected that could facilitate interpretation of the experimental data by addressing issues such as spectral overlap, better sensitivity to small perturbations of the environments of substituted aromatic side chains, and, most importantly, novel spectral windows. To date, five noncanonical analogs of Trp were reported to be incorporated into proteins: (4-F)Trp, (5-F)Trp, (6-F)Trp, (7-Aza) Trp, and (5-OH)Trp (Soumillion et al. 1995; Ross et al. 1997; Minks et al. 1999). All these Trp analogs bring about the lowest possible level of structural alterations, that is, single atom exchanges such as H→F or =CH– →– NH–, thus providing "atomic mutations" for studying protein folding, activities, dynamics, and stability (Budisa et al. 1998a; Minks et al. 1999).
With β-(thienopyrrolyl)alanines incorporated into proteins, two notable spectroscopic properties are achieved: altered absorption profiles, at least when the Trp contribution is dominant as in the case of the barstar mutant (Fig. 4 ▶), and an efficient static fluorescence quenching. Besides the two β-(thienopyrrolyl)alanines as isosteric analogs of Trp reported in this study, (4-F)Trp was the only known nonfluorescent Trp analog incorporated into proteins in vivo (Bronskill and Wong 1988). Thus, the repertoire of "silent" fluorophores as protein building blocks for in vivo translation is increased as needed for investigating protein–DNA interactions or multiprotein assemblies.
Structural stability of β-(thienopyrrolyl)alanine proteins
The aromatic amino acids Phe, Tyr, and Trp harbor in their chemical structure two properties, hydrophobicity, being composed of hydrocarbon units, and polarity, the ability to bind ions through cation–π interactions, that are often considered to be mutually exclusive (Dougherty 1996). Because of these properties, the aromatic residues are generally placed in the interior of proteins or interact with cell membranes. For example, Trp187 of AxV is closely packed in the hydrophobic niche of the domain III (Fig.1 ▶), and on addition of Ca2+ in the presence of lipid bilayers, it undergoes large local conformation changes with insertion into the membrane (Concha et al. 1993; Sopkova et al. 1994; Liemann and Huber 1997).
When Trp187 of AxV is replaced with β-(thienopyrrolyl)alanines, the secondary structure of the mutants is identical to that of the parent protein, although the near-UV CD spectra differ (Fig. 5B ▶). The differences may originate from coupled-oscillator interactions between the aromatic side chains as is often observed with other proteins (Woody and Dunker 1999). Indeed, the neighboring Phe194 that is in van der Waals contact with the C4 position of Trp187 (Huber et al. 1990) could be involved in such interactions. Alternatively, the observed effects could also result just from the intrinsic properties of [3,2]Tpa.
Both [3,2]Tpa-AxV and [2,3]Tpa-AxV crystallize preferentially in the molecular form, in which the mutated side chain at position 187 is exposed to the surface ("open form"), like some other AxV mutants (Berendes et al. 1993) and rat AxV (Concha et al. 1993) (data not reported). On the other hand, wild-type AxV crystallizes preferentially in the "closed form," in which Trp187 is buried in a hydrophobic core (Huber et al. 1990). Thus, the relatively small differences in the unfolding profiles between native AxV and mutants could be explained as follows: On unfolding of the wild-type AxV, the indole moiety of Trp187 undergoes a transition from a hydrophobic to a water-solvent environment, while the β-(thienopyrrolyl)alanine residues are already in contact with the bulk water. Indeed, it is well known that only buried residues contribute significantly to experimental folding parameters (Dill and Shortle 1991).
Keeping this in mind, it is not surprising that in b* mutants thermodynamic parameters are changed to greater extents than in AxV (Table 2, Fig. 6C ▶). Namely, b* residue 44 is partially and residue 53 completely buried into the protein interior. Aromatic residues in proteins are often found to form a network of three or more interacting side chains, and these interactions are supposed to serve as nucleation sites in protein folding pathways and as main stabilizing forces of the tertiary structures (Nath and Udgaonkar 1997). In b*, this clustering occurs around Trp53, which is sandwiched between Phe56 and Phe74. Site-directed mutagenesis showed that Trp53 is contributing predominantly to the absorption and fluorescence of barstar upon unfolding (Nath and Udgaonkar 1997). The near-UV CD spectrum of the [3,2]Tpa mutant (Fig. 5C ▶) would indicate an exceptionally strong coupled-oscillator interaction between the [3,2]Tpa residue and the neighboring Phe side chains. On the other hand, it is difficult to conceive large structural rearrangements upon the substitution because it is well documented that the environment of Trp53 is rigid and devoid of any flexibility (Nath and Udgaonkar 1997). As expected, the X-ray analysis of the crystals of a β-(selenolo[3,2-b]pyrrolyl)alanine-containing b* mutant analog fully confirmed these observations (detailed X-ray structure of this b* mutant will be reported elsewhere).
