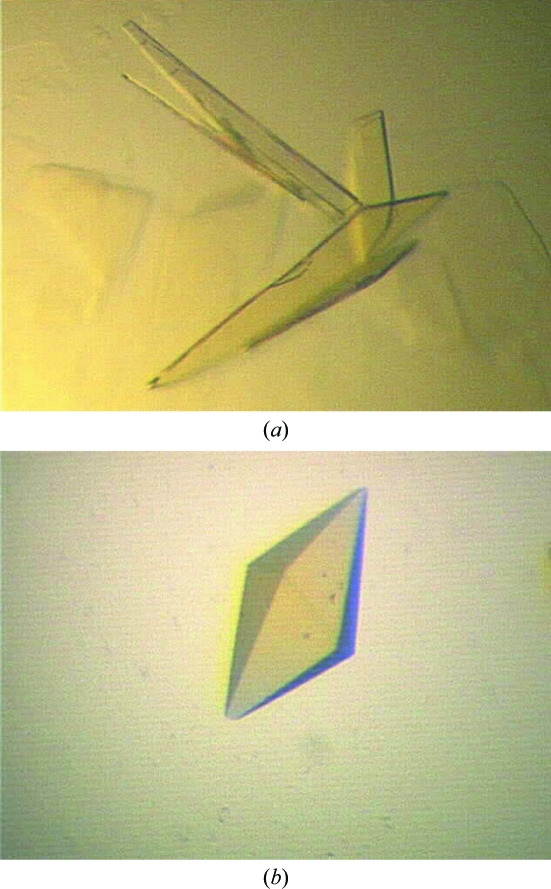
Crystals of A. niger monoamine oxidase variants display P21 or P41212/P43212 symmetry, with eight or two molecules in the asymmetric unit, respectively.
Keywords: monoamine oxidase, Aspergillus niger, directed evolution, enantioselectivity
Abstract
Monoamine oxidase from Aspergillus niger (MAO-N) is an FAD-dependent enzyme that catalyses the conversion of terminal amines to their corresponding aldehydes. Variants of MAO-N produced by directed evolution have been shown to possess altered substrate specificity. Crystals of two of these variants (MAO-N-3 and MAO-N-5) have been obtained; the former displays P21 symmetry with eight molecules per asymmetric unit and the latter has P41212 or P43212 symmetry and two molecules per asymmetric unit. Solution of these structures will help shed light on the molecular determinants of improved activity and high enantioselectivity towards a broad range of substrates.
1. Introduction
Monoamine oxidase (MAO) catalyses the oxidative deamination of terminal amines and is found in a wide range of organisms from bacteria and fungi to humans. In vertebrates, the substrates of the enzyme include several important neurotransmitters such as serotonin and dopamine. MAO was first isolated from Aspergillus niger in 1995 and was found to be present in two forms (Schilling & Lerch, 1995 ▶). Type 1 was a previously identified copper-dependent enzyme and type 2 was a novel flavin-dependent enzyme named MAO-N. The type 2 enzyme is an homogenous tetramer in solution, with subunit molecular weight 55 617 Da, each monomer being 495 amino acids in length.
There has been considerable interest in MAO-N, firstly because it has some sequence homology to the two forms of MAO (MAO-A and MAO-B) in humans and because it displays a similar response towards the highly selective inhibitors of MAO-A and MAO-B. The X-ray structures of both human MAO-A and MAO-B have been solved and a range of ligand-bound structures have been published (Binda et al., 2002 ▶, 2003 ▶; De Colibus et al., 2005 ▶). These structures led to suggestions that MAO-N could be an evolutionary precursor of vertebrate MAOs (Schilling & Lerch, 1995 ▶). The second area of interest is focused on exploiting the catalytic properties of the enzyme for application in biotransformations. During catalysis, free imine intermediates are generated that may be intercepted by chemical reductants, which together with the enzyme form an oxidation–reduction cycle that can be used for the quantitative deracemization of chiral amines. The wild-type enzyme, which was shown to be weakly active towards benzylamine and simple aliphatic amines (Sablin et al., 1998 ▶), was subjected to rounds of in vitro evolution experiments, resulting in a catalyst that was selective for oxidation of the (S)-enantiomer of α-methylbenzylamine when challenged with the racemic substrate (Alexeeva et al., 2002 ▶).
MAO-N variant N336S/M348K/I246M (MAO-N-3) exhibited improved activity towards a range of amine substrates compared with the wild-type enzyme, including chiral secondary amines (Carr et al., 2005 ▶). A second variant was later identified with the same three mutations as MAO-N-3, but with two additional mutations T384N/D385S. This new variant (MAO-N-5) showed improved activity and enantioselectivity towards a broad range of tertiary amines (Dunsmore et al., 2006 ▶).
