Abstract
We have designed a heterodimerizing leucine zipper system to target a radionuclide to prelocalized noninternalizing tumor-specific antibodies. The modular nature of the leucine zipper allows us to iteratively use design rules to achieve specific homodimer and heterodimer affinities. We present circular-dichroism thermal denaturation measurements on four pairs of heterodimerizing leucine zippers. These peptides are 47 amino acids long and contain four or five pairs of electrostatically attractive g ↔ e′ (i, i′ +5) interhelical heterodimeric interactions. The most stable heterodimer consists of an acidic leucine zipper and a basic leucine zipper that melt as homodimers in the micro (Tm = 28°C) or nanomolar (Tm = 40°C) range, respectively, but heterodimerize with a Tm >90°C, calculated to represent femtamolar affinities. Modifications to this pair of acidic and basic zippers, designed to destabilize homodimerization, resulted in peptides that are unstructured monomers at 4 μM and 6°C but that heterodimerize with a Tm = 74°C or Kd(37) = 1.1 × 10−11 M. A third heterodimerizing pair was designed to have a more neutral isoelectric focusing point (pI) and formed a heterodimer with Tm = 73°C. We can tailor this heterodimerizing system to achieve pharmacokinetics aimed at optimizing targeted killing of cancer cells.
Keywords: Leucine zipper, dimerization, electrostatic interactions, radioimmunotherapy, salt bridges, heterodimer
Leucine zipper–containing proteins have been adapted for a variety of purposes, but in the natural setting they are only known to function within cells. Heterodimerizing leucine zippers have been used previously to bring chimeric proteins together in solution (Scott et al. 1996), within cells (Katz et al. 1998), and extracellularly (Behncken et al. 2000). We intend to use designed heterodimerizing leucine zippers to deliver radionuclides to the surface of cancer cells. This allows a pretargeting approach in which a tumor-specific antibody appended to one leucine zipper monomer is first bound to the cancer cells. After removal of nonspecifically bound and circulating antibody, the complementary zipper containing a radionuclide can be introduced.
Presently, investigators are using the streptavidin/biotin system to introduce a radionuclide to a prelocalized cancer antibody (Axworthy et al. 2000; Breitz et al. 2000; Weiden et al. 2000). In this three-step procedure, streptavidin-conjugated antibody first is injected and allowed to accumulate on the tumor surface. After peak tumor uptake has occurred, the unlocalized antibody is removed from the circulation with a clearing agent. Finally, a small molecular weight biotin conjugate, radiolabeled to high specific activity, is injected. The radiolabeled biotin either binds to streptavidin on the antibody or is rapidly excreted in the urine, thereby minimizing its toxic effects. Although this method has shown promise in clinical trials, it has several disadvantages. First, biotin occurs naturally in human plasma thus blocking streptavidin sites on the conjugated antibody. Second, both avidin and streptavidin are immunogenic.
We present a heterodimerizing leucine zipper system that overcomes both limitations with the streptavidin/biotin system and offers potential advantages. First, heterodimeric leucine zippers derived from human B-ZIP proteins are unlikely to be immunogenic. Second, there are no known leucine zippers circulating in the blood to complicate the function of a heterodimerizing leucine zipper system. Third, as the dimerization affinity of both homo- and heterodimers can be experimentally manipulated, we have the ability to select heterodimerizing leucine zipper pairs with the optimal pharmacokinetics.
The structural rules used in our design (Alber 1992; Baxevanis and Vinson 1993; Hodges 1996; Lupas 1996) are derived from studies of troponin C (Zhou et al. 1994), the yeast GCN4 leucine zipper (Harbury et al. 1993), and vertebrate leucine zippers (O'Shea et al. 1992; Krylov et al. 1998;). These heterodimerizing zippers are variants of VBP, a B-ZIP protein extensively studied in this laboratory. The VBP homodimer contains four pairs of attractive g ↔ e′ interhelical electrostatic interactions. Previously, we mutated the last two heptads of VBP to produce either an acidic zipper, EE34 (Tm = 22°C, Kd(37) = 8.1 × 10−4 M) or a basic zipper, RR34 (Tm = 27°C, Kd(37) = 3.1 × 10−5 M). These leucine zippers have two pairs of attractive and two pairs of repulsive g ↔ e′ salt bridges in the homodimer, but the heterodimer has four pairs of attractive g ↔ e′ salt bridges and similar stability to the natural VBP homodimer (Kd = ∼10−8 M). Mutating all four salt bridges to produce EE1234 and RR1234 maintains four pairs of salt bridges in the heterodimer, but homodimers have four pairs of repulsive interactions. Consistent with this, homodimers are unstructured monomers at 6°C but heterodimers have a Tm of 51°C, identical to the original VBP homodimer (O'Shea et al. 1992; Krylov et al. 1998).