As in the case of AxV, the differences in the near-UV CD spectral properties of the barstar variants correspond well with the unfolding profiles. The [3,2]Tpa mutant is significantly less stable in terms of Tm than the [2,3]Tpa mutant and even less than the parent b* (Table 1). Taking into account the dominant role of Trp53 for b* folding and assuming that the local geometry is not changed significantly on substitution, the packing interactions might be responsible for the observed differences. Such reasoning is based on the assumption that two isosterically shaped moieties may occupy a cavity in a different mode because of their different van der Waals interactions that may exert a strong impact on the protein-restricted internal architecture. Previous thermodynamic studies on crystallographically isomorphous proteins with Met and Trp isosteric analogs confirmed this as well (Budisa et al. 1998a; Minks et al. 1999). Because both [2,3]Tpa and [3,2]Tpa are mutually isosteric, the most conceivable explanation for the differences observed in related protein mutants might derive from their differentiated physico-chemical properties. These differences are caused by the stereochemical position of the sulfur atom relative to the protonated nitrogen in the thienopyrrolyl moiety. Indeed, theoretical molecular orbital calculations indicate that the stabilities of these thienopyrrole positional isomers should differ significantly (Milun and Trinajstic 1977). Thus, by cotranslational incorporation of these Trp surrogates into proteins, their different properties are transmitted, integrated, and modulated into the structures of the related mutants.
Biophysical properties of thieno-surrogates of Trp
All benzene-based amino acids (Phe, Tyr, Trp) show strong quadrupole moments that arise from the nonspherical charge distribution; thus, introduction of heteroatoms into these systems results in novel properties not found in the parent molecule. For example, the sulfur atom in thiophene has an unshared pair of electrons in a p-orbital conjugated with the carbon–carbon double bonds, and, unlike the carbon, nitrogen, and oxygen, it has vacant d-orbitals in the outer shell and can therefore act as an electron acceptor. The fused pyrroles are more permissive to interacting with nearby charges with induction of dipoles in the system as well as additional dispersion forces, polarizabilites, exciplex formation, resonance energy transfer, or with formation of charge transfer complexes. Other properties that differentiate them from benzene include enhanced hyperpolarizability and differences in aromatic delocalization energies (benzene: 36 kcal/mol, thiophene: 29 kcal/mol, thiazole: 25 kcal/mol) (Bird 1985).
It is therefore expected that the replacement of the benzene ring of indole with a thiophene results in altered interactions responsible for the structure of biological macromolecules and for mediating processes such as receptor–ligand interactions, enzyme–substrate binding, and antigen–antibody recognition. This was well exemplified in the studies of structure–activity relationships with the enzyme tryptophan-indole lyase. For [2,3]Tpa a kcat/KM value of one order of magnitude higher (8.6 × 103 M−1s−1) than that of [3,2]Tpa (1.2 × 103 M−1s−1) was determined, and this was attributed to the differences in electronic composition (Sloan and Phillips 1996).
It is also not surprising that thiophene-based substances have attracted widespread interests in material science, as they may show many useful properties such as improved optical transparency or good thermal stability that result from inductive effects of the electron-rich sulfur (Kothakota et al. 1995; Breitung et al. 2000). The rather unusual optical and thermodynamic properties of the proteins containing β-(thienopyrrolyl)alanines may therefore be better explained and understood if more information from suitable model peptides was available.
