Abstract
Glutathione S-transferase (GST) from Schistosoma japonicum has been prepared in both normal protiated (pGST) and fully deuteriated (dGST) form by recombinant DNA technology. Electrospray mass spectrometry showed that the level of deuteriation in dGST was 96% and was homogeneous across the sample. This result is attributed to the use of a deuterium-tolerant host Escherichia coli strain in the preparation of the protein. 10 heteroatom-bound deuteriums (in addition to the carbon-bound deuteriums) were resistant to exchange when dGST was incubated in protiated buffer. The physicochemical and biological properties of the two proteins were compared. dGST was relatively less stable to heat denaturation and to proteolytic cleavage than was pGST. The midpoint transition temperature for pGST was 54.9°C, whereas that for dGST was 51.0°C. Static light-scattering measurements revealed that the association behavior of dGST is also different from that of pGST. The perdeuteriated enzyme shows a tendency to associate into dimers of the fundamental dimer. This is in contrast with results that have been obtained for other perdeuteriated proteins in which perdeuteriation has been shown to promote dissociation of aggregates. dGST showed a similar Km to pGST; similar results had been obtained previously with bacterial alkaline phosphatase. However, whereas the alkaline phosphatase showed a reduced rate of catalysis on deuteriation, dGST exhibited a slightly higher rate of catalysis than pGST. It is clear that the bulk substitution of deuterium for protium has significant effects on the properties of proteins. Until many more examples have been studied, it will be difficult to predict these effects for any given protein. Nevertheless, deuteriation represents an intriguing method of preparing functional analogs of recombinant proteins.
Keywords: Deuteriation, glutathione S-transferase, deuteriated proteins, physicochemical properties
Perdeuteriation, that is, the substitution of nonexchangeable, carbon-bound protons for deuterons, is a procedure that has been used for many years in the preparation of proteins for nuclear magnetic resonance (NMR) and neutron diffraction studies. In both these techniques, protium and deuterium behave rather differently; the contrast has been exploited in NMR structure determinations of ligand-protein complexes (Petros and Fesik 1994) and an unfolded protein (Mok et al. 1999), and in neutron diffraction investigations of proteins and macromolecular assemblies (Serdyuk et al. 1994). Perdeuteriated proteins are obtained by growing simple organisms, usually algae or bacteria, on fully deuteriated growth media, such as d4-sodium succinate based on D2O.
An obvious requirement for the validity of isotopic substitution in structural studies is that the normal and isotopically enriched protein species do not differ significantly in their properties. It is generally assumed that the replacement of hydrogen by deuterium has little effect on the structure and biological activity of proteins. The crystal structures of two different proteins, a small 149–amino acid single domain Staphylococcal nuclease (Gamble et al. 1994) and the larger 3-domain, 393–amino acid elongation factor Tu from Escherichia coli (Cooper et al. 1998) have been solved for both protiated and perdeuteriated forms (96% and 95% replacement, respectively). Perdeuteriation does not affect the overall fold of these proteins. Deuteriation is, however, atypical of isotopic substitutions. A perdeuteriated protein typically has a mass of 105% of the corresponding protiated protein, the vibrational motion of the C–D bond is much slower than that of the C–H bond, and hydrophobic interactions are reported to be affected (Hattori et al. 1965b). Thus, the physicochemical properties of perdeuteriated proteins are unlikely to be identical to those of the parent protiated proteins. Differences have been shown, most notably by the group of Crespi and Katz, who pioneered and perfected the mass culture of perdeuteriated blue-green algae and used these as a source of the photosynthetic pigment phycocyanin. Perdeuteriated phycocyanin was more sensitive to thermal denaturation than was normal phycocyanin (Berns et al. 1963), and it had an increased tendency to dissociation of subunits (Hattori et al. 1965a, b). However, in a more recent study, no such decrease in stability of a SH3 domain from the Drosophila drk protein was observed on perdeuteriation (Mok et al. 1999).
There has been a relatively recent increase in activity in the preparation of perdeuteriated proteins, which has been fueled by interest in the structures of large macromolecular complexes, such as the bacterial ribosome and complexes of GroEL (Svergun et al. 1997; Nierhaus et al. 1998; Stegmann et al. 1998). The success of these investigations depends on the ability of perdeuteriated macromolecules to adopt and maintain native conformations. Conversely, if perdeuteriated proteins have different properties from their protiated parents, it is possible that these properties may be exploitable in science or even, in the case of therapeutic proteins, in medicine. We decided, therefore, to conduct a systematic comparison of the properties of a perdeuteriated protein with those of its protiated counterpart.
For this study, we have chosen glutathione S-transferase (GST) a homodimeric (26 kD/subunit) protein from Schistosoma japonicum that is involved in the Phase-II detoxification pathway (Walker et al. 1993).
GSTs catalyse the glutathione conjugation of a large range of electrophilic metabolites, thereby rendering them more water soluble. These proteins can also bind many hydrophobic molecules extremely tightly, which lowers their circulatory levels. We present a comparison of a range of physicochemical properties for the protiated and perdeuteriated proteins as well as a comparison of the enzymatic activities of the two proteins.
Results
Preparation and isolation of protiated GST (pGST) and perdeuteriated GST (dGST)
Normal, protiated GST (pGST) and perdeuteriated GST (dGST) were prepared by overexpression of the pGEX-2T plasmid in E. coli. pGST was expressed in E. coli strain BL21, as recommended by the suppliers. dGST was expressed in strain MRE600D (Bloor 1992), a strain selected for tolerance to deuterium. Isolation of the proteins was by a single-step affinity chromatography procedure, which resulted in both proteins being ∼95% pure (as assessed by electrospray ionization mass spectrometry (ESI-MS) (see below)).
Growth in perdeuteriated minimal medium resulted in a drastic decrease in expression with expression levels of ∼10 mg L−1 compared with ∼60 mg L−1 for cells grown on rich protiated medium; however, this decrease is associated with the change from rich to minimal medium, not with deuteriation (Bloor 1992).