We used EE1234 and RR1234 as a starting point for the leucine zipper protein designs presented here. By altering residues in the leucine zipper a, d, g, and e positions, we have designed heterodimerizing pairs with varying pIs and with stabilities in the femta- to nanomolar range. Designing heterodimers with a range of thermodynamic properties will allow us to select pairs with the appropriate pharmacokinetics for the delivery of radionuclides to antigen-specific antibodies prelocalized to cancer cells.
Results
Stabilizing the heterodimer (Tm > 90°C): EE1234L and RR1234L
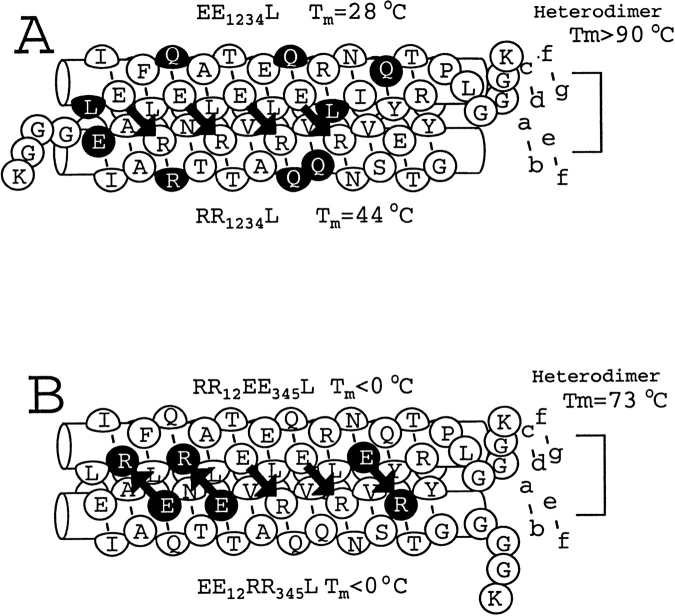
To increase heterodimer stability of EE1234/RR1234 from Tm= 51.5°C, ΔG = −11.6 kcal/mole (Kd(37) = 7 × 10−9 M), we altered the hydrophobic interface by mutating the first and fifth d positions to leucine, replacing an isoleucine and cysteine, respectively. Previous work indicates that a cysteine to leucine change in the fifth d position of the VBP leucine zipper contributes 7.2 kcal/mole/dimer to stability. An isoleucine to leucine change contributes 6.0 kcal/mole/dimer to stability (Moitra et al. 1997). Several amino acids in the b, c, and f positions also were altered to eliminate charged residues that may interfere with attractive g ↔ e′ salt bridges (see Fig. 1A ▶ for schematic). All lysine residues, except those at the end of the N-terminal or C-terminal extensions, were replaced to allow specific attachment of the radionuclide (or antibody) at the termini via lysine chemistry. We denote these new peptides EE1234L and RR1234L with the L indicating the stabilized hydrophobic interface.
Fig. 1.
Schematic of two heterodimerizing leucine zipper pairs. The coiled coil region is presented, without the supercoiling, with the relative placement of the seven amino acids in the coiled coil heptad presented by circles. To the right is the identification of the a, b, c, d, e, f, and g positions of the coiled coil. (Solid arrows) The potential electrostatic interactions between the g position of one helix and the following e′ position of the opposite helix (we refer to this interaction as the g ↔ e′ or the i, i′ +5 interaction). (Black circles with white lettering) Changes from the previous construct in the series. (A) Heterodimer of EE1234L and RR1234L. These constructs are altered from EE1234 and RR1234. (B) Heterodimer of RR12EE345 and EE12RR345. These are derived from EE1234L and RR1234L.