Chemistry and pharmacology of thienopyrroles
Bioisosteres are isosteric molecules that have near-equal shapes and volumes, approximately the same distribution of electrons, and show similar or antagonistic properties in biological systems. Such compounds are found abundantly in nature, such as, in alkaloid-bearing plants (Burger 1991). We have attempted to produce the indole isosteres in which the imino group of the indole moiety is replaced with other heteroatoms (sulfur or oxygen). Unfortunately, these experiments resulted in compounds that did not predictably retain biological properties analogous to their indole counterpart. Firstly, these planar systems were not activated in the enzymatic condensation reaction with serine by Trp-synthase (N. Budisa, L. Moroder, and R. Huber, unpubl.). Secondly, even if the related Trp analogs would have been synthesized, probably they would not act as substrates for activation by tryptophanyl-tRNA synthetase (TrpRS) in protein translation. Finally, even changes at the neighboring positions of the protonated nitrogen of indole were disturbing because the introduction of an additional nitrogen into the indole position 2 leads to (2-Aza)Trp, which is not recognized as a substrate for protein synthesis. Conversely, introduction of a nitrogen in position 7 of the indole results in (7-Aza)Trp, which is recognized by cellular TrpRS and incorporated into proteins. These findings illustrate the universal biological significance of the indole imino function and thus indicate that the indole benzene ring is a much better target for chemical transformations that might lead to biologically interesting isosteric compounds.
The biomedical potentials of aromatic systems consisting of a pyrrole nucleus fused to a thiophene are fully recognized because of their similarities with indoles (Gronowitz et al. 1976). Indeed, it is well established that thieno[3,2-b]-pyrrole and thieno[2,3-b]-pyrrole are bioisosteric analogs of the hallucinogen and serotonin agonist N,N-′dimethyltryptamine (Blair et al. 1999). In this context, a step further for their wider utility in biomedicine could result from their incorporation into suitable proteins, because pharmaceutically active substances could convert protein mutants into useful therapeutic or even diagnostic tools, as proposed recently (Budisa et al. 1998b). Namely, recombinant proteins that contain such pharmaceutically active amino acids could act as specific "shuttles" or even "magic bullets" because of their potential ability of selective delivery and targeting in the human body (Minks et al. 2000b).
"Second code" and "tailored-to-fit" proteins
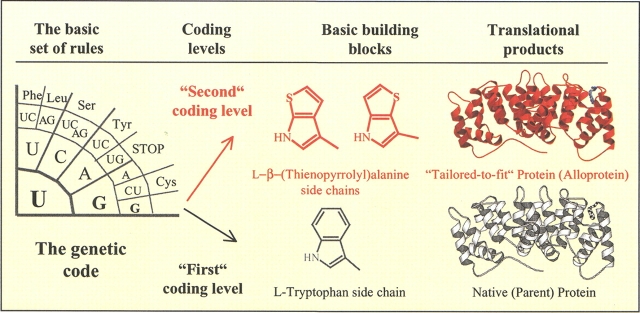
The expansion of an amino acid repertoire in vivo is certainly a novel form of protein engineering, as such "new" protein engineering is not performed by classical codon manipulation at the DNA level (oligonucleotide-directed mutagenesis), but rather by codon reassignment at the level of ribosome-directed protein translation under efficient selective pressure. Moreover, at the level of the genetic code these codon reassignments represent new possibilities to gain an additional ("second") coding level in template-directed protein synthesis, as outlined in Figure 7 ▶. The potentials of this approach are rather novel possibilities for rational approaches in design and engineering of proteins "tailored-to-fit" for specific purposes, as synthetic amino acids built into proteins may generate desirable new physico-chemical properties, functions, and functional relationships transcending those found in nature.
Fig. 7.
Schematic representation of the basic principles behind the "tailored-to-fit" protein production. The genetic code as a basic set of rules in the template-directed protein biosynthesis prescribes which codon (or codon family) is assigned to which amino acid. This is exemplified here by a UGG codon, which in ribosome-mediated protein synthesis is a signal for the canonical (coded) amino acid Trp. However, in the context of SPI methodology it is possible to accommodate (reassign) additional noncanonical amino acids into proteins as a response to a UGG codon. This possibility is labeled here as the "second" coding level in the frame of universal genetic code, achieved under the strictly defined experimental conditions. Because the proteins expressed in this way contain artificial amino acids (i.e., not usually found in nature), they are named "alloproteins" (Koide et al. 1988). In the context of rational protein design they can be regarded as "tailored-to-fit," as they are created with tailored functions capable of performing specific roles in user-defined environments.
