The first crystallization of a W. pipientis protein, α-DsbA1, was achieved using hanging-drop and sitting-drop vapour diffusion.
Keywords: DsbA homologues, Wolbachia pipientis
Abstract
α-DsbA1 is one of two DsbA homologues encoded by the Gram-negative α-proteobacterium Wolbachia pipientis, an endosymbiont that can behave as a reproductive parasite in insects and as a mutualist in medically important filarial nematodes. The α-DsbA1 protein is thought to be important for the folding and secretion of Wolbachia proteins involved in the induction of reproductive distortions. Crystals of native and SeMet α-DsbA1 were grown by vapour diffusion and belong to the monoclinic space group C2, with unit-cell parameters a = 71.4, b = 49.5, c = 69.3 Å, β = 107.0° and one molecule in the asymmetric unit (44% solvent content). X-ray data were recorded from native crystals to a resolution of 2.01 Å using a copper anode and data from SeMet α-DsbA1 crystals were recorded to 2.45 Å resolution using a chromium anode.
1. Introduction
The Gram-negative α-proteobacterium Wolbachia pipientis is an obligate intracellular endosymbiont that infects a wide range of insects, spiders and mites amongst others (Jeyaprakash & Hoy, 2000 ▶; Werren & O’Neill, 1997 ▶; Breeuwer & Jacobs, 1996 ▶; Riegler & O’Neill, 2007 ▶). Wolbachia live inside a vacuole of host origin, mainly in the reproductive tissues of infected hosts, where they induce a variety of reproductive distortions that increase the proportion of infected females in the population, thereby favouring Wolbachia’s own maternal transmission. Wolbachia-induced traits in insects are being investigated for the control of insect pest populations and for the control of mosquito-borne diseases such as malaria and dengue (Zabalou et al., 2004 ▶; Brownstein et al., 2003 ▶). Wolbachia bacteria also infect medically important filarial nematodes (Taylor & Hoerauf, 1999 ▶) that cause lymphatic filariasis, elephantiasis and African river blindness. In these associations, Wolbachia act as mutualists in that the nematodes rely on Wolbachia for the production of metabolic coenzymes that are essential for proper larval development (Foster et al., 2005 ▶). As a consequence of this dependence, clinical approaches are being developed that target the symbiont rather than the disease-causing host (see reviews by Johnston & Taylor, 2007 ▶; Pfarr & Hoerauf, 2006 ▶).
The molecular basis of Wolbachia–host communication remains largely unknown, but it is likely that Wolbachia proteins that are secreted into the host cell, possibly through a type IV secretion system (Wu et al., 2004 ▶), play a crucial role by affecting the expression of host genes or by interacting with host cellular components. Several Wolbachia proteins have been proposed to be involved in reproductive distortions of the infected hosts (Wu et al., 2004 ▶; Iturbe-Ormaetxe et al., 2005 ▶), but no clear candidates have yet emerged.
In general, secreted proteins face a harsh environment, so that additional structural bracing in the form of disulfide bonds is usually required. In Gram-negative bacteria, the formation of disulfide bridges takes place in the periplasm. The pathways for disulfide-bond formation have been described in detail for Escherichia coli (Messens & Collet, 2006 ▶; Kadokura et al., 2003 ▶), but remain largely unknown for other bacteria. A key player in the process is DsbA, a strongly oxidizing protein that introduces disulfides into unfolded proteins. The genome of the W. pipientis strain (wMel) that infects Drosophila melanogaster (Wu et al., 2004 ▶) encodes two DsbA homologues that share low sequence homology with each other (21%) and with E. coli DsbA (α-DsbA1, 12%; α-DsbA2, 11%; Kurz et al., 2008 ▶).
Here, we describe the crystallization of one of the two DsbA homologues from W. pipientis: α-DsbA1. This is the first report, to our knowledge, of the crystallization of a Wolbachia protein and the first report of the crystallization of a DsbA protein from the α-class of Proteobacteria.