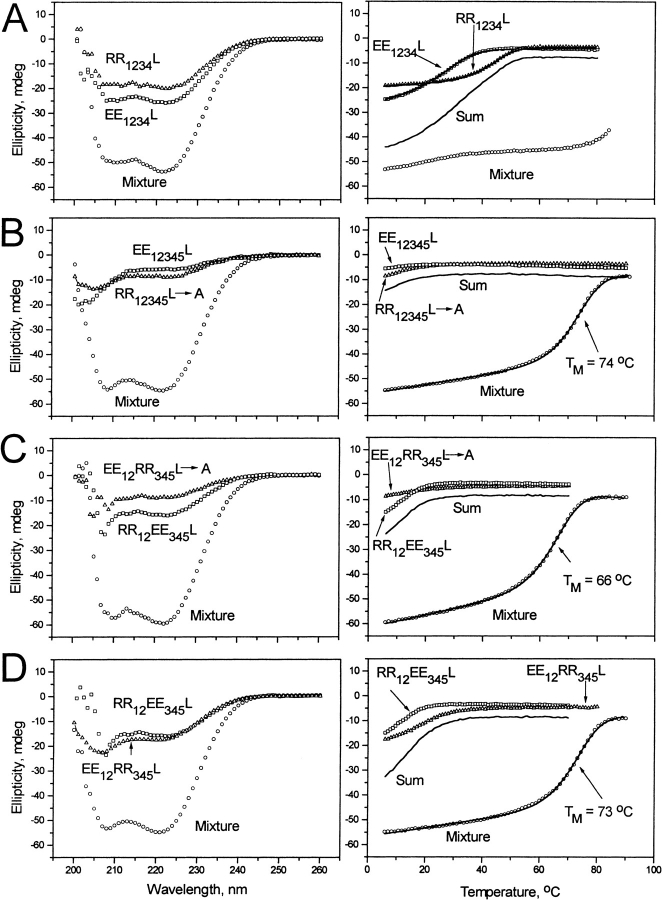
The EE1234L and RR1234L peptides have increased stability as either homodimers or heterodimers (Fig. 2A ▶; Table 1). Homodimer stability was increased from unstructured at 6°C to Tm = 28°C for EE1234L and Tm = 40°C for RR1234L. We have shown that a R ↔ R salt bridge is 0.47 kcal/mole more stabilizing than a E ↔ E salt bridge (Krylov et al. 1998). Thus, we calculate that RR1234L should be 3.76 kcal/mole more stable than EE1234L (Krylov et al. 1994), in good agreement with the 3.2 kcal/mole observed difference in stability.
Fig. 2.
CD spectral data (left) and thermal denaturation curves (right) for heterodimerizing leucine zippers. (Circles) Heterodimer; (squares) acidic monomer/homodimer; (triangles) basic peptide. CD denaturation curves are recorded at 222 nm. (Solid line) Expected thermal denaturation if the two proteins do not interact. The line through each denaturation curve is a fitted curve for a two-state transition as described in Materials and Methods. (A) EE1234L/RR1234L; (B) EE12345L/RR12345L → A; (C) EE12RR345L → A/RR12EE345L; (D) EE12RR345L/RR12EE345L.
Table 1.
Thermodynamic parameters of heterodimerizing zippers
| Protein (homodimer) Θ, Tm(ΔG, ΔH)Kd(37) | Protein (homodimer) Θ, Tm(ΔG, ΔH)Kd(37) | Protein (heterodimer) Θ, Tm(ΔG, ΔH)Kd(37) | Heterodimer MW (kD) calculated (experimental) |
| EE1234L (pI = 3.7) −25, 28 (−6.2, −45) 4.4 × 10−5 | RR1234L (pI = 12.4) −19, 44(−9.4, −66) 2.5 × 10−7 | EE1234L/RR1234L −53, >90, N.D., N.D., ∼10−15a | 10,870 (11.8 ± 0.8) |
| EE12345L (pI = 3.6) Tm < 0 | RR12345L → A (pI = 12.7) Tm < 0 | EE12345L/RR12345L → A −55, 74 (−15.5, −75) 1.1 × 10−11 | 10,844 (11.6 ± 1.0) |
| RR12EE345L (pI = 4.7) Tm < 0 | EE12RR345L → A (pI = 11.0) Tm < 0 | RR12EE345L/EE12RR345L → A −59, 66 (−14.6, −81) 4.7 × 10−11 | N.D. |
| RR12EE345L (pI = 4.7) Tm < 0 | EE12RR345L (pI = 10.9) Tm < 0 | RR12EE345L/EE12RR345L −57, 73 (−15.4, −74) 1.3 × 10−11 | 10,886 (11.1 ± 1.2) |
The table presents ellipticity at 6°C (Θ), the melting temperature (Tm, °C), ΔG (in kcal/mole), ΔH (kcal/mole), and Kd (molar) at 37°C in circular dichroism thermal melts for homodimer and heterodimer samples, and molecular weights of mixtures at 6°C as determined by analytical ultracentrifugation (Krylov et al. 1994).
a Calculated from previously derived thermodynamic data (Krylov et al. 1998) as described in the text.(MW) molecular weight; (N.D.) not determined.