Assessment of isotopic incorporation by electrospray mass spectrometry
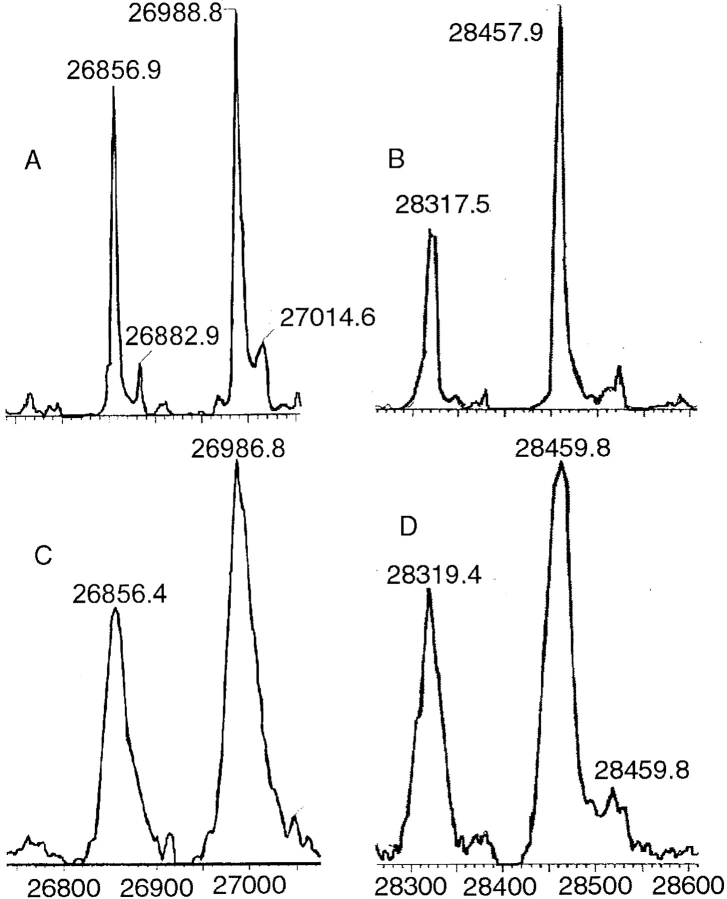
After maximum entropy deconvolution of the recorded m/z spectra (Ferrige et al. 1992), two peaks could be seen in the electrospray mass spectrum for each species. For pGST the higher mass was recorded as 26986.1 Da (sample 1) and 26988.8 Da (sample 2). GST has a theoretical average molecular mass of 26986.4 Da. The lower mass was 26855.0/26856.9 Da, corresponding to the loss of the N-terminal methionine from the protein. This is a common posttranslational modification for both prokaryotic and eukaryotic proteins, especially when the second position in the polypeptide chain is occupied by a small side-chained amino acid, such as serine in S. japonicum GST (SjGST) (Hirel 1989). For dGST, the two peaks appeared at 28456.6/28457.9 Da and 28316.2/28317.5 Da, the mass difference corresponding to the loss of perdeuteriated methionine.
One sample of dGST was treated under mildly denaturing conditions for 12 hr. The molecular weight of 28486.6 Da decreased over a period to 28454.2 Da. This corresponds to the loss of 32 deuteriums. dGST was isolated in normal H2O-based buffer, so most of the exchangeable deuterons had already exchanged. When pGST was incubated in the same denaturing buffer based on D2O, the mass increased over a period of 12.25 hr to 27381.7 Da. This corresponds to a mass increase of 395.6 Da, which is equivalent to 393 deuteriums.
Inspection of the sequence (GenBank accession no. u13850) showed that there are 1901 protons in GST, of which 403 are attached to heteroatoms (nitrogen, oxygen, sulphur) and are, at least in principle, exchangeable. Therefore, under these (denaturing) conditions 10 'exchangeable' protons are resistant to exchange. The dGST, when isolated, therefore contains 10 heteroatom-bound deuterons that are essentially nonexchangeable, plus 32 deuterons that do not exchange during the normal aqueous workup, but which can be exchanged under mildly denaturing conditions.
The average number of deuterons in dGST after denaturation was (28454.2–26986.4)/1.0063 (the difference between the mass of protium and the mass of deuterium) = 1459. The theoretical maximum is 1498 nonexchangeable + 10 resistant to exchange = 1508 deuterons. The average level of deuteriation was, therefore, 96.75%.
The mass spectrum (Fig. 1 ▶) showed that there was a narrow range of deuteriation across the sample. When the experimental data were transformed to a mass scale, the peak width at half height for pGST was 29 Da, reflecting principally the width of the natural isotope distribution; the peak width for dGST was only slightly greater at 35 Da. This contrasts with the results of Gamble et al. (1994). The peak width they observed for perdeuteriated Staphylococcal nuclease was three times the width of that of the protiated nuclease. The narrow mass range we observe probably arises from the strain-development work we carried out on the producing organism. Not only did we adapt the organism to growth on deuterium in the usual way, but we also subjected it to two colony-selection steps. These steps were designed to give a homogeneous strain with excellent deuterium tolerance. In unadapted cultures, bacterial cells will select protium from the medium in the early stages of growth, so that the deuterium content of a protein molecule depends on the time at which it was synthesized. We believe that our deuterium-tolerant strain has very little preference for protium over deuterium and that the protein molecules all reflect the isotopic mix of the culture medium.
Fig. 1.
Mass spectra of pGST (A) and dGST (B) after 12 hr in mildly denaturing conditions; the data were subjected to maximum entropy processing. Spectra C and D represent the same data; in these cases, m/z data were subjected to transformation to a mass scale without maximum entropy processing.