The SPI method can fairly be said to have emerged as the method of choice for such a purpose because, being a strictly in vivo approach, it has at least two advantages: (1) a high level of protein production (usually comparable to native proteins) and (2) the simplicity of the methodology. It is therefore a reasonable alternative to various in vitro approaches because it should make possible industrial-scale production of protein mutants. However, at this stage of development only a relatively low number of noncanonical amino acids can be incorporated in a residue-specific manner (Budisa et al. 1999), which has certainly impeded wider practical promise of the SPI method in the field of protein engineering and design. To break through these limits, any future development of the method should include further genetic changes in the host cell genome, that is, the engineering of the protein translation components, presumably including aminoacyl-tRNA synthetase and optimization of the codon usage prescribed by the genetic code.
Materials and methods
Chemicals, expression host strain, and growth media
The noncanonical amino acids [2,3]Tpa and [3,2]Tpa were enzymatically synthesized according to protocols described elsewhere (Phillips et al. 1995). All other chemicals were purchased from Sigma or Aldrich unless stated otherwise. Trp-auxotrophic E. coli strain from ATCC (catalog number 49980 [genotype: WP2 uvrA]) served as host for expression experiments with AxV and b* as model proteins. Transformed host cells ATCC49980 were grown in NMM (Budisa et al. 1995, Minks et al. 1999), which contains 22 mM KH2PO4, 50 mM K2HPO4, 8.5 mM NaCl, 7.5 mM (NH4)2SO4, 1 mM MgSO4, 20 mM glucose, 1 μg/mL Ca2+, 1 μg/mL Fe2+, 0.001 μg/mL trace elements (Cu2+, Zn2+, Mn2+, MoO42−), 10 μg/mL thiamin, 10 μg/mL biotin, and the appropriate antibiotics (100 μg/mL ampicillin and 70 μg/mL kanamycin). The stock solutions (10 mg/mL) of both thia-analogs were kept in the dark at −20°C and routinely used for expression and incorporation at a concentration of 0.25 mM (50 mg/L) without special precautions. The color of the culture after overnight fermentation with thia-analogs remained faint yellow like normal Trp-containing culture without any sign of degradation of β-(thienopyrrolyl)alanines.
Fermentation and expression of β-(thienopyrrolyl)alanine-containing proteins
All fermentation and expression experiments were performed in NMM. Thereby, a T5 promoter/polymerase expression system inducible with IPTG was used (Qiagen). The protein expression host E. coli ATCC49980 was routinely cotransformed with two plasmids: ampicillin-resistant pQE-60-PP4 harboring the AxV gene sequence (NcoI-HindII fragment) under the control of T5 promoter, and kanamycin resistant pREP4 containing a repressor gene lacIq. The plasmid with the gene for b* was constructed as follows: an EcoRI-HindIII fragment from pKK223-3 plasmid (Pharmacia) containing a ribosome binding site and the b* (C40A/C82A/P27A/W38F) DNA sequence was inserted into the pQE-30 vector (Qiagen), resulting in pQIA-30b*, which is transformed into the expression host together with pREP4.
The incorporation experiments were performed using cultures grown in NMM in the presence of 100mg/L ampicillin and 70mg/L kanamycin and 0.015 mM Trp as the optimal limiting concentration of the native substrate at 30°C. The culture fermentation after the induction of protein synthesis with 1 mM IPTG was 4 h or overnight at 30°C for AxV and at 26°C for b*.
The mutant proteins of AxV and b* were purified as for the wild-type forms (Budisa et al. 1995; Golbik et al. 1999). The purity of the recombinant proteins was checked by SDS-PAGE (Coomassie and silver staining) and HPLC-profile analyses.
Analytical and spectroscopic methods
Mass spectrometry
The quantitative replacement of the native Trp residues by its noncanonical thia-containing surrogates ([3,2]Tpa and [2,3]Tpa) was confirmed routinely by electrospray mass spectrometric analyses (ESI-MS) as described earlier (Budisa et al. 1995).