The EE1234L/RR1234L heterodimer denaturation is incomplete, melting above 90°C, which prevents determination of its thermodynamic parameters from these data. The EE1234L/RR1234L heterodimer has four E ↔ R and four R ↔ E salt bridges compared with eight R ↔ R salt bridges found in the RR1234L homodimer. We have shown that E ↔ R is 1.2 kcal/mole more stable than R ↔ R and R ↔ E is 1.5 kcal/mole more stable than R ↔ R (Krylov et al. 1998). Thus, we calculate that the heterodimer should be 10.8 kcal/mole more stable than RR1234L, giving an expected Kd(37) of 5.1 × 10−15 M. A similar calculation relative to the EE1234L homodimer gives a Kd(37) of 1.4 × 10−15 M, numbers consistent with the heterodimer denaturation curve. These values are similar to the dissociation constant for steptavidin/biotin that has Kd(37) = 1 × 10−15 M (Axworthy et al. 2000).
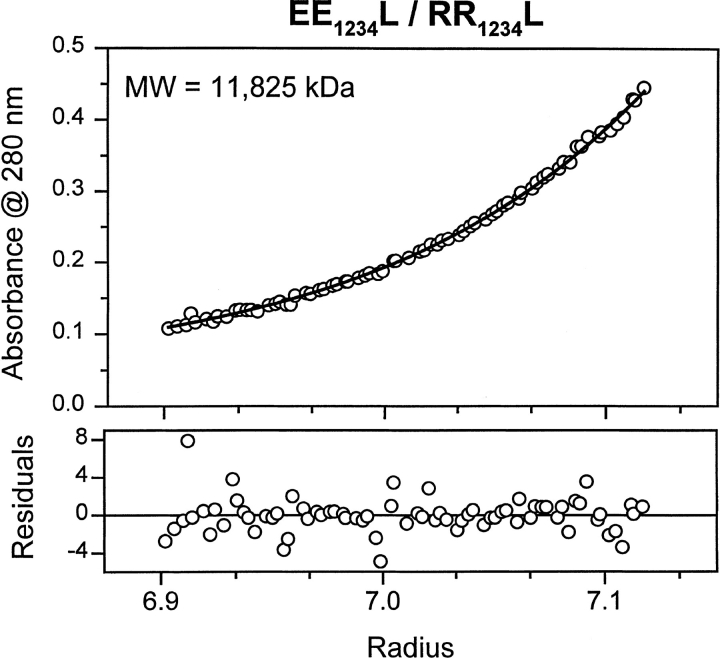
Sedimentation equilibrium experiments confirm that the EE1234L and RR1234L mixture is dimeric (Fig. 3 ▶; Table 1), making this the most stable dimeric leucine zipper described to date.
Fig. 3.
Molecular weight (MW) determination by sedimentation equilibrium of EE1234L/RR1234L at 6°C. The sample was in 12.5 mM potassium phosphate at pH 7.4, 150 mM KCl, 0.25 mM EDTA. (Solid line) Theoretical curve for dimer molecular weight; (circles) plotted actual data. (Bottom panel) The residuals for fitting the experimental data to a dimer model. No systematic error was observed.
Destabilizing the homodimers: EE12345L and RR12345L ↔ A
Although we had achieved exceptional heterodimer stability, we were concerned that homodimerization in vivo would interfere with heterodimer formation. To decrease homodimer stability, we introduced an additional repulsive electrostatic g ↔ e′ pair in both zippers. In EE1234L, we changed the fifth g ↔ e′ pair from I ↔ E to E ↔ E to form EE12345L. Similarly, in RR1234L, we changed I ↔ E to R ↔ R to form RR12345L.