Circular dichroism spectroscopy
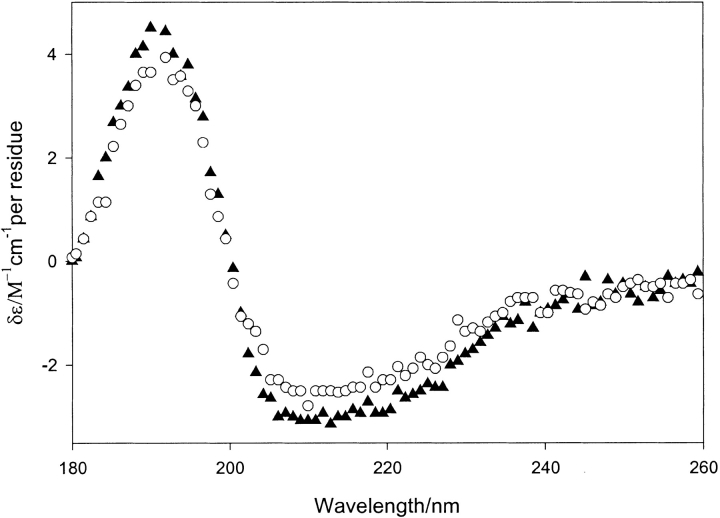
Circular dichroism spectra for both pGST and dGST are shown in Fig. 2 ▶. The two spectra are very similar, showing predominately α-helical structure with a small amount of β-sheet. The differences that are observed are only those caused by a small difference in concentration between the two samples. Sample concentrations are calculated using the extinction coefficient estimated for pGST (Gill and von Hippel 1989).
Fig. 2.
Circular dichroism spectra of protiated glutathione-S-transferase (closed triangles) and deuteriated glutathione-S-transferase (open circles) at concentrations of 1.4 mg mL−1.
Enzyme activity
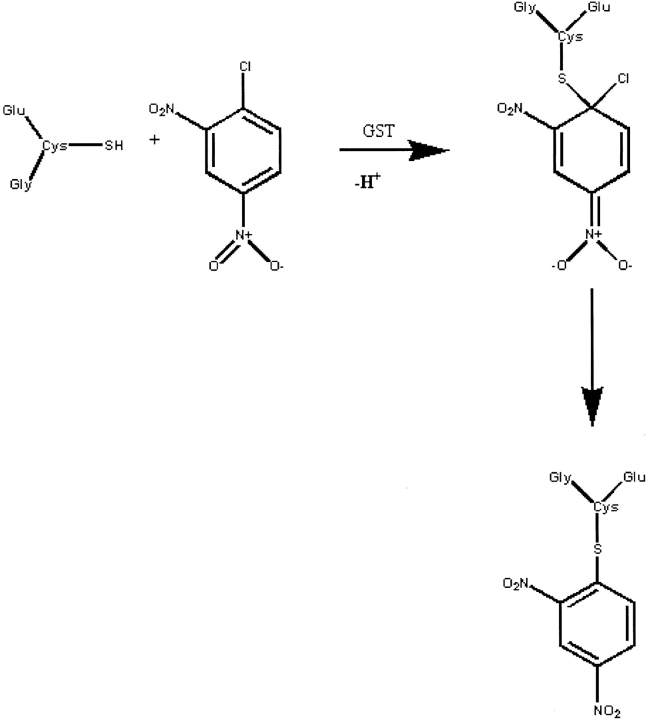
GST catalyzes the combination of glutathione with many different substrates, including the model substrate 1-chloro-2,4-dinitrobenzene (CDNB). The reaction is shown in Fig. 3 ▶. The glutathione conjugate formed in this reaction absorbs maximally at 340 nm, well separated from the λmax of the two starting materials. The Michaelis constants (Km) for pGST and dGST were measured for CDNB in the presence of excess glutathione by determining the initial rates of reaction at seven different substrate concentrations. The raw data are shown in Table 1. The Km values derived in this way were 0.95 ± 0.06 mM for pGST and 1.1 ± 0.11 mM for dGST. These values are closely similar, although a small increase in Km on perdeuteriation cannot be excluded. Vmax was 14.0 ± 0.87 × 10−5 mM s−1 for pGST and 19.5 ± 0.79 × 10−5 mM s−1 for dGST. The data indicate that kcat/Km in this instance is slightly increased by perdeuteriation of the enzyme.
Fig. 3.
The reaction between glutathione and CDNB, catalyzed by glutathione-S-transferase.
Table 1.
Enzyme kinetics of pGST and dGST—raw data
| Initial velocity (A340 min−1) | |||||
| CDNB conc. (mM) | pGST sample 1 | PGST sample 2 | pGST sample 3 | dGST sample 1 | dGST sample 2, |
| 5 | 0.0059 | 0.0062 | 0.0074 | 0.0096 | 0.0081 |
| 10 | 0.0109 | 0.0111 | 0.0136 | 0.0166 | 0.0155 |
| 20 | 0.0215 | 0.0211 | 0.0245 | 0.0258 | 0.0250 |
| 30 | 0.0274 | 0.0294 | 0.0323 | 0.0358 | 0.0354 |
| 40 | 0.0304 | 0.0361 | 0.0389 | 0.0441 | 0.0422 |
| 50 | 0.0362 | 0.0380 | 0.0431 | 0.0488 | 0.0490 |
| 60 | 0.0392 | 0.0433 | 0.0478 | 0.0546 | NC |
NC, Data not collected.
Inactivation of pGST and dGST by heat
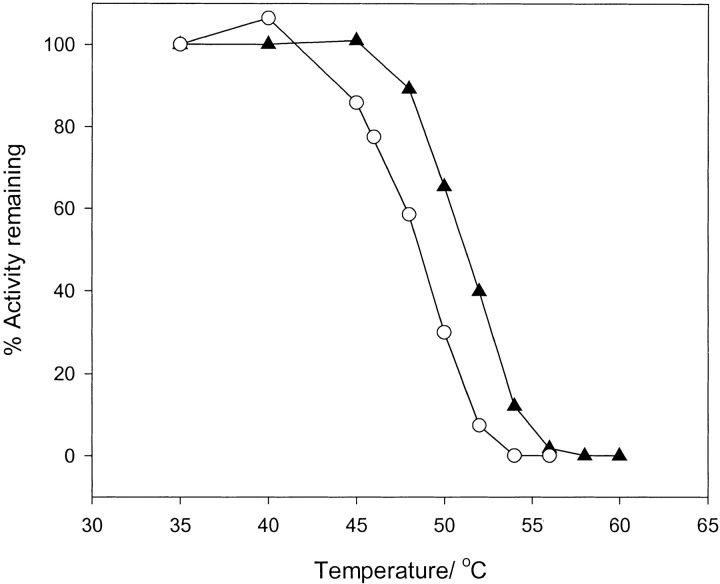
Samples of pGST and dGST were treated at different temperatures for 10 min. The residual enzymatic activities were measured using the glutathione-CDNB reaction and are shown graphically for each species in Fig. 4 ▶. pGST retained 50% activity at 51.2 ± 0.3°C (n = 3). This figure is in agreement with published data using Sj26GST derived from the same plasmid (Kaplan et al. 1997).