UV/VIS spectroscopy
UV-absorption spectra of proteins and amino acids in buffer solutions were routinely recorded with a Perkin-Elmer Lambda 17 UV/VIS spectrophotometer. Extinction coefficients for native and substituted proteins (Table 1) were determined from quantitative amino acid analysis of the acid hydrolysates (6 M HCl, 24 h, 110°C). Extinction coefficients for the amino acids in the zwitterionic form (Table 1) were determined in 50 mM sodium dihydrogen phosphate (pH 8.0) at concentrations of 100 μM for L-Trp, 82 μM for [3,2]Tpa, and 54 μM for [2,3]Tpa at 20°C.
Fluorescence
Fluorescence spectra were recorded on a Perkin-Elmer spectrometer (LS50B) equipped with digital software. Protein probes prepared in PBS (1.0 μM AxV) or in 50 mM sodium dihydrogen phosphate (pH 8.0) (0.45 μM b*) were excited at 280 nm (slit 2.5 nm), and the emission spectra were recorded in the 300–450 nm range.
Circular dichroism
Far-UV and near-UV CD spectra were recorded at 20°C on a JASCO J-715 spectrometer in a configuration described by JASCO hardware manual P/N:0302-0265A (1995). Secondary-structure determination spectra were performed with protein concentrations of 0.08 mg/mL AxV in PBS and 0.2 mg/mL b* in 50 mM sodium dihydrogen phosphate (pH 8.0). The spectra were measured in quartz Hellma 110-QS cells with a 0.1-cm optical path length. A sufficient signal-to-noise ratio is achieved by recording four accumulations for the far-UV CD spectra. Near-UV CD spectra were recorded at protein concentrations of 0.5 mg/mL in PBS (AxV) or in 50 mM Na-phosphate (pH 8.0) (b*) in quartz Hellma 110-QS cells with a 1.0-cm optical path length. Ten scans were accumulated per spectrum and raw data were processed using the "processing method" in the JASCO software package (Software manual P/N:0302-0266A, 1995).
Thermal denaturation
For thermal unfolding measurement experiments, the JASCO spectrometer equipped with a Peltier type FDCD attachment, model PFD-350S/350L, was applied and probes were pipetted in rectangular 110-QS Hellma quartz cells with an optical path of 0.1 cm. The melting curves of wild type as well as AxV mutants in PBS were measured (heating rate of 30°/h) by monitoring the changes in dichroic intensity at 222 nm as a function of temperature change. Thermal unfolding for wild-type b* and its substituted forms was recorded at 222 nm at a protein concentration of 0.2 mg/mL using a temperature gradient of 50 °/h as described elsewhere (Budisa et al. 1998a; Golbik et al. 1999).
Thermodynamic parameters
The midpoint of denaturation (melting temperature or Tm value) as well as the van't Hoff enthalpy (ΔHm) were determined using essentially the same methods reported previously (Golbik et al. 1999; Minks et al. 1999).
Biological assays
Both native and substituted AxV and b* were tested quantitatively for their biological activity. b* and its variants were tested for inhibitory activity against the barnase (Golbik et al. 1999) whereas AxV and its mutants were tested for the ability to bind effectively at phospholipid-containing membranes in the presence of high calcium concentrations (Berendes et al. 1993).
Acknowledgments
We thank Mrs. Elisabeth Weyher for the mass spectrometric analysis and Jörg Auerhemimer for his skilful assistance in the experimental work. Mrs. Waltraud Wenger, Petra Birle, and Tatjana Krywcun are acknowledged for their excellent technical assistance. We thank Prof. Edith Miles (National Institute of Health, Bethesda, MD, USA) who kindly provided us with E. coli strain CB149 with plasmids pSTB7 and pEBA10 for tryptophan synthase production.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
AxV, annexin V
b*, pseudo wild-type barstar (b*C40A/C82A/P27A/W38F)
ɛM, molar extinction coefficient
ΔHm, van't Hoff enthalpy of unfolding
IPTG, isopropylthio-β-D-galactoside
NMM, New Minimal Medium
OD600, absorption at 600 nm
PBS, phosphate buffered saline
SPI, selective pressure incorporation method
