Abstract
The coiled coil is a ubiquitous protein-folding motif. It generally is accepted that coiled coils are characterized by sequence patterns known as heptad repeats. Such patterns direct the formation and assembly of amphipathic α-helices, the hydrophobic faces of which interface in a specific manner first proposed by Crick and termed "knobs-into-holes packing". We developed software, socket, to recognize this packing in protein structures. As expected, in a trawl of the protein data bank, we found examples of canonical coiled coils with a single contiguous heptad repeat. In addition, we identified structures with multiple, overlapping heptad repeats. This observation extends Crick's original postulate: Multiple, offset heptad repeats help explain assemblies with more than two helices. Indeed, we have found that the sequence offset of the multiple heptad repeats is related to the coiled-coil oligomer state. Here we focus on one particular sequence motif in which two heptad repeats are offset by two residues. This offset sets up two hydrophobic faces separated by ≈150°–160° around the α-helix. In turn, two different combinations of these faces are possible. Either similar or opposite faces can interface, which leads to open or closed multihelix assemblies. Accordingly, we refer to these two forms as α-sheets and α-cylinders. We illustrate these structures with our own predictions and by reference to natural variants on these designs that have recently come to light.
Keywords: α-Cylinder, α-sheet, amphipathic α-helix, coiled coil, colicin Ia, knobs-into-holes packing, TolC, variant surface glycoproteins
Understanding macromolecular assemblies is a key issue in modern-day structural biology. Arguably, the α-helical coiled coil is one of the simpler and more readily understood building blocks of small, medium, and large protein complexes (O'Shea et al. 1989; Sutton et al. 1998; Wigge et al. 1998). The accepted hallmark of the coiled coil is the seven-residue heptad repeat. This is more of a sequence pattern than a motif, as the traditional requirement is for hydrophobic side chains alternately separated by three and four residues. This average is one hydrophobic residue for every 3.5 residues, which falls short of the 3.6-residue repeat of the α-helix. Thus, the pattern sets up a hydrophobic seam that slowly winds around the α-helix in an opposite sense to the helix. The association of these seams produces coiled coils. Because of the twisting of the seam, the helices wrap (i.e., supercoil) around each other to keep the hydrophobic residues in contact and away from solvent. For heptad-based coiled coils, the supercoil is left handed.
By convention, heptads are assigned abcdefg, with the hydrophobic residues at a and d. After Crick (1953), these residues form a seam of knobs on one helix, which dock into complementary holes of a neighboring helix. We developed software (socket) to recognize this packing and assign heptad registers in protein structures.
Full details of the socket algorithm will be given elsewhere (J. Walshaw and D. Woolfson, in prep.). Briefly, SOCKET uses Crick's original postulates (Crick 1953) to locate and analyze clusters of residues that constitute knobs-into-holes interactions. A residue is termed a knob if the center of its side chain simultaneously lies within a packing-cutoff distance from the centers of four other side chains; we describe the latter as the sides of the hole. We classify each knob as Type 1, 2, 3, or 4. Type 1 and 3 knobs lie across the holes, whereas Type 2 and 4 knobs lie inside them. An insertion cutoff distinguishes these variations. More importantly, Type 1 and 2 knobs occur in isolated knobs-into-holes interactions, whereas Type 3 and 4 knobs occur with what we term pairwise or cyclic complementarity. Two helices, X and Y, show pairwise-complementary knobs-into-holes interactions if (1) a knob on X fits into a hole on Y, (2) one side of the hole on Y is itself a knob that fits back into a hole on X, or (3) the first mentioned knob on X forms a side of the hole that accepts the knob from Y. This is the classic knobs-into-holes interaction as observed in the leucine zipper (O'Shea et al. 1991) and illustrated in Figure 1 ▶, A and B1. Cyclically complementary knobs occur in coiled-coil structures with more than two interacting helices. In these cases, knobs-into-holes interactions occur in tandem. For instance, in an XYZ trimer, a knob on X fits into a hole on Y; the symmetry-related knob on Y fits into a hole on Z; the corresponding knob on Z fits back into a hole on X. In this case, the complementarity is cyclic and is completed by the first mentioned knob on X forming part of the hole that accepts the knob from Z. This case is illustrated in Figure 1C ▶. Similarly, this description extends to describe knobs-into-holes interactions in tetramers and pentamers.