Fig. 4.
Profile of residual activity of protiated glutathione-S-transferase (closed triangles) and deuteriated glutathione-S-transferase (open circles) after heating for 10 min at various temperatures.
dGST, in contrast, had lost 50% activity at 48.5 ± 0.4°C (n = 2), a reduction of 2.7°C relative to the protiated protein.
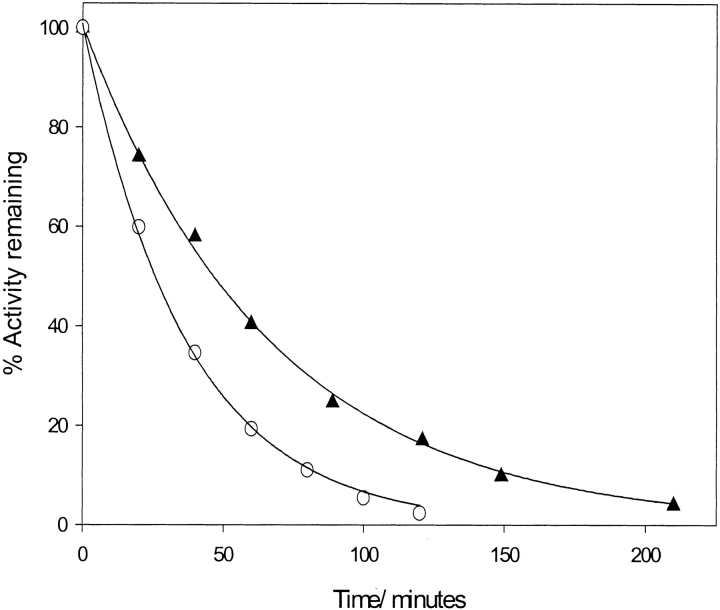
Comparison of rates of proteolytic degradation of pGST and dGST
Fig. 5 ▶ shows the effect of perdeuteriation on the rate of proteolytic cleavage of GST by chymotrypsin. Samples of GST were incubated in the presence of chymotrypsin at 20°C. Small samples were removed, and the residual enzyme activity was tested using the CDNB model reaction. The results for both proteins were well described by single exponential functions with half lives of 46.5 min and 26.4 min for pGST and dGST, respectively.
Fig. 5.
The effect of perdeuteriation on the rate of proteolytic degradation of protiated glutathione-S-transferase (closed triangles) and deuteriated glutathione-S-transferase (open circles). Lines show fit to a single exponential decay.
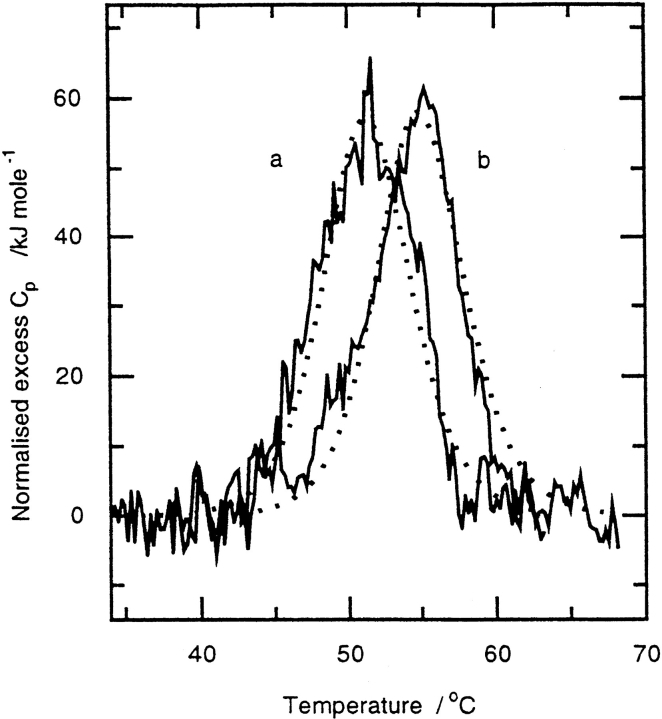
Differential scanning microcalorimetry
Differential scanning calorimetry (DSC) was used to monitor the global thermal unfolding of the proteins and to obtain quantitative thermodynamic data on the effect of perdeuteriation on unfolding, its cooperativity, and stoichiometry. Fig. 6 ▶ shows DSC curves for pGST and dGST, analysis of which indicates that pGST had a midpoint transition temperature (Tm) of 54.9 ± 0.1°C, compared with a value of 51.0 ± 0.1°C for dGST under the same conditions. The destabilizing effect of perdeuteriation was, therefore, 3.8°C, which corresponds to a decrease in standard Gibbs free energy of unfolding, ΔΔG°unf, of ∼5 kJ mol−1 under these conditions. Although these transitions are not reversible on rescan, they are consistent (within experimental uncertainty) with simple two-state cooperative unfolding of the GST dimer with similar unfolding enthalpies, ΔH°unf = 450 ± 40 kJ mol−1.
Fig. 6.
Differential scanning calorimetry scans of the thermal denaturation of (a) deuteriated glutathione-S-transferase and (b) of protiated glutathione-S-transferase (∼0.25 mg mL−1) in phosphate buffer at pH 7.6. The dotted lines show theoretical two-state transition curves using the data given in the text.