2. Materials and methods
2.1. Protein production
The α-DsbA1 coding sequence (WD1055 Genbank accession No. AE017196.1) was amplified by PCR from genomic wMel W. pipientis DNA and cloned into pET42 (Kurz et al., 2008 ▶). The construct used to produce α-DsbA1 for crystallization studies corresponded to residues 2–216 of the mature protein, with residue 2 replaced by Met (Val in the native sequence) and with an octahistidine tag (LEHHHHHHHH) at the C-terminus. Expression was carried out by autoinduction (Studier, 2005 ▶) and the protein was purified by metal-affinity and size-exclusion chromatography, as described in detail elsewhere (Kurz et al., 2008 ▶). The protein in 25 mM 4-(2-hydroxyethyl)-1-piperazine-N-2-ethanesulfonic acid (HEPES) pH 7, 150 mM NaCl was concentrated for crystallization experiments (Amicon Ultra centrifugal filter devices, 10 kDa cutoff, Millipore, Billerica, Massachusetts, USA). The protein concentration was estimated from the absorbance measured at 280 nm (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, Delaware, USA) and the purity was assessed by SDS–PAGE analysis (Nu-PAGE novex bis-Tris gel, 4–12%; Invitrogen, Carlsbad, California, USA).
To produce selenomethionine-labelled (SeMet) α-DsbA1, we followed previously described protocols (Heras et al., 2003 ▶). Briefly, a pre-culture (100 ml) was inoculated with a freshly transformed single colony and grown overnight at 310 K in LB medium containing 50 µg ml−1 kanamycin. The next day, 2 l defined M63 minimal media containing 0.05 mg ml−1 dl-selenomethionine was inoculated in a 1:50 ratio with the pre-culture and grown at 310 K until the OD600 of the cell culture was ∼0.8. Expression was then induced with isopropyl β-d-1-thiogalactopyranoside (IPTG; 1 mM final concentration) for a further 3.5 h and SeMet α-DsbA1 was then purified as described above for the native protein. SeMet was incorporated at all four methionines (mass-spectrometric data not shown).
After purification of the proteins, the active-site cysteines of either native or SeMet α-DsbA1 were oxidized using copper(II) (1,10-phenanthroline) (CuIIPhen3) following the protocol described previously for E. coli DsbA (Martin et al., 1993 ▶). Briefly, a fourfold molar excess of the reagent was added (to a final concentration of 1.7 mM) to the protein sample and the mixture was then incubated at 277 K for 1 h. CuIIPhen3 was then removed by buffer exchange with 25 mM HEPES pH 7, 150 mM NaCl using a PD-10 column (GE Healthcare, Piscataway, New Jersey, USA).
2.2. Crystallization
The sitting-drop and hanging-drop vapour-diffusion methods were used for crystallization of native α-DsbA1 with the commercially available screens Crystal Screens 1 and 2, PEG/Ion Screen (Hampton Research, San Diego, California, USA), JBScreen 2, JBScreen 3 and Precipitant Synergy Screen (Jena Bioscience, Jena, Germany) and a screen prepared in-house based on that reported by Page & Stevens (2004 ▶). Sitting-drop plates were set up manually using a multi-channel pipette (Matrix Impact, Thermo Fisher Scientific, Waltham, Massachusetts, USA). Each drop consisted of 1 µl protein solution and 1 µl well solution and a range of protein concentrations were evaluated (8–15 mg ml−1). The crystallization plates were incubated at 293 K in a temperature-controlled room. Small crystals of native α-DsbA1 were obtained within a week in a condition from the in-house screen comprising 22% polyethylene glycol of average molecular weight 3350 Da (PEG 3350), 0.1 M ammonium citrate pH 5.5. A grid screen was then set up on the basis of this condition in a VDXm 24-well hanging-drop plate. The pH was varied from 5.1 to 5.9 in increments of 0.2 pH units and the concentration of PEG 3350 was varied from 18 to 24% in increments of 2%. Siliconized cover slips (18 mm, Hampton Research, San Diego, California, USA) were used and each cover slip held two 2 µl drops containing protein (11 mg ml−1) and well solution in two different ratios (1:1, 1 µl and 1 µl, or 1:2, 0.65 µl and 1.35 µl, respectively). To increase the crystal size, larger volume sitting drops were set up (5 µl, with a protein-to-well solution ratio of 1:1) using the optimized condition.
Crystallization of SeMet α-DsbA1 was undertaken after the unsuccessful structure determination of native α-DsbA1 by molecular replacement. SeMet α-DsbA1 crystals were grown from the same protein concentration and crystallization conditions as native α-DsbA1 crystals and appeared within 5 d from 5 µl sitting drops (1:1 ratio).