Tm, melting temperature
[3,2]Tpa, β-(thieno[3,2-b]pyrrolyl)-L-alanine
[2,3]Tpa, β-(thieno[2,3-b]pyrrolyl)-L-alanine
[θ]M, molar dichroic ellipticity
[θ]R, residual dichroic ellipticity
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/
References
- Berendes, R., Voges, D., Demange, P., Huber, R., and Burger, A. 1993. Structure-function analysis of the ion channel selectivity filter in human annexin V. Science 262 427–430. [DOI] [PubMed] [Google Scholar]
- Bird, C.W. 1985. A new aromaticity index and its application to five-member ring heterocycles. Tetrahedron 41 1409–1414. [Google Scholar]
- Blair, J., Marona-Lewicka D., Kanthasamy, A., Lucaites, V., Nelson, D., and Nichols, D. 1999. Thieno[3,2-b]- and Thieno[2,3-b]-pyrrole bioisosteric analogues of the hallucinogen and serotonin agonist N,N-dimethyltrytamine. J. Med. Chem. 42 1106–1111. [DOI] [PubMed] [Google Scholar]
- Breitung, E.M., Shu, C.F., and McMahon, R.J. 2000. Thiazole and thiophene analogues of donor-acceptor stilbenes: Molecular hyperpolarizibilities and structure-property relationships. J. Am. Chem. Soc. 122 1154–1160. [Google Scholar]
- Bronskill, P.M. and Wong, J.T. 1988. Suppression of fluorescence of tryptophan residues in proteins by replacement with 4-fluorotryptophan. Biochem. J. 249 305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa, N., Steipe, B., Demange, P., Eckerskorn, C., Kellermann, J., and Huber, R. 1995. High level biosynthetic substitution of methionine in proteins by its analogues 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem. 230 788–796. [DOI] [PubMed] [Google Scholar]
- Budisa, N., Huber, R., Golbik, R., Minks, C., Weyher, E., and Moroder, L. 1998a. Atomic mutations in annexin V. Theromdynamic study of isomorphous protein variants. Eur. J. Biochem. 53 1–9. [DOI] [PubMed] [Google Scholar]
- Budisa, N., Minks, C., Medrano, F.J., Lutz, J., Huber, R., and Moroder, L. 1998b. Residue-specific bioincorporation of non-natural, biologically active amino acids into proteins as possible drug carriers: Structure and stability of the per-thiaproline mutant of annexin V. Proc. Natl. Acad. Sci. USA 95 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa, N., Minks, C., Alefelder, S., Wenger, W., Dong, F., Moroder, L., and Huber, R. 1999. Toward the experimental codon reassignment in vivo: Protein building with an expanded amino acid repertoire. FASEB J. 13 41–51. [DOI] [PubMed] [Google Scholar]
- Burger, A. 1991. Isosterism and bioisosterism in drug design. Prog. Drug. Res. 37 287–381. [DOI] [PubMed] [Google Scholar]
- Cohen, G.N. and Cowie, D.B. 1957. Remplacement total de la methionine par la selenomethionine dans les proteines d'Escherichia coli. Cr. Hebd. Acad. 244 680–683. [PubMed] [Google Scholar]
- Concha, N.O., Head, J.F., Kaetzel, M.A., Dedman, J.R., and Seaton, B.A. 1993. Rat annexin V crystal structure: Ca2+-induced conformational changes. Science 261 1321–1324. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. and Shortle, D. 1991. Denatured states of proteins. Annu. Rev. Biochem. 60 795–825. [DOI] [PubMed] [Google Scholar]
- Dougherty, D.A. 1996. Cation-π interactions in chemistry and biology: A new view of benzene, Phe, Tyr and Trp. Science 271 163–168. [DOI] [PubMed] [Google Scholar]
- Golbik, R., Fischer, G., and Fersht, A.R. 1999. Folding of barstar C40/C82A/P27A and catalysis of the peptydil-prolyl cis/trans isomerisation by human cytosolic cyclophilin (Cyp18). Prot. Sci. 8 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronowitz, S., Westerlund, C., and Hörnfeldt, A.B. 1976. The synthetic utility of heteroaromatic azido compounds: Preparation of some furo-, thieno-, and selenolo[3,2-b]pyrroles. Acta Chem. Scand. B30 391–395. [Google Scholar]