Fig. 1.
Pairwise and cyclically complementary knobs-into-holes interactions. (A) A view into the interface of a parallel, dimeric coiled coil. The central line of four residues represents a layer. Side chains are contributed from both helices: Those in white are from the furthermost helix shown as a ribbon, and thus, point toward the viewer; those shaded grey are from the near helix and point into the paper. Dotted lines link the centers of mass of the side chains, which are indicated by black circles. (B) A schematic representation of part A using the same shading and highlighting the knob (k) and hole (h) residues. (C) A layer of cyclically complementary knobs-into-holes interactions in a trimeric coiled coil. The view is from above the layer (i.e., down the superhelical axis). Each strand is shaded differently. The leucine residues are the knobs and the bases of the holes are highlighted by straight lines. The ribbon diagrams shown here and in subsequent figures were created using molscript (Kraulis 1991).
Using standard Brookhaven Protein Data Bank (PDB) coordinate files (Berman et al. 2000) and corresponding DSSP (Definition of Secondary Structure of Proteins) format files (Kabsch and Sander 1983), socket identifies knobs and holes for an input structure, assigns knob type, and searches for complementary knobs-into-holes interactions. Using these data, socket performs the following analyses: (1) structure-based assignment of heptad register to the coiled-coil sequences, (2) calculation of the number of helices in, and the topologies of, intact coiled-coil units (e.g., parallel dimer, antiparallel dimer, etc.), and (3) calculation of core-packing angles , that is, the angle at which a knob points into its associated hole (O'Shea et al. 1991; Harbury et al. 1993).
socket was parameterized (that is, optimal values for the packing and insertion cutoffs were calibrated, each to 7 Å) using a set of coiled-coil structures known from literature. This analysis and our first trawl of the entire PDB using socket revealed that the distinction between isolated and complementary knobs-into-holes interactions was important; many isolated knob-into-hole interactions were returned for noncoiled-coil structures. To avoid such examples, socket was set to require either one layer of cyclically complementary knobs-into-holes, or two or more layers with pairwise complementarity before classing a structure as coiled-coil positive.
We used socket to identify coiled-coil containing structures in release 87 of the PDB. At a 7Å packing cutoff, 202 of the 9255 structures tested positive for coiled-coil motifs longer than two full heptad repeats. These were grouped into 43 nonhomologous sequence families and 63 distinct coiled-coil motifs (some intact structures contained more than one coiled-coil domain). We found examples of two-, three-, four-, and five-stranded coiled coils. For all of these oligomer states except the five-stranded motif, which is a unique parallel structure (Malashkevich et al. 1996), both parallel and antiparallel examples were observed. In addition to these structures, which hitherto would be regarded as classic coiled-coil assemblies, we found a number of borderline and unusual examples. Borderline examples included a variety of four-helix bundles, which showed contiguous layers of knobs-into-holes packing, but without cyclic complementarity. Essentially, these were examples of multiple, two-stranded coiled coils. More unusual structures included the core of gp41 from HIV (Chan et al. 1997), which consists of six helices with a homotrimeric, supercoiled coiled-coil hub, symmetrically abutted by three helices, effectively producing a trimer of heterotrimers. Full details of our analysis of the PDB will appear elsewhere (J. Walshaw and D. Woolfson, in prep.) and will be made available as a resource on the World Wide Web (http://www.biols.susx.ac.uk/coiledcoils).