To further decrease the stability of RR1234L, the more stable homodimer, we mutated the fourth d position from leucine to alanine to create RR12345L → A. Previous work indicates that this should decrease stability of the homodimer by 9.2 kcal/mole (Moitra et al. 1997). Both homodimers show the beginning of a transition at 6°C in the 4-μm sample indicating a Tm below 6°C (Fig. 2B ▶). The EE12345L/RR12345L → A mixture is a dimer with a Tm of 74°C, corresponding to ΔG = −15.5 kcal/mole and a dissociation constant of Kd(37) = 1.1 × 10−11 M (Table 1). The low stability of the homodimers prevents us from determining heterodimer specificity. However, in previous work we determined that the heterodimer between EE34 and RR34 is 6.8 kcal/mole more stable than EE34 homodimer. This corresponds to a ratio of 5.7 × 104 for Kd(37) homodimer/Kd(37) heterodimer, or a 240-fold difference in Kd/heptad. For the RR34 homodimer, heterodimerization with EE34 is favored 50-fold/heptad (Krylov et al. 1994). Assuming five pairs of independent attractive interhelical salt bridges, we calculate that EE12345L prefers to heterodimerize with RR12345L → A by 505- or 3 × 108-fold. Likewise, RR12345L → A prefers to heterodimerize with EE12345L by 2405- or 8 × 1011-fold.
Attaining a more neutral pI: EE12RR345L → A and RR12EE345L
A potential clinical problem is the extreme acidity of EE12345L and the extreme basicity of RR12345L → A. Discounting the lysine at the termini that will be chemically modified in the chimeric proteins, EE12345L has 12 glutamates and 2 arginines with a calculated pI of 3.6. RR12345L → A has 13 arginines and 2 glutamates (pI = 12.7). These highly charged peptides may participate in nonspecific interactions in vivo. To make a peptide with more neutral overall charge, we changed the four amino acids in the first two g and e positions from glutamate to arginine to create RR12EE345L. RR12EE345L now has eight glutamates and six arginines with a pI = 4.7. Similarly, four amino acids were changed from arginine to glutamate in the first two salt bridges of RR12345L → A to create EE12RR345L → A. This protein has six glutamates and nine arginines (pI = 11.0).
Figure 2C ▶ and Table 1 present thermal denaturation data for these peptides monitored at 222 nm by circular-dichroism (CD) spectroscopy. The homodimers were unstructured at 6°C whereas the heterodimer between EE12RR345L → A and RR12EE345L has a Tm of 66°C with a ΔG of −14.6 kcal/mole (Kd(37) = 4.7 × 10−11 M), which is 0.9 kcal/mole less stable than EE12345L/RR12345L → A. This decrease in stability is presumably because of repulsive interactions between the second and third heptads. There will be one E ↔ E and one R ↔ R e ↔ g′ or i, i′ +2 repulsive interaction in the EE12RR345L → A and RR12EE345L heterodimer that may account for the 0.9-kcal/mole decrease in stability. A similar decrease in stability has been observed in the C/EBP leucine zipper when a salt bridge is reversed (Moll et al. 2000). The sharpness of the melting transition reflected in the high enthalpies (ΔH) suggests that the EE12RR345L → A/RR12EE345L heterodimer is a homogeneous sample, able to populate only a limited number of protein conformations. There was some concern that the monomer would fold onto itself producing intramolecular electrostatic interactions that may be observed in a less cooperative thermal denaturation, but this does not appear to be the case.
To restabilize the heterodimer, we changed EE12RR345L → A to EE12RR345L by changing the fourth d position from alanine to leucine. We also moved the lysine linker from the N terminus to the C terminus to facilitate the chemical linking of this peptide to the antibody (Fig. 1B ▶). The EE12RR345L and RR12EE345L mixture is a dimer with a Tm of 73°C, an increase of 7°C (Fig. 2D ▶; Table 1). Because of a decrease in the measured ΔH, we calculate an increase in stability of only 0.8 kcal/mole. From previous studies, we would expect an L ↔ L d position interaction to be >4.6 kcal/mole more stable than an L ↔ A interaction. This discrepancy may reflect moving the lysine linker from the N terminus to the C terminus.
Discussion
We designed a series of heterodimerizing leucine zipper peptides that have a greatly expanded range of homo- and heterodimer stabilities and pIs. This system could be widely applicable in bringing together a diverse array of molecules. Our aim is to use these heterodimerizing leucine zippers as an alternative to the streptavidin/biotin system that has been used in human cancer patients to deliver a radionuclide to prelocalized tumor antibodies (Axworthy et al. 2000; Breitz et al. 2000; Weiden et al. 2000). These heterodimerizing leucine zippers may overcome some of the experimental problems associated with the avidin/biotin system.