- Huber, R., Römisch, J., and Paques, E.P. 1990. The crystal and molecular structure of human annexin V, an anticoagulant protein that binds to calcium and membranes. EMBO J. 9 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide, H., Yokoyama, S., Kawai, G., Ha, J.M., Oka, T., Kawai, S., Miyake, T., Fuwa, T., and Miyazawa, T. 1988. Biosynthesis of a protein containing a nonprotein amino acid by Escherichia coli. Proc. Natl. Acad. Sci. USA 85 6237–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothakota, S., Mason, T., Tirrell, D.A., and Fournier, M.J. 1995. Biosynthesis of a periodic protein containing 3-thienylalanine: A step toward genetically engineered conducting polymers. J. Am. Chem. Soc. 117 536–537. [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24 946–950. [Google Scholar]
- Liemann, S. and Huber, R. 1997. Three-dimensional structure of annexins. Cell. Mol. Life Sci. 53 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D.R. and Schultz, P.G. 1999. Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl. Acad. Sci. USA 96 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaul, C.P. and Ludescher, R.D. 1999. Room temperature phosphoroscence from tryptophan halogenated analogs in amorphous sucrose. Photochem. Photobiol. 70 166–171. [Google Scholar]
- Martin, C., Hartley, R.W., and Mauguen, Y. 1999. X-ray structural analysis of compensating mutations at the barnase-barstar interface. FEBS Lett. 452 128–132. [DOI] [PubMed] [Google Scholar]
- Milun, M. and Trinajstic, N. 1977. On the aromatic stability of positional isomers consisting of bicyclic systems composed entirely of five-membered heterocycles. Croat. Chem. Acta. 49 107–113. [Google Scholar]
- Minks, C., Huber, R., Moroder, L., and Budisa, N. 1999. Atomic mutations at the single tryptophan residue of human recombinant annexin V: Effects on structure, stability and activity. Biochemistry 38 10649–10659. [DOI] [PubMed] [Google Scholar]
- ———. 2000a. Non-invasive tracing of recombinant proteins with "fluorophenylalanine-fingers.". Anal. Biochem. 284 29–34. [DOI] [PubMed] [Google Scholar]
- Minks, C., Alefelder, S., Huber, R., Moroder, L., and Budisa, N. 2000b. Towards new protein engineering: In vivo building and folding of protein shuttles for drug delivery and targeting by the selective pressure incorporation (SPI) method. Tetrahedron 56 9431–9442. [Google Scholar]
- Nath, U. and Udgaonkar, J.P. 1997. Folding of tryptophan mutants of barstar: Evidence for an initial hydrophobic collapse on the folding pathway. Biochemistry 36 8602–8610. [DOI] [PubMed] [Google Scholar]
- Phillips, R.S., Cohen, L.A., Annby, U., Wensbo, D., and Gronowitz, S. 1995. Enzymatic synthesis of thia-L-tryptophans. Bioorg. & Med. Chem. Lett. 5 1133–1134. [Google Scholar]
- Rennert, O.M. and Anker, H.S. 1963. On the incorporation of 5′5′5′-Trifluoroleucine into protein of Escherichia coli. Biochemistry 2 471–476. [DOI] [PubMed] [Google Scholar]
- Ross, J.B., Szabo, A.G., and Hogue, C.W. 1997. Enhancement of protein spectra with tryptophan analogues: Fluorescence spectroscopy of protein–protein and protein–nucleic acid interactions. Method. Enzymol. 278 151–190. [DOI] [PubMed] [Google Scholar]
- Sopkova, J., Gallay, J., Vincent, M., Pancoska, P., and Lewit-Bentley, A. 1994. The dynamic behaviour of annexin V as a function of calcium ion binding: A circular dichroism, UV absorption, and steady-state and time-resolved fluorescence study. Biochemistry 33 4490–4499. [DOI] [PubMed] [Google Scholar]
- Soumillion, P., Jespers, L., Vervoort, J., and Fastrez, J. 1995. Biosynthetic incorporation of 7-azatryptophan into the phage lambda lysozyme. Prot. Eng. 8 451–456. [DOI] [PubMed] [Google Scholar]
- Sloan, M. and Phillips, R.S. 1996. Effects of α-deuteration and of aza and thia analogs of L-tryptophan and formations of intermediates on the reaction of Escherichia coli tryptophan idole-lyase. Biochemistry 35 16165–16173. [DOI] [PubMed] [Google Scholar]
- Woody, R.W. and Dunker, K. 1996. Aromatic and cystine side-chain circular dichroism in proteins. In Circular dichroism and the conformational analysis of biomolecules (ed. G.D. Fasman), pp. 109–157. Plenum, New York.