Inspection of the structures highlighted by socket and the resulting structure-based heptad assignments revealed an interesting feature common in high-order coiled-coil assemblies (i.e., those above dimer). Whereas isolated dimers showed classic heptad repeats leading to a single (a/d) hydrophobic seam and an interface with pairwise-complementary knobs-into-holes, higher order structures showed peripheral knobs-into-holes interactions in addition to those centered on the a and d sites. In other words, for the high order structures, the helix-helix interfaces extended further around the individual helices. Effectively, this is achieved by sequences showing more than one heptad repeat per helix. We note that others have alluded to this for specific protein assemblies (Harbury et al. 1993; Kim et al. 1999; Koronakis et al. 2000). We refer to such sequences as having overlapping heptad repeats and to the resulting helices as multifaceted. For two overlapping heptad repeats, there are three possible sequence offsets of one, two, and three residues. We found that many natural coiled-coil trimers can be considered as having two heptad repeats offset by three residues, whereas many tetramers and pentamers are variations on a one-residue offset. A full treatment of these ideas is beyond the scope of this communication. This is because (1) the concept draws together coiled-coil trimers, tetramers, and pentamers, (2) it relates to the structural organisation of the aforementioned four-helix bundles and unusual assemblies like those observed in gp41, and (3) it plays a role in helical repeat structures such as those formed by the armadillo and HEAT motifs and tetratricopeptide repeats (Groves and Barford 1999).
Here we focus on overlapping heptad repeats offset by two residues. Compared to the one- and three-residue offsets, this leads to two clearly defined and distinct hydrophobic seams in each helical segment. This has unusual structural consequences for helical assemblies that may be treated separately from the other cases. First, we outline a general theory for the possible assemblies. Second, we describe how this is realized in examples of natural protein structures.
In the standard heptad assignment, a two-residue sequence offset places hydrophobic residues at a, c, d, and f, with c and f effectively making up the a and d positions of the second, offset repeat. Canonical α-helices have 3.6 residues per turn, and therefore, successive residues arc out 100° around a helical wheel. Thus in this representation, a two-residue sequence offset manifests as an angular separation between the two hydrophobic seams (a/d and c/f) of 200°, which is equivalent to 160° (θ) when measured in the opposite direction around the wheel. To accommodate the seven-residue sequence repeat, helical-wheel diagrams for coiled coils are traditionally drawn in supercoil space, with successive residues separated by ≈103°. Effectively, this makes all the a residues equivalent and so on. In this case, θ is altered to ≈154°. These ideas are illustrated for a seven-residue-based helical wheel in Figure 2A ▶.
Fig. 2.

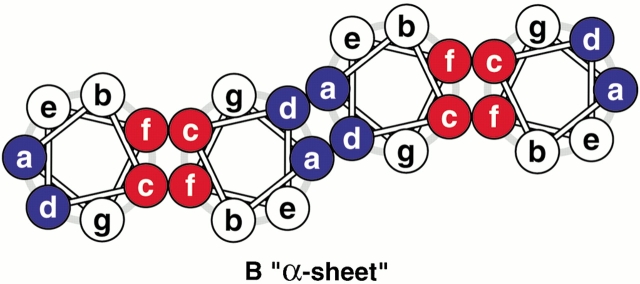
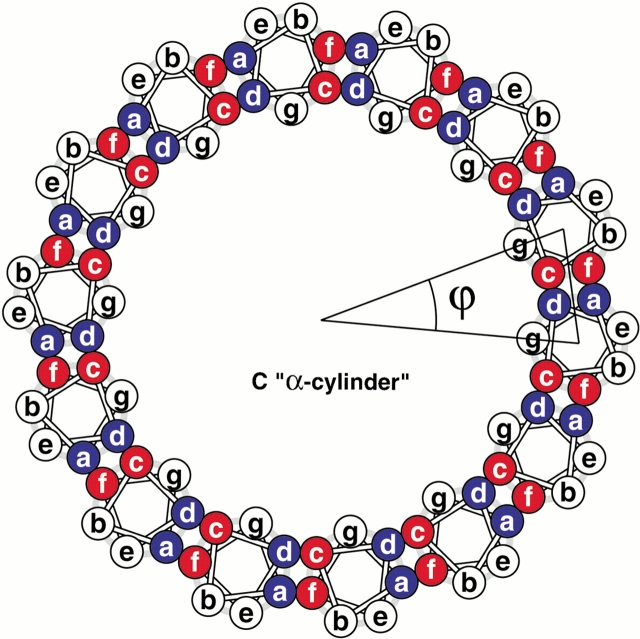
Construction of coiled-coil protein assemblies from bifaceted α-helices. (A) The lower part shows two heptad repeats aligned with a sequence offset of two residues. The two hydrophobic seams are highlighted and distinguished by the letters X and Y. The upper panel shows a 3.5-residue-per-turn helical wheel for the combined sequence. The resulting angular offset between the two hydrophobic seams, represented by the curved arrow and labelled θ, is ≈154°. (B) A syntypic assembly in which the a/d and c/f seams self-associate to form α-sheets. (C) An antitypic assembly in which the a/d and c/f faces associate. Such structures may close to form α-cylinders. The arc swept out by each helix (ϕ) is related to θ (ϕ = 180 − θ) and the number of helices required to close the cylinder (n) is given by n = 360 ÷ (180 − θ).