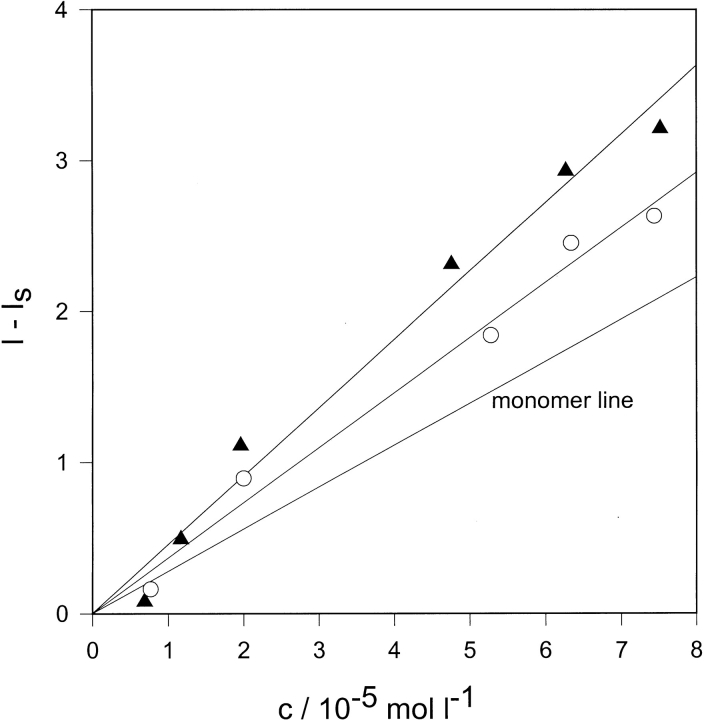
Static light scattering
Light-scattering data are presented in Fig. 7 ▶ as plots of the scattering intensity of the protein solutions, I, in excess of that from solvent, Is, as a function of protein concentration c. The basis for the analysis of the light scattering data was the Rayleigh-Gans-Debye equation in the form
Fig. 7.
Static light scattering. Plots of excess light scattering intensity from solutions of of protiated glutathione-S-transferase (closed triangles) and deuteriated glutathione-S-transferase (open circles) as a function of protein concentration. Monomer line represents scattering from nonassociated protein molecules.
 |
where Mw is the mass-average molecular weight of the solute, A2 is the second virial coefficient and K* is the appropriate optical constant.
Comparison of the scattering intensities of the protein solutions with that calculated assuming ideality for nonassociated molecules suggests limited self-association extending to high dilution. Masses of 68.0 kD for pGST and 113.4 kD for dGST were calculated by application of the Rayleigh-Gans-Debye equation, corresponding to weight average aggregation numbers of 1.26 and 2.10 dimeric units, respectively.
Discussion
This work builds on important early work of the Crespi group (Hattori et al. 1965a, b), and modern methodology and equipment have allowed us to move on in this investigation of dGST. In view of the increasing importance of deuteriated macromolecules in NMR and neutron-scattering studies, such an investigation is called for.
We have shown, first, that the use of the MRE600D strain of E. coli (Bloor 1992) allows us to obtain perdeuteriated protein in which the level of deuteriation is very precise, varying little between different molecules. We believe that this is because the strain does not change significantly during cultivation, having been rigorously preadapted to growth on deuterium. This is potentially important in neutron-scattering studies, as constant levels of deuteriation across the whole sample will increase the precision of the data obtained.
Most importantly, we have shown a number of differences in the physico-chemical properties of pGST and dGST.
The stability of the protein is affected quite significantly by perdeuteriation. dGST is relatively less stable to heat, as shown by both the effect of heat on enzymatic activity and by differential scanning microcalorimetry. dGST is also more sensitive to proteolytic cleavage than is pGST. Thus it would seem that both the global unfolding equilibrium (as revealed by DSC) and the activation barriers for transient local unfolding fluctuations (as indicated by proteolysis studies, for which global unfolding is presumably not a necessary requirement) are reduced on perdeuteriation. Hattori et al. (1965a) found similar effects in their work with deuteriated phycocyanins. They pointed out that C–D bonds have a reduced steric requirement relative to C–H bonds, owing to the reduced vibrational amplitudes of the C–D bond. In a sterically crowded transition state, deuteriation will therefore be favored, resulting in a lower activation energy of unfolding. In contrast, 42 residual heteroatom-bonded deuterons in dGST survive the aqueous work–up; of these, 10 are highly resistant to exchange, which implies higher than average protection factors. Many of these deuterons would be expected to take part in hydrogen bonding, which would be expected to be more stabilizing than hydrogen bonding that involves protium, although it should be noted that the whole issue of the stabilizing effect of core hydrogen bonding is still unclear. (Dill 1990; Shirley et al. 1992; Bowers and Klevit 1996). In the case of GST, the destabilizing effect of the side-chain deuterons outweighs the stabilizing effect of the core hydrogen bonding, and dGST is more sensitive to heat than is pGST. It is not, however, surprising that Mok et al. (1999) observed no significant difference in thermal stability between protiated and deuteriated drk protein SH3 domain. The stability of perdeuteriated proteins depends on the balance of several factors, including steric crowding of the transition state, exchangeability of the heteroatom-bound hydrogens, and perhaps even the speed of isolation. The SH3 domain has some unusual features, notably a small core and an unusual, destabilizing Thr–Gln in place of the usually conserved Gly–Glu or Gly–Asp sequence in a turn region. This destabilization may be caused by steric crowding, which would be reduced on deuteriation.
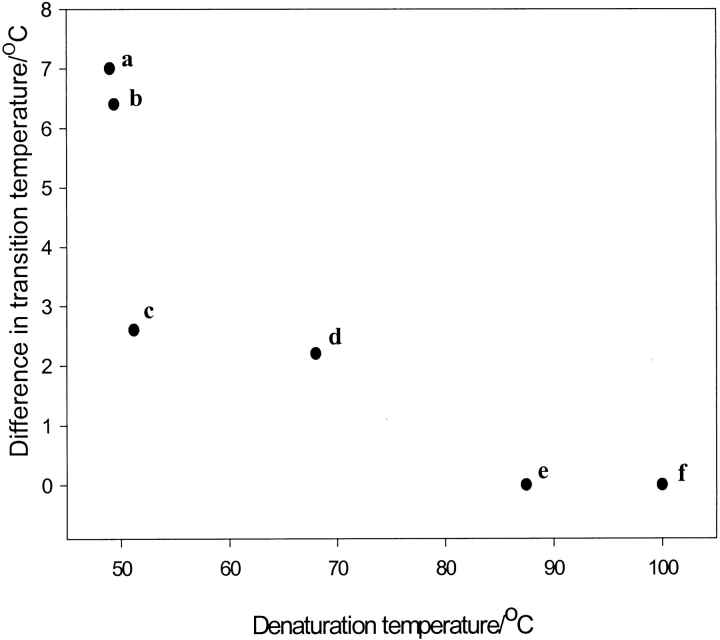
One further point of interest is that, to generalize from the rather small number of examples in the literature, the stability of stable proteins is unaffected by deuteriation, whereas proteins with low denaturation temperatures are profoundly destabilized by deuteriation. This is illustrated in Fig. 8 ▶.