Whilst random mutagenesis has been shown to improve enzyme properties such as enantioselectivity up to a point, the accumulated evidence suggests that a combination of structure-based selection of amino-acid targets coupled with randomization at these positions will offer the best opportunities for enzyme improvement (Morley & Kazlauskas, 2005 ▶). The solution of the X-ray structures of MAO-N and its variants will clarify why random mutations have had the observed effects on the catalytic properties of the enzyme and will also aid in the optimization of the redesign of MAO for improved performance.
2. Experimental
2.1. Cloning
The genes encoding MAO-N (EC 1.4.3.4) and its variants were subcloned from previously used pET-16b constructs (Carr et al., 2005 ▶; Dunsmore et al., 2006 ▶) into the vector pETYSBLIC-3C, which endows the expressed gene products with a 3C protease-cleavable N-terminal hexahistidine tag with sequence MGSSHHHHHHSSGLEVLFQGPA. Genes were amplified from pET-16b constructs using the following PCR primers: forward, 5′-CCAGGGACCAGCAATGACCTCCCGAGACGGATACCAG-3′, and reverse, 5′-GAGGAGAAGGCGCGTTETCACAAACGAGCCTTCACCTCC-3′. Cloning was then performed using a ligation-independent cloning (LIC) method (Bonsor et al., 2006 ▶).
2.2. Overexpression and purification
The purified plasmid was transformed into Escherichia coli Rosetta 2 (DE3) and cells were cultured in auto-induction medium type ZY (Studier, 2005 ▶) at 310 K for 18 h. Cells were harvested by centrifugation at 5000g for 20 min and the resulting pellet was resuspended in 50 mM HEPES, 100 mM KCl pH 7.0 with 1 mM AEBSF. Cells were disrupted by sonication and the debris was collected by centrifugation at 15 000g for 30 min. The cell-free extract was loaded onto a 5 ml Hi-trap Ni-affinity column (GE Healthcare) and eluted with a gradient from 0 to 500 mM imidazole. Fractions were analysed by SDS–PAGE and those containing a 55 kDa protein were pooled and then buffer-exchanged into 50 mM HEPES, 100 mM KCl pH 7.0; 3C protease was then added in a 100:1 protein:protease ratio. The His-tag cleavage reaction was incubated at 277 K for 36 h before analysing the cleaved protein by SDS–PAGE. The volume of the protein was reduced to approximately 2 ml using ultrafiltration centrifugation before loading onto a HiLoad 16/60 Superdex 200 gel-filtration column equilibrated with 50 mM HEPES, 100 mM KCl, 5 mM dithiothreitol pH 7.0. Fractions containing the eluted protein were pooled and concentrated to 10 mg ml−1 for crystallization.
2.3. Crystallization
MAO-N and variants were screened for crystallization using the sitting-drop vapour-diffusion method in 96-well MRC-Wilden plates with a variety of commercially available screens (Hampton Research and Molecular Dimensions). Drops consisting of 150 nl protein at 10 and 5 mg ml−1 mixed with 150 nl mother liquor were equilibrated against 54 µl well volume. Screens were set up using a TTP Mosquito nanopipetting robot and were incubated at 292 K. Conditions from the screens that produced crystals were optimized and scaled-up to hanging-drop format using 24-well plates and a protein concentration of 10 mg ml−1. The well volume was 1 ml and the drops were formed from 1 µl protein solution and 1 µl mother liquor; the plates were incubated at 292 K. Prior to data collection, crystals were transferred into a cryoprotectant solution containing the appropriate mother liquor with 20%(v/v) ethylene glycol in place of an equivalent volume of water. Crystals were soaked in the cryoprotectant for approximately 30 s before mounting in a cryoloop and flash-cooling in liquid nitrogen. The crystals were transferred to a storage dewar and transported to the European Synchrotron Radiation Facility (ESRF) for data collection.
2.4. Data collection, processing and analysis
Data were integrated and scaled using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶). Merged data were converted from SCALEPACK format to CCP4 format using SCALEPACK2MTZ (Collaborative Computational Project, Number 4, 1994 ▶). Unit-cell contents were analysed using the Matthews coefficient (Matthews, 1968 ▶) and the self-rotation function was calculated using MOLREP (Vagin & Teplyakov, 1997 ▶). Molecular-replacement techniques were applied to the data using MOLREP, AMoRe (Navaza, 1994 ▶) and Phaser (McCoy et al., 2007 ▶).
3. Results and discussion
The genes encoding MAO-N-3 and MAO-N-5 were cloned into pET-YSBLIC-3C and the proteins were expressed and purified. Initial screening of MAO-N-3 with 384 conditions produced small crystals or microcrystalline precipitate in 29 of these conditions. All hits were obtained with the protein at 10 mg ml−1, with screens at 5 mg ml−1 only producing clear drops. The conditions generating the most promising crystals of MAO-N-3, with the His tag cleaved with 3C protease, were optimized. A fine grid method was used, with 72 different conditions set up at a 1:1 protein:precipitant ratio. The largest crystals were obtained from 20%(w/v) PEG 3350, 0.2 M ammonium sulfate, 0.1 M HEPES pH 7.5 as clusters of thin plates from which it proved difficult to isolate a single crystal.