Whether viewed in Cartesian or supercoil space, the two hydrophobic seams are distinct, that is, they do not merge. This wide separation of two hydrophobic seams produces bifaceted helices and sets up different opportunities for α-helical assemblies. Considering all-parallel structures, there are two possibilities for helix-helix packing: In what we term syn-facing assemblies, two like faces from neighboring helices combine to produce an open α-sheet (Fig. 2B ▶). Effectively, the angular offsets (θ) of successive helices cancel. In antifacing assemblies, however, a/d faces pair with c/f and the resulting structures may close to form an α-cylinder (Fig. 2C ▶). The number of helices (n) required to close the cylinder is related to θ (n = 360 ÷ [180 − θ]).
This relationship can be understood as follows: The locus described by the axes of the n helices is a circle (360°); strictly speaking, it is the points of an n-sided polygon. Therefore, each helix turns through 360° ÷ n (ϕ in Fig. 2C ▶) around the circle. Successive helices also rotate about their own axes by a constant angle with respect to the previous helix. This rotation is equal to 180° − θ. Because helix n + 1 and helix 1 effectively superimpose, the overall axis rotation for n helices must be 360° (i.e., the rotation of each helix about its own axis is also 360° ÷ n). Thus, 360° ÷ n = 180° − θ, or n = 360° ÷ (180° − θ).
Substituting numerical values into this equation is illuminating: If θ were 180°, then the helix axes would describe a straight line, and the axes of the helices would not rotate. However, with θ <180°, the circle closes. For heptad-based supercoiled helices, θ is ≈154°, but it is exactly 360° ×(3 ÷ 7). Using the precise value gives n = 14 exactly. Thus, the theoretical α-cylinder should have exactly 14 helices in the ring.
For antiparallel helix pairs, the principles outlined above do not change, but the details do: Syntypic association should lead to cylinders, whereas sheets should be formed from antitypic interfaces.
The various possibilities for assemblies based on bifaceted helices occurred to us following the first trawl of the PDB with socket, which returned two examples of three-helix α-sheet structures (Freymann et al. 1990; Blum et al. 1993; Wiener et al. 1997). With the ideas outlined above and the theory developed out of our observations. However, the recently described structure of TolC (Koronakis et al. 2000), which was not in release 87 of the PDB, provides the first natural example of an α-cylinder, which is referred to as an α-helical barrel by the investigators. However, as described below, the structure is not a uniform or an ideal cylinder.
The examples of α-sheets were in the variant surface glycoproteins (VSGs) from the trypanosome that causes sleeping sickness (Freymann et al. 1990; Blum et al. 1993) (Figs. 3A,B ▶), and in colicin Ia from Escherichia coli (Wiener et al. 1997). In both cases, each α-sheet comprises three helices with the central helix harboring the double-heptad repeats. Otherwise, these examples show differences: In the VSGs, the adjacent helices are in mixed parallel/antiparallel arrangements, whereas in colicin Ia, adjacent helices are all antiparallel (Figs. 3A,C ▶). Second, although the seams identified by socket in the central helices (helix A) of the VSGs do overlap, those in the structurally similar helix (helix T2) of colicin Ia do not (Fig. 3C ▶). Finally, in the VSG structures, two sheets make contact through the edge helices to form an oval barrel-like motif, although the intermolecular packing is not mediated by knobs-into-holes interactions.