Fig. 8.
The effect of perdeuteriation on the denaturation temperature of several proteins. The difference in inactivation temperature between protiated and deuteriated forms of each protein is plotted against the inactivation temperature of the protiated form of each protein. aphycocyanin from Plectonema calothricoides (Hattori et al. 1965a, b), bphycocyanin from Phormidium luridum (Hattori et al. 1965a, b), cglutathione-S-transferase from Schistosoma japonicum, dphycocyanin from Synechococcus lividus (Hattori et al. 1965a, b), ealkaline phosphatase from Escherichia coli (Rokop et al. 1969), fribonuclease from Fremyella diplosiphon (Rokop et al. 1969).
The light-scattering results indicate a difference in behavior between deuteriated phycocyanins and dGST. Hattori et al. (1965b) showed, by sedimentation measurements, that deuteriated phycocyanins have a lower tendency to association than do their deuteriated counterparts. For GST, we observe the opposite, pGST showing a lower tendency for association compared with dGST, which exists as dimers of the fundamental dimeric unit over the measured concentration range. Such association of dGST must, however, be relatively weak, seeing that DSC data for both pGST and dGST could be modeled in terms of cooperative unfolding of a simple (fundamental) dimer. Probably the higher-order aggregates of dGST dissociate gradually with increasing temperature, before global unfolding, during the DSC scan.
Hattori et al. (1965b) suggested that their results could be explained in terms of the reduced size of the deuteriated side chains leading to reduced intermolecular (as well as intramolecular) hydrophobic interactions. In dGST we see increased intermolecular interactions, but these may also be attributed to the reduced size of the deuteriated side chains. The general case is that deuteriated side chains are hydrophobic. Deuteriated methyl groups have been shown to interact more favorably with hydrophobic solvents than do the corresponding protiated methyl groups (Szydlowski and Milewska 1999), and there is substantial anecdotal evidence that deuteriated proteins and peptides are difficult to solubilize in water (e.g., S. Radford, pers. comm.). GST is typical: concentrated solutions of the deuteriated protein are unstable and precipitate on storage; the protein is also resistant to resolubization after freeze drying. The difference in association behavior between dGST and the deuteriated phycocyanins studied by Hattori et al. (1965b) may, nevertheless, be easily reconciled. Proteins have evolved in protium, and specific interactions are likely to be optimal for protiated side chains. On perdeuteriation, the side chains become less bulky and interact less favorably; in some cases, folding is even impaired (Yokogaki et al. 1995). The reduced steric requirement of the side chains facilitates other nonspecific interactions. These may be with organic solvents or with other side chains. In dGST such effects lead to an apparent dimerization of the dimeric units.
Rokop et al. (1969) measured Kms of protiated and perdeuteriated bacterial alkaline phosphatases and plant ribonucleases and found no significant change on perdeuteriation. Our results for GST were also closely similar. Rokop et al. (1969) reported an apparent reduction in the rate of catalysis for deuteriated alkaline phosphatase. Interestingly, dGST shows a small (25%) increase in the rate of catalysis relative to pGST; kcat/Km is also apparently increased on deuteriation, which suggests that dGST is actually a (slightly) more efficient enzyme than pGST. It seems that perdeuteriation can affect rates of catalysis either positively or negatively.
Our data, together with those from the literature, indicate that, in general, perdeuteriated proteins are biochemically similar to their protiated counterparts. Their physicochemical properties may, however, be quite different, although the differences are not yet easy to predict. We suggest, therefore, that there may be roles for perdeuteriated proteins as functional analogs of their protiated counterparts. One possible area would be in the catalysis of biotransformations in which high tolerance of organic solvents and low substrate specificity are desirable.
Materials and methods
Expression and purification of natural abundance and isotopically labeled Sj26GST
The pGEX-2T plasmid (Pharmacia Biotech) was used to express both types of Sj26GST. pGST was overexpressed in E. coli BL21 grown in Luria Bertani medium. An overnight culture of BL21pGEX-2T with 50 μg mL−1 ampicillin was used to inoculate Luria Bertani medium (Sambrook et al. 1989) containing 100 μg mL−1 ampicillin. These cells were then cultured to an A550 of ∼0.75 and induced with IPTG to a final concentration of 1 mM. After a further 5 hr, the cells were harvested by centrifugation and stored at −20°C until required. dGST was expressed in MRE600D (Bloor 1992), a strain selected for tolerance to deuterium. An overnight culture of MRE600DpGEX-2T cultured in d4-sodium succinate medium with 50 μg mL−1 ampicillin was used to inoculate d4-sodium succinate medium in 99.9% D2O containing 100 μg mL−1 ampicillin. These cells were induced at an optical density of 3 and harvested 8 hr later.
pGST and dGST were purified in an identical manner. Cell pellets were resuspended in 100 mL PBS buffer pH 7.6 containing 10 μM phenylmethylsulphonyl fluoride (PMSF) and 1 mM sodium azide, and lysed by the addition of lysozyme in the presence of EDTA (15 mg and 6 mg, respectively, per 10 g cell paste). After gentle mixing at room temperature for 10 min, 5 mL 4% sodium deoxycholic acid (per 10 g cell paste) was added and the mixture left for a further 10 min at room temperature. DNA was digested by addition of Dnase I (200 μL, 1 mg mL−1) in the presence of MgCl2 (100 mg; both per 10 g cell paste) followed by incubation at room temperature for 20 min. The cell lysate was then incubated on ice for 20 min with 10 mL 1% Triton X. After centrifugation for 20 min at 10,000 rpm (9600 g, Sorvall, GSA rotor) the supernatant was then subjected to ultracentrifugation (45,000 rpm, Dupont Sorvall Ultracentrifuge OTD 65B, TFT 65.13 fixed angle rotor 135,500 g, 90 min) to remove ribosomes. GST was then isolated by glutathione affinity chromatography (Smith and Johnson 1988) using Glutathione Sepharose 4B resin (Pharmacia Biotech) and the pure protein stored at −20°C until required.