Previous work has shown that heterodimerizing leucine zippers can be designed by having one amphipathic α helix with glutamates in the e and g positions and a second amphipathic α helix with either lysines or arginines in the e and g positions. The homodimer is destabilized by repulsive interhelical g ↔ e′ interactions whereas the heterodimer is stabilized by attractive interhelical g ↔ e′ interactions. The stability of leucine zippers also can be regulated by changes in the a and d positions of the hydrophobic core (Moitra et al. 1997; Wagschal et al. 1999; Tripet et al. 2000). These two simple design rules have been used to successfully build four pairs of heterodimerizing peptides with the desired properties. The experimental versatility of this system will allow us to screen a series of heterodimerizing leucine zippers with a range of homo- and heterodimer stabilities for a pair that has the best pharmacokinetics.
Two experimental concerns were addressed in these designs. We wanted to attach the radionuclide to the acidic peptide because acidic peptides are rapidly excreted by the kidney, thus limiting toxicity resulting from circulating radionuclide. We also wanted the zipper bound to the antibody to be unstable. We expect that the zipper–antibody chimera will be constrained by diffusion on the cell surface after binding of the antibody, potentially leading to homodimerization at micromolar Kd and preventing heterodimerization.
In the first phase, EE1234L and RR1234L were designed to increase heterodimer stability as compared with EE1234 and RR1234 by placing leucine residues in the hydrophobic interface. The heterodimer was extremely stable with a calculated dissociation constant of ∼1 × 10−15, which is similar to that reported for the streptavidin/biotin system. The homodimers, however, had stabilities in the micro- to nanomolar range, which could kinetically preclude heterodimer formation in vivo.
In the second phase, we increased the number of attractive salt bridges in the heterodimer and repulsive interactions in the homodimers by adding an additional heptad of salt bridge interactions to produce RR12345L and EE12345L. We thereby maximized ΔΔG between hetero- and homodimers and increased the range of affinities attainable. Introduction of a destabilizing alanine residue into the fourth d position to create RR12345L → A made this homodimer unstable even at 6°C, and only moderately destabilized the heterodimer.
The first two design phases comprise an acid-base delivery system that may be useful in facilitating excretion of unincorporated radionuclide. We were concerned, however, that proteins with such extremely basic and acidic pIs may bind nonspecifically to host molecules. In the third design, we addressed this potential problem by altering the order of the g ↔ e′ salt bridges to make peptides with more neutral overall charge. The 0.9-kcal/mole decrease in stability in the heterodimer of the more neutral peptide EE12RR345L → A/RR12EE345L (compared with EE12345L/RR12345L → A) is likely due to repulsive i, i′ +2 interactions between the second and third heptads in the heterodimer (Kohn et al. 1995; Moll et al. 2000). To once again increase heterodimer stability, we reintroduced leucine in the fourth d position to produce EE12RR345 and RR12EE345. This change increased the stability 7°C.
We have a range of possibilities to further increase heterodimer stability, if necessary. The a positions are A1N2V3V4V5. The first a position alanine could be changed to valine or leucine to increase stability without affecting dimerization. The asparagine in the second a position is critical for two reasons. It limits oligomerization to dimers (Harbury et al. 1993), and it regulates the parallel orientation of the monomer, preventing the peptides with mixed salt bridges from homodimerizing in an antiparallel manner (Oakley and Kim 1998). We may, however, be able to replace the a position valines with leucine to further increase both homo- and heterodimer stability. We also can elevate heterodimer stability by increasing the length of the leucine zippers to incorporate more hydrophobic interface and attractive g ↔ e′ salt bridges.
Radioimmunotherapy is a promising method to fight cancer. The advantage of the heterodimerizing leucine zippers is that their peptide sequences can be manipulated to produce homo- and heterodimers of any desired stability to optimize the pharmacokinetic properties of the antibody–leucine zipper–radionuclide chimeras.