The MAO-N-5 variant with the His tag cleaved was screened with 192 different conditions, with 18 of these producing small crystals or microcrystalline precipitate. As for MAO-N-3, only a protein concentration of 10 mg ml−1 generated crystals. Optimization was carried out using fine grid screen of 48 conditions around the initial hits with a 1:1 protein:precipitant ratio. The largest tetragonal bipyramid-shaped crystals were obtained from 22%(w/v) PEG 3350, 0.2 M sodium sulfate, 0.1 M bis-tris propane pH 7.0. Even though MAO-N-3 and MAO-N-5 only differ at two residues and were crystallized under similar conditions, the crystal morphology is very different (Fig. 1 ▶). The MAO-N-5 crystals were found to be extremely fragile and crumbled easily when being mounted in cryoloops. In order to improve the stability of the crystals, the protein was purified without the removal of the His tag. After re-screening in 192 conditions, larger and more robust crystals were obtained from either 10%(w/v) PEG 3350, 0.2 M proline, 0.1 M HEPES pH 7.5 or 10%(w/v) PEG 5K MME, 5%(v/v) tacsimate, 0.1 M HEPES pH 7.0, with no difference in diffraction quality between the crystals from the two conditions.
Figure 1.
(a) Crystals of MAO-N-3, space group P21, with approximate dimensions 450 × 270 × 5 µm. (b) Crystal of MAO-N-5, space group P41212 or P43212, with approximate dimensions 120 × 120 × 210 µm.
Data were collected from crystals of MAO-N-3 and MAO-N-5 on beamlines ID14-EH4 and ID29, respectively, at the ESRF (Table 1 ▶). Data for MAO-N-3 were collected to 2.4 Å resolution; the crystals belonged to space group P21 with unit-cell parameters a = 103.4, b = 132.6, c = 187.2 Å, β = 90.1°. The Matthews coefficient (Matthews, 1968 ▶) indicated that there were likely to be eight molecules in the asymmetric unit, corresponding to a solvent content of 57.8%. As the enzyme forms tetramers in solution, it was probable that there were two tetramers in the asymmetric unit. This seems to be supported by the self-rotation function, which was calculated using MOLREP. Data for MAO-N-5 were collected to 1.85 Å resolution; the crystals belonged to space group P41212 or P43212 with unit-cell parameters a = 108.3, c = 235.7 Å. The Matthews coefficient for this variant indicated the presence of only two molecules per asymmetric unit with a solvent content of 60.2%, which again seemed to be confirmed by the self-rotation function. Peaks produced by noncrystallographic symmetry were displayed in the self-rotation functions for both MAO-N-3 and MAO-N-5.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution bin.
| MAO-N-3 | MAO-N-5 | |
|---|---|---|
| Beamline | ID14-EH4 | ID29 |
| Wavelength (Å) | 0.9765 | 0.9762 |
| Space group | P21 | P41212 or P43212 |
| Unit-cell parameters (Å, °) | a = 103.4, b = 132.6, c = 187.2, β = 90.1 | a = 108.3, c = 235.7 |
| Matthews coefficient VM (Å3 Da−1) | 2.91 | 3.09 |
| Solvent content (%) | 57.8 | 60.2 |
| No. of molecules per ASU | 8 | 2 |
| Resolution range (Å) | 50.0–2.45 (2.54–2.45) | 30.0–1.85 (1.92–1.85) |
| No. of unique reflections | 184776 | 66613 |
| No. of observed reflections | 179801 (16723) | 117844 (11446) |
| Completeness (%) | 97.3 (90.8) | 99.8 (98.5) |
| Redundancy | 2.1 (2.1) | 17.2 (8.1) |
| 〈I/σ(I)〉 | 15.0 (5.2) | 28.2 (1.8) |
| Rmerge† | 4.6 (15.8) | 9.4 (66.1) |
R
merge = 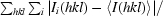
 .
.