Assessment of isotopic incorporation
ESI-MS spectra were collected using a Micromass Quattro mass spectrometer, equipped with an ESI source and upgraded to Quattro II specifications. The instrument was calibrated using myoglobin (2 pmol μL−1). GST samples were infused into the ESI source at 5 μL min−1. The instrument was scanned from m/z 700–1300 in 6 s.
Approximately 20 scans were summed to yield the recorded spectrum. ESI-MS data were processed using simple transformation and maximum-entropy software (Ferrige et al. 1992) assuming protonation and deuteronation as appropriate.
pGST and dGST were dialyzed overnight in sodium phosphate buffer pH 7.8 with 2 mM dithiothreitol, 10 μM PMSF and 1 mM sodium azide. To assess the number of deuterons incorporated at exchangeable positions 26 μL of dGST at 0.75 mg mL−1 was diluted with 260 μL 0.1% formic acid in solvent consisting of 10% acetonitrile : 90% H2O, and the mixture was applied to a protein trap (Michrom Bioresources). We applied 300 μL of the same solvent to the trap to wash the column before elution of the protein by the addition of 100 μL of 0.1% formic acid in 80% acetonitrile : 20% H2O. The eluate was diluted to 2 pmol μL−1 using the same solvent before MS analysis. pGST was prepared in an analogous manner but replacing D2O for H2O in each buffer, and the concentration was 4 pmol μL−1.
Enzyme activity assay
The enzymatic activity of each GST species was assessed at 20°C by monitoring the reaction of the model substrate 1-chloro-2,4-dinitrobenzene (CDNB) with glutathione, spectrophotometrically at 340 nm (Habig et al. 1974). Enzyme was added to 0.1 M potassium phosphate buffer pH 6.5 containing reduced glutathione (2 mM) and CDNB at seven different substrate concentrations (see Table 1). The uncatalyzed reaction has a measurable rate, so a control reaction without enzyme was set up in the reference cell. The reaction was carried out in triplicate for pGST and in duplicate for dGST (dGST was available in much smaller quantity).
Comparison of rates of proteolytic degradation
pGST and dGST were desalted into 66 mM sodium phosphate buffer at pH 7.8. We incubated 125 μL protein samples (0.2 mg mL−1) at 20°C for 170 min before adding 20 μL α-chymotrypsin (2 mg mL−1). Enzyme activity was assessed spectrophotometrically at intervals using 1-chloro-2,4-dintrobenzene as a model substrate, as described above. Each species was tested in triplicate.
Differential scanning microcalorimetry
DSC measurements were performed using a Microcal MC-2D instrument fitted with an EM Electronics N2a nanovolt preamplifier at a routine scan rate of 60°C h−1 employing standard methodology (Cooper and Johnson 1994) and a protein concentration of 9 μM in sodium phosphate pH 7.6. Sample and reference buffer solutions were degassed for ∼1 min under a vacuum with gentle stirring before loading and were held under 2–3 atm N2 pressure during DSC to suppress bubble formation. Buffer baselines were collected separately under identical conditions and were subtracted from experimental thermograms before concentration normalization and analysis using Microcal Origin software.
Heat denaturation
We heated 2–3 μM solutions (100 μL) of each protein in sodium phosphate buffer pH 7.6 for 10 min at incremental temperatures between 35°C and 60°C using a Hybaid thermal reactor. The residual enzyme activity of a 10–20 μL aliquot was then assessed spectrophotometrically, as described previously. Each temperature point was repeated in triplicate for pGST and duplicate for dGST.
Circular dichroism spectroscopy
All circular dichroism measurements were made on station 3.1 of the CLRC Daresbury Laboratory synchrotron radiation source. Samples in Helma '124' plate cells with a path length of 0.1 mm were scanned between 175 nm and 260 nm with 3 s per point and a step size of 0.5 nm All experiments were performed at a concentration of 1.4 mg mL−1 (calculated as above) in 60 mM sodium phosphate buffer pH 7.8, 1 mM sodium azide, 10 μM PMSF and 2 mM dithiothreitol.
Static light scattering
Static light-scattering intensities were measured at a scattering angle of 90° by means of a Malvern PCS 100 instrument using vertically polarized incident light of wavelength 488 nm supplied by a 2 W argon-ion laser (Coherent Innova 90). The intensity scale was calibrated against benzene. Protein solutions (0.3 mg–4 mg mL−1) were prepared in 20 mM Tris HCl pH 7.8, 2 mM dithiothreitol, 10 μM PMSF, and 1 mM sodium azide. Solutions were clarified by filtering through 0.22 μm Millipore Millex filters (Triton free) directly into scattering cells that previously had been cleaned with condensing acetone vapor. Values of the specific refractive index increment were determined using an Abbé 60/ED precision refractometer (Bellingham and Stanley Ltd.).
Acknowledgments
We thank The Wellcome Trust for a Sir Henry Wellcome Commemorative Award for Innovative Research, the UK CVCP for the award of an Overseas Research Studentship to LY and Professors J.R. Helliwell and K.T. Douglas for helpful discussions. We thank Drs. Gareth Jones and David Clarke (CLRC Daresbury Laboratory) for help with the CD spectroscopy work. The biological microcalorimetry facility in Glasgow is funded by BBSRC and EPSRC.
The publication costs of this article were defrayed in part by payment of page charges.This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.46001.