Materials and methods
Proteins
The name and then the protein sequence of the peptides, grouped into g,a,b,c,d,e,f heptads are as follows:
The RR1234L and EE1234L peptides were made using the Impact T7 system from New England Biolabs (Chong et al. 1997). The genes encoding these peptides were inserted into the multicloning site (NdeI/SapI) of the pTYB1 vector to create an in-frame fusion between their C terminus and the N terminus of the gene encoding intein, a protein splicing element from Saccharomyces cerevesiae. At the C terminus of the intein gene is DNA encoding a chitin-binding domain. After T7 overexpression, the crude extract is bound to a chitin column. The chimera is induced to undergo an intein-mediated self-cleavage by overnight incubation at 4°C in the presence of 30 mM DTT, causing the specific elution of the leucine zipper. The remaining peptides were generated by peptide synthesis from Genosys. The molar concentrations were calculated using the molar extinction at 230 nm of the peptide bond (300/bond) and the contribution of tyrosine (4980/residue) at 230 nm (Cantor and Schimmel 1980).
Circular dichroism
CD studies were performed using a Jasco J-720 spectropolarimeter with a 5-mm rectangular CD cuvette and heated as described previously (Krylov et al. 1994). All protein stock solutions were in 12.5 mM potassium phosphate at pH 7.4, 150 mM KCl, 0.25 mM EDTA.
Thermodynamic calculations
Melting temperature (Tm) and enthalpy (ΔH) values were determined from denaturation curves assuming a two-state equilibrium dissociation of α-helical dimers into unfolded monomers using ΔCp of −1.2 kcal/mole−1 (Krylov et al. 1994). ΔG values are reported at 37°C.
Equilibrium sedimentation
Equilibrium sedimentation measurements were performed using a Beckman XL-A Optima Analytical Ultracentrifuge equipped with absorbance optics and a Beckman An-60Ti rotor. Samples were loaded at three concentrations, 20, 40, and 60 μM into a six-hole centerpiece and spun at 28,000 rpm for 28 h. Nine data sets for three concentrations were jointly fit for a singular molecular weight. Compositional partial specific volumes for the proteins were calculated according to Zamyatnin (1984). All scans were performed at 6°C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Table t.
| def | gabcdef | gabcdef | gabcdef | gabcdef | gabcdef | gabcd | |
| EE1234L: | LEI | EAAFLEQ | ENTALET | EVAELEQ | EVQRLEN | IVSQYET | RYGPLGGGK |
| RR1234L: | KGGGLEI | RAAFLRR | RNTALRT | RVAELRQ | RVQRLRN | IVSQYET | RYGPL |
| EE12345L: | LEI | EAAFLEQ | ENTALET | EVAELEQ | EVQRLEN | EVSQYET | RYGPLGGGK |
| RR12345L→A: | KGGGLEI | RAAFLRR | RNTALRT | RVAELRQ | RVQRARN | RVSQYRT | RYGPL |
| RR12EE345L: | LEI | RAAFLRQ | RNTALRT | EVAELEQ | EVQRLEN | EVSQYET | RYGPLGGGK |
| EE12RR345L→A: | KGGGLEI | EAAFLER | ENTALET | RVAELRQ | RVQRARN | RVSQYRT | RYGPL |
| EE12RR345L: | LEI | EAAFLER | ENTALET | RVAELRQ | RVQRLRN | RVSQYRT | RYGPLGGGK |
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.39401.
References
- Alber, T. 1992. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2 205–210. [DOI] [PubMed] [Google Scholar]
- Axworthy, D.B., Reno, J.M., Hylarides, M.D., Mallett, R.W., Theodore, L.J., Gustavson, L.M., Su, F., Hobson, L.J., Beaumier, P.L., and Fritzberg, A.R. 2000. Cure of human carcinoma xenografts by a single dose of pretargeted yttrium-90 with negligible toxicity. Proc. Natl. Acad. Sci. 97 1802–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis, A. and Vinson, C. 1993. Interactions of coiled coils in transcription factors: Where is the specificity? Curr. Opin. Genet. Dev. 3 278–285. [DOI] [PubMed] [Google Scholar]
- Behncken, S.N., Billestrup, N., Brown, R., Amstrup, J., Conway-Campbell, B., and Waters, M.J. 2000. Growth hormone (GH)-independent dimerization of GH receptor by a leucine zipper results in constitutive activation. J. Biol. Chem. 275 17000–17007. [DOI] [PubMed] [Google Scholar]
- Breitz, H.B., Weiden, P.L., Beaumier, P.L., Axworthy, D.B., Seiler, C., Su, F.M., Graves, S., Bryan, K., and Reno, J.M. 2000. Clinical optimization of pretargeted radioimmunotherapy with antibody-streptavidin conjugate and 90Y-DOTA-biotin. J. Nucl. Med. 41 131–140. [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry. W.H. Freeman, San Francisco.