Solution of the crystal structure of both variants by molecular replacement was attempted using the programs MOLREP, Phaser and AMoRe. The dimeric monoamine oxidase B from Homo sapiens (MAO-B; PDB code 1oja; Binda et al., 2003 ▶) has the highest homology of all available structures to MAO-N, with 42% similarity but only 24% identity. Molecular-replacement trials were carried out using this structure both at full length and with varying levels of truncation and as a monomer or dimer. Both polyalanine and polyserine models were used, with truncations carried out using CHAINSAW (Schwarzenbacher et al., 2004 ▶). In order to aid the satisfactory detection of rotation and translation functions, another search model was created based on MAO-B. The sequence was truncated to leave only the highly conserved regions, with each monomer subdivided into three domains, each as a separate chain. Self-rotation function peaks were also included in molecular-replacement searches with the model as both a monomer and a dimer, but no acceptable packing solutions were obtained. A variety of other related structures were tried such as human MAO-A (PDB code 2bxr, 23% identity; De Colibus et al., 2005 ▶), polyamine oxidase from Zea mays (PDB code 1b37, 20% identity; Binda et al., 1999 ▶) and MAO-A from Rattus norvegicus (PDB code 1o5w, 23% identity; Ma et al., 2004 ▶), with all the structural alterations and truncations applied to MAO-B. Models were also prepared both with and without inhibitor/substrate and FAD. The presence of these molecules has very little effect on the structures of these proteins and therefore did not affect the outcomes of molecular-replacement trials.
Despite the availability of the structures of other amine oxidases, the low level of sequence homology of MAO-N made molecular replacement challenging and no solution has yet been obtained. A significant factor affecting the likelihood of obtaining a solution by molecular replacement was the likely new and unique oligomeric states of the MAO-N variants. MAO-N exists as a tetramer in solution, with the X-ray data for MAO-N-3 indicating two tetramers in the asymmetric unit and the X-ray data for MAO-N-5 indicating a dimer or half a tetramer in the asymmetric unit. In contrast, all existing models of MAOs are dimers (or trimers in the polyamine oxidase case). The fact that MAO-N is a tetramer may result in differences in the monomer–monomer interface and packing, hence hampering molecular-replacement solutions. The low sequence homology between MAO-N and all available models could also be reflected in subtle but significant and systematic changes in the structure within each molecule, subsequently affecting the monomer surfaces of MAO-N variants involved in the tetramer/dimer packing. Attempts to obtain phases for the structure using selenomethionine and a range of heavy-atom derivatives are currently under way.
Acknowledgments
We would like to thank the Biotechnology and Biological Sciences Research Council (BBSRC) UK and GlaxoSmithKline for funding (KA and RR, respectively). We would also like to thank Johan Turkenburg for his help with data collection and the staff and facilities at the European Synchrotron Radiation Facility (ESRF).
References
- Alexeeva, M., Enright, A., Dawson, M. J., Mahmoudian, M. & Turner, N. J. (2002). Angew. Chem. Int. Ed.41, 3177–3180. [DOI] [PubMed]
- Binda, C., Coda, A., Angelini, R., Federico, R., Ascenzi, P. & Mattevi, A. (1999). Struct. Fold. Des.7, 265–276. [DOI] [PubMed]
- Binda, C., Li, M., Hubalek, F., Restelli, N., Edmondson, D. E. & Mattevi, A. (2003). Proc. Natl Acad. Sci. USA, 100, 9750–9755. [DOI] [PMC free article] [PubMed]
- Binda, C., Newton-Vinson, P., Hubalek, F., Edmondson, D. E. & Mattevi, A. (2002). Nature Struct. Biol.9, 22–26. [DOI] [PubMed]
- Bonsor, D., Butz, S. F., Solomons, J., Grant, S., Fairlamb, I. J. S., Fogg, M. J. & Grogan, G. (2006). Org. Biomol. Chem.4, 1252–1260. [DOI] [PubMed]
- Carr, R., Alexeeva, M., Dawson, M. J., Gotor-Fernandez, V., Humphrey, C. E. & Turner, N. J. (2005). Chembiochem, 6, 637–639. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- De Colibus, L., Li, M., Binda, C., Lustig, A., Edmondson, D. E. & Mattevi, A. (2005). Proc. Natl Acad. Sci. USA, 102, 12684–12689. [DOI] [PMC free article] [PubMed]
- Dunsmore, C. J., Carr, R., Fleming, T. & Turner, N. J. (2006). J. Am. Chem. Soc.128, 2224–2225. [DOI] [PubMed]
- Ma, J., Yoshimura, M., Yamashita, E., Nakagawa, A., Ito, A. & Tsukihara, T. (2004). J. Mol. Biol.338, 103–114. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Morley, K. L. & Kazlauskas, R. J. (2005). Trends Biotechnol.23, 231–237. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sablin, S. O., Yankovskaya, V., Bernard, S., Cronin, C. N. & Singer, T. P. (1998). Eur. J. Biochem.253, 270–279. [DOI] [PubMed]
- Schilling, B. & Lerch, K. (1995). Biochim. Biophys. Acta, 1243, 529–537. [DOI] [PubMed]
- Schwarzenbacher, R., Godzik, A., Grzechnik, S. K. & Jaroszewski, L. (2004). Acta Cryst. D60, 1229–1236. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.