References
- Berns, D.S., Crespi, H.L., and Katz, J.J. 1963. Isolation, amino acid composition and some physico-chemical properties of the protein deuterio-phycocyanin. J. Am. Chem. Soc. 85 8–14. [Google Scholar]
- Bloor, D.J. 1992. Production of deuteriated elongation factor Tu for use in NMR studies. Ph.D. thesis University of Manchester.
- Bowers, P.M. and Klevit, R.E. 1996. Hydrogen bonding and equilibrium isotope enrichment in histidine-containing proteins. Nat. Sruct. Biol. 3 522–531. [DOI] [PubMed] [Google Scholar]
- Cooper, S.J., Brockwell, D., Raftery, J., Attwood, D., Barber, J., and Helliwell, J.R. 1998. The X-ray crystal structures of perdeuteriated and protiated enzyme elongation factor Tu are very similar. J. Chem. Soc. Chem. Commun. 1063–1064.
- Cooper, A. and Johnson, C.M. 1994. Differential scanning calorimetry. Methods in Molecular Biology 22 125–136. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. 1990. Dominant forces in protein folding. Biochemistry 29 7133–7155. [DOI] [PubMed] [Google Scholar]
- Ferrige, A.G., Seddon, M.J., Green, B.N., Jarvis, S.A., and Skilling, J. 1992. Disentangling electrospray spectra with maximum entropy. Rapid Commun. Mass Spectrom. 6 707–711. [Google Scholar]
- Gamble, T.R., Clauser, K.R., and Kossiakoff, A.A. 1994. The production and X-ray structure determination of perdeuterated Staphylococcal nuclease. Biophys. Chem. 53 15–26. [DOI] [PubMed] [Google Scholar]
- Gill, S. and von Hippel, P. 1989. Calculation of protein extinction coefficients from amino-acid sequence data. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Habig, W.H., Pabst, M.J., Jakoby, and W.B. 1974. Glutathione S-transferases the first energetic step in mercapturic acid formation. J. Biol. Chem. 249 7130–7139. [PubMed] [Google Scholar]
- Hattori, A., Crespi, H.L., and Katz, J.J. 1965a. Effect of side-chain deuteration on protein stability. Biochemistry 4 1213–1225. [DOI] [PubMed] [Google Scholar]
- ———. 1965b. Association and dissociation of Phycocyanin and the effects of deuterium substitution on the processes. Biochemistry 4 1225–1238. [Google Scholar]
- Hirel, P. 1989. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. 86 8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, W., Hüsler, P., Klump, H., Erhardt, J., Sluis-Cremer, N., and Dirr, H. 1997. Conformational stability of pGEX expressed Schistosoma japonicum glutathione S-transferase: A detoxification enzyme and fusion affinity tag. Protein Sci. 6 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, Y.K., Kay, C.M., Kay, L.E., and Forman-Kay, J. 1999. NOE data demonstrating a compact unfolded state for an SH3 domain under non-denaturing conditions. J. Mol. Biol. 289 619–638. [DOI] [PubMed] [Google Scholar]
- Nierhaus, K., Wadzack, J., Burkhardt, N., Junemann, R., Meerwinck, W., Willumeit, R., and Stuhrmann, H. 1998. Structure of the elongating ribosome: Arrangement of the two tRNAs before and after translocation. Proc. Natl. Acad. Sci. 95 945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros, A.M. and Fesik, S.W. 1994. Nuclear-magnetic-resonance methods for studying protein-ligand complexes. Methods Enzymol. 239 717–739. [DOI] [PubMed] [Google Scholar]
- Rokop, S., Gajda, L., Parkmerter, S., Crespi, H.L., and Katz, J.J. 1969. Purification and characterization of fully deuterated enzymes. Biochem. Biophys. Acta 191 707–715. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 3: A.1.
- Serdyuk, I.N., Pavlov, M.Y., Rublevskaya, I.N., Zaccai, G., and Leberman R. 1994. The triple isotopic substitution method in small-angle neutron scattering. Biophys. Chem. 53 123–130. [DOI] [PubMed] [Google Scholar]
- Shirley, B.A., Stanssens, P., Hahn, U., and Pace, C.N. 1992. Contribution of hydrogen to the conformational stability of phage ribonuclease T1. Biochemistry 31 725–732. [DOI] [PubMed] [Google Scholar]
- Smith, D.B. and Johnson, K.S. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67 31–40. [DOI] [PubMed] [Google Scholar]
- Stegmann, R., Manakova, E., Rossle, M., Heumann, H., Nieba-Axmann, S., Pluckthun, A., Hermann, T., May, R., and Wiedenmann, A. 1998. Structural changes of the Escherichia coli GroEL-GroES chaperonins on complex formation in solution: A neutron small angle scattering study. J. of Struct. Biol. 121 30–40. [DOI] [PubMed] [Google Scholar]
- Svergun, D., Burkhardt, N., Pedersen, J., Koch, M., Volkov, V., Kozin, M., Meerwink, W., Stuhrmann, H., Diedrich, G., and Nierhaus, K. 1997. Solution scattering structural analysis of the 70 S Escherichia coli ribosome by contrast variation. 2. A model of the ribosome and its RNA at 3.5 nm resolution. J. Mol. Biol. 271 602–618. [DOI] [PubMed] [Google Scholar]
- Szydlowski, J. and Milewska, A. 1999. Deuterium isotope effect on miscibility of some binary mixtures containing acetonitrile or nitromethane. Fluid Phase Equilibria 162: 181–191. [Google Scholar]
- Walker, J., Crowley, P., Moreman, A.D., and Barrett, J. 1993. Biochemical properties of cloned glutathione S-transferase from Schistosoma mansoni and Schistosoma japonicum. Mol. Biochem. Parasitol. 61 255–264. [DOI] [PubMed] [Google Scholar]
- Yokogaki, S., Unno, K., Oku, N., and Okada, S. 1995. Chaparonin-reparable subtle incompleteness of protein assembly induced by a substitution of hydrogen with deuterium: Effect of GroE on deuterated ribulose 1,5-bisphosphate carboxylase. Plant Cell Physiol. 36 419–423. [Google Scholar]