- Chong, S., Mersha, F.B., Comb, D.G., Scott, M.E., Landry, D., Vence, L.M., Perler, F.B., Benner, J., Kucera, R.B., Hirvonen, C.A., et al. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192 271–281. [DOI] [PubMed] [Google Scholar]
- Harbury, P.B., Zhang, T., Kim, P.S., and Alber, T. 1993. A switch between two-, three-, and four-stranded coiled cils in GCN4 leucine zipper mutants. Science 262 1401–1407. [DOI] [PubMed] [Google Scholar]
- Hodges, R.S. 1996. Boehringer Mannheim award lecture 1995. La conference Boehringer Mannheim 1995. De novo design of α-helical proteins: Basic research to medical applications. Biochem. Cell Biol. 74 133–154. [DOI] [PubMed] [Google Scholar]
- Katz, B., Krylov, D., Aota, S., Vinson, C., and Yamada, K. 1998. Green fluorescent protein labeling of cytoskeletal structures—A novel targeting approach based on leucine zippers. Biotechniques 25 298–303. [DOI] [PubMed] [Google Scholar]
- Kohn, W.D., Kay, C.M., and Hodges, R.S. 1995. Protein destabilization by electrostatic repulsions in the two-stranded α-helical coiled-coil/leucine zipper. Protein Sci. 4 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov, D., Mikhailenko, I., and Vinson, C. 1994. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 13 1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov, D., Barchi, J., and Vinson, C. 1998. Inter-helical interactions in the leucine zipper coiled coil dimer: pH and salt dependence of coupling energy between charged amino acids. J. Mol. Biol. 279 959–972. [DOI] [PubMed] [Google Scholar]
- Lupas, A. 1996. Coiled coils: New structures and new functions. Trends Biochem. 21 375–382. [PubMed] [Google Scholar]
- Moitra, J., Szilák, L., Krylov, D., and Vinson, C. 1997. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry 36 12567–12573. [DOI] [PubMed] [Google Scholar]
- Moll, J.R., Olive, M., and Vinson, C. 2000. Attractive interhelical electrostatic interactions in the proline- and acidic-rich region (PAR) leucine zipper subfamily preclude heterodimerization with other basic leucine zipper subfamilies. J. Biol. Chem. 275 34826–34832. [DOI] [PubMed] [Google Scholar]
- Oakley, M.G. and Kim, P.S. 1998. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry 37 12603–12610. [DOI] [PubMed] [Google Scholar]
- O'Shea, E.K., Rutkowski, R., and Kim, P.S. 1992. Mechanism of specificity in the fos-jun oncoprotein heterodimer. Cell 68 699–708. [DOI] [PubMed] [Google Scholar]
- Scott, C.A., Garcia, K.C., Carbone, F.R., Wilson, I.A., and Teyton, L. 1996. Role of chain pairing for the production of functional soluble IA major histocompatibility complex class II molecules. J. Exp. Med. 183 2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet, B., Wagschal, K., Lavigne, P., Mant, C.T., and Hodges, R.S. 2000. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position "d." J. Mol. Biol. 300 377–402. [DOI] [PubMed] [Google Scholar]
- Wagschal, K., Tripet, B., Lavigne, P., Mant, C., and Hodges, R.S. 1999. The role of position a in determining the stability and oligomerization state of α-helical coiled coils: 20 amino acid stability coefficients in the hydrophobic core of proteins. Protein Sci. 8 2312–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiden, P.L., Breitz, H.B., Press, O., Appelbaum, J.W., Bryan, J.K., Gaffigan, S., Stone, D., Axworthy, D., Fisher, D., and Reno, J. 2000. Pretargeted radioimmunotherapy (PRIT) for treatment of non-Hodgkin's lymphoma (NHL): Initial phase I/II study results. Cancer Biother. Radiopharm. 15 15–29. [DOI] [PubMed] [Google Scholar]
- Zamyatnin, A. 1984. Amino acid, peptide, and protein volume in solution. Annu. Rev. Biophys. Bioeng. 13 145–165. [DOI] [PubMed] [Google Scholar]
- Zhou, N., Kay, C., and Hodges, R. 1994. The net energetic contribution of interhelical electrostatic attractions to coiled-coil stability. Protein Eng. 7 1365–1372. [DOI] [PubMed] [Google Scholar]