Fig. 3.
Natural examples of α-sheets. (A and B) Orthogonal views of the S, A, and B helices (left to right, respectively) of the helical bundle of the VSG MITat 1.2 (Freymann et al. 1990). A kinemage has been produced to display this structure in three dimensions and to highlight knobs-into-holes packing. (C) The three-helix sheet of colicin 1a (Wiener et al. 1997). Knob side chains highlighted by socket using a 7 Å cutoff are shown in ball-and-stick representation. Parallel and antiparallel helix orientations are distinguished by shading; the light helices have the N-terminal end at the top.
TolC has two α-barrel-like domains (Koronakis et al. 2000) as shown in Figures 4 ▶, A and B. Both have 12 helices contributed by three monomers. In the lower barrel, each helix pairs with another from the same protomer to form separate supercoiled, antiparallel coiled coils. Knob residues located by socket for one TolC protomer are highlighted as balls-and-sticks in Figure 4C ▶. The lower part of the figure shows extended runs of knobs, which conform to a canonical heptad pattern, in two distinct and long, antiparallel coiled-coil motifs. In contrast, the helices of the upper barrel appear to pack more uniformly, albeit with a slant, to describe a type of α-cylinder. The socket output for this part of the structure (upper part of Fig. 4C ▶), however, revealed markedly fewer knobs-into-holes interactions than found in the lower barrel. Nevertheless, on this structural basis, we unambiguously were able to assign heptad registers for the helices of the upper barrel. This revealed knobs at relative a, c, d, and f positions, and syn-faced association of two seams between adjacent antiparallel helices, which is fully consistent with the theory outlined above.
Fig. 4.

TolC, a natural variant on the α-cylinder structure. (A and B) Orthogonal views of the TolC trimer showing the barrel-like structures (Koronakis et al. 2000). The three protomer chains are distinguished by shading. (C) The two helical domains (upper and lower) of a TolC monomer, highlighting the many knobs-into-holes of the two-stranded antiparallel coiled coils of the lower barrel compared with the small number of such interactions in the upper barrel. The orientation of this figure is the same as that used in Figures 1B and 1C ▶ of the original paper describing the TolC structure (Koronakis et al. 2000). Thus, from left to right in the upper barrel, the helices are H7, H6, H3, and H2. Otherwise, the legend is the same as for Figure 3 ▶. A kinemage has also been prepared for this structure.
In more detail, residues Arg-82 (assigned by socket as an f position) to Asp-101 (a d position) in helix 3 (H3) form a bifaceted coiled coil. Knobs at Gln-87 and Tyr-98 fall at d and a positions, respectively, in this register, and interact with H2; whereas, residues Leu-86 and Val-96 form knobs at c and f positions, respectively, and interact with H6. However, knobs-into-holes interactions were not contiguous around the upper cylinder. In particular, there were more between helices of the same monomer than between the helices at the monomer-monomer interfaces (see TolC kinemage). For example, for chain A of the upper barrel, eight intraprotomer knobs were located by socket (Fig. 4C ▶), which, as described above, cemented the H2-H3 and H3-H6 coiled-coil interfaces. In contrast, none of the interprotomer, helix-helix interfaces were classed as coiled coils by socket. The program found only two knob residues, Glu29 of H2 and Gln-304 of H7 (see kinemage), which formed complementary, but isolated knobs-into-holes interactions in the interface. Incidentally, relaxing the packing cutoff to 7.5Å revealed new, albeit poorer, knobs-into-holes interaction between H6 and H7. However, no more interactions came to light at the protomer-protomer interfaces.
We predicted a barrel with 14 helices and not 12 as observed. There are several possible reasons for this difference. First, our prediction was for parallel helices, whereas adjacent helices in TolC are antiparallel. However, as argued above, this should only alter the details of the helix-helix packing. Second, the ≈154° angle assumes helix supercoiling. However, helices cannot supercoil in two directions simultaneously as would be required for both the a/d and c/f seams to participate in canonical coiled-coil interfaces. Thus, to maintain packing at both interfaces needs either some helical distortion or some relaxation of the knobs-into-holes interactions. socket analysis of the PDB revealed structural precedents for the former, that is, tight knobs-into-holes packing may be maintained in distorted helices. For instance, the central helices of the three-helix α-sheets are straightened (Fig. 3B ▶). In TolC, both mechanisms occur: As described above, the knobs-into-holes interactions are not as extensive in the upper barrel as compared with the lower barrel. In addition, the helices of the upper barrel slant rather than supercoil (Koronakis et al. 2000). Slanting, straightening, and other distortions of the helices of the cylinder will alter θ, ϕ, and ultimately n. For example, for straight nonsupercoiled helices θ = 160°, ϕ = 20°, and n = 18. Thus, variations in helix number in natural and designer α-cylinders should be expected.
In summary, the TolC barrel represents a variation on the α-cylinders that we propose. Important to note, however, is that this structure, together with the natural α-sheets, shows that more unusual coiled-coil assemblies are possible. We anticipate additional natural examples and even designer peptide-based sheets and nanotubes to follow.
Electronic supplemental material
Kinemages are available showing the α-sheets and α-cylinders and the knobs-into-holes interactions that occur within these structures.
Acknowledgments
We thank the Medical Research Council of the UK for funding this work. We thank Chris Davies, Richard Sessions, and Jenny Shipway for helpful discussions and comments on the manuscript.The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.36901.
Supplemental material: See www.proteinscience.org.
References
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, M.L., Down, J.A., Gurnett, A.M., Carrington, M., Turner, M.J., and Wiley, D.C. 1993. A structural motif in the variant surface glycoproteins of Trypanosoma Brucei. Nature 362 603–609. [DOI] [PubMed] [Google Scholar]
- Chan, D.C., Fass, D., Berger, J.M., and Kim, P.S. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89 263–273. [DOI] [PubMed] [Google Scholar]
- Crick, F.H.C. 1953. The packing of alpha-helices: Simple coiled-coils. Acta. Crystallogr. 6 689–697. [Google Scholar]
- Freymann, D., Down, J., Carrington, M., Roditi, I., Turner, M., and Wiley, D. 1990. 2.9 Å resolution structure of the N-terminal domain of a variant surface glycoprotein from Trypanosoma Brucei. J. Mol. Biol. 216 141–160. [DOI] [PubMed] [Google Scholar]
- Groves, M.R. and Barford, D. 1999. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9 383–389. [DOI] [PubMed] [Google Scholar]
- Harbury, P.B., Zhang, T., Kim, P.S., and Alber, T. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262 1401–1407. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kim, K.K., Yokota, H., and Kim, S.H. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400 787–792. [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., and Hughes, C. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405 914–919. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. molscript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Malashkevich, V.N., Kammerer, R.A., Efimov, V.P., Schulthess, T., and Engel, J. 1996. The crystal structure of a five-stranded coiled coil in COMP: A prototype ion channel? Science 274 761–765. [DOI] [PubMed] [Google Scholar]
- O'Shea, E.K., Rutkowski, R., and Kim, P.S. 1989. Evidence that the leucine zipper is a coiled coil. Science 243 538–542. [DOI] [PubMed] [Google Scholar]
- O'Shea, E.K., Klemm, J.D., Kim, P.S., and Alber, T. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254 539–544. [DOI] [PubMed] [Google Scholar]
- Sutton, R.B., Fasshauer, D., Jahn, R., and Brunger, A.T. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution [see comments]. Nature 395 347–353. [DOI] [PubMed] [Google Scholar]
- Wiener, M., Freymann, D., Ghosh, P., and Stroud, R.M. 1997. Crystal structure of colicin Ia. Nature 385 461–464. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Jensen, O.N., Holmes, S., Soues, S., Mann, M., and Kilmartin, J.V. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]