2.3. X-ray diffraction
Native and SeMet α-DsbA1 crystals were cryoprotected by soaking for 5 min in a solution comprising the crystallization condition plus 20% PEG 400 before cryocooling in the nitrogen-gas stream (100 K) generated from a Cryo Industries CryoCool LN2 (Cryo Industries, Manchester, New Hampshire, USA). Diffraction data for native α-DsbA1 were measured using a high-brilliance FR-E generator operating at 45 kV and 45 mA with a copper anode, Osmic Confocal HiRes2 optics and an R-AXIS IV++ imaging-plate area detector (Rigaku Americas, Houston, Texas, USA). Diffraction data from an SeMet α-DsbA1 crystal were measured using an RU-H2R generator with a chromium anode, chromium Osmic MaxFlux optics and an R-AXIS IV++ imaging-plate area detector (Rigaku Americas, Houston, Texas, USA). The crystal-to-detector distances used were 150 mm for α-DsbA1 and 102 mm for SeMet α-DsbA1 and oscillation images were collected every 0.5° over a total of 180° with an exposure time of 2 min for native α-DsbA1 or 5 min for SeMet α-DsbA1. Diffraction data were processed and scaled using the HKL-2000 program package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
Small plate-shaped crystals (0.2 × 0.2 × 0.05 mm) of native α-DsbA1 were observed after one week using an in-house crystallization screen condition containing 22% PEG 3350, 0.1 M ammonium citrate pH 5.5. To optimize the condition, a 24-well grid screen was set up around this condition. Plate-shaped crystals of native α-DsbA1 appeared after 4 d in several conditions and single crystals were observed with 22% PEG 3350, 0.1 M ammonium citrate pH 5.7 or pH 5.9. These conditions produced diffraction-quality crystals of native α-DsbA1. To increase the crystal size, 5 µl sitting drops (1:1 ratio) were prepared using the same condition and a protein concentration of 11 mg ml−1. Plate-shaped crystals of dimensions 0.32 × 0.2 × 0.05 mm grew under these conditions after 4–5 d (Fig. 1 ▶).
Figure 1.
Crystals of native α-DsbA1. Plate-shaped crystals of native α-DsbA1 grew from 22% PEG 3350, 0.1 M ammonium citrate pH 5.7 or pH 5.9. The scale bar corresponds to 100 µm.
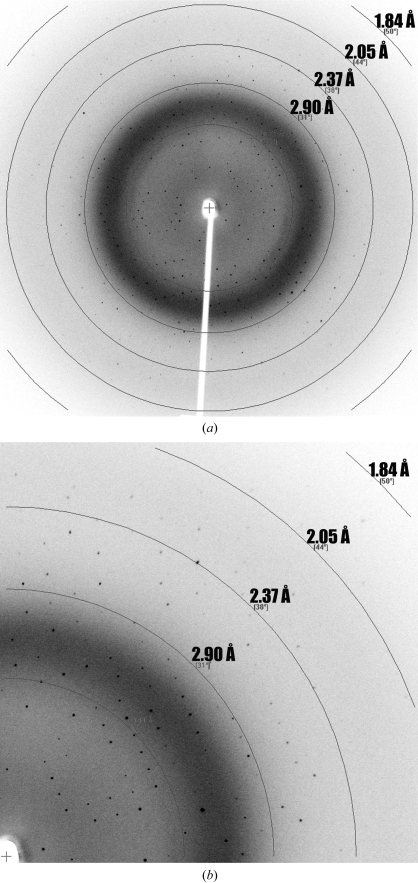
A 2.01 Å resolution data set was measured from a native α-DsbA1 crystal (Table 1 ▶, Fig. 2 ▶). The crystals belong to the C-centred monoclinic space group C2, with one molecule in the asymmetric unit and unit-cell parameters a = 71.4, b = 49.5, c = 69.3 Å. We attempted to solve the structure by molecular replacement using Phaser (Storoni et al., 2004 ▶) and the structure of E. coli DsbA (PDB code 1fvk; Guddat et al., 1997 ▶). However, no solution was found, which is perhaps not surprising given the low sequence identity between the two proteins (∼12%).
Table 1. Summary of X-ray diffraction data.
Values in parentheses are for the outermost resolution shell.
| Native α-DsbA1 | SeMet α-DsbA1 | |
|---|---|---|
| Wavelength (Å) | 1.5418 | 2.2909 |
| Resolution range (Å) | 50–2.01 (2.08–2.01) | 50–2.45 (2.54–2.45) |
| Space group | C2 | C2 |
| Unit-cell parameters | ||
| a (Å) | 71.4 | 71.1 |
| b (Å) | 49.5 | 49.7 |
| c (Å) | 69.3 | 69.2 |
| β (°) | 107.0 | 106.8 |
| No. of observations | 53805 | 43345 |
| No. of unique reflections | 15438 (1514) | 15044 (1599) |
| Redundancy | 3.5 (3.3) | 2.9 (2.4) |
| Completeness (%) | 99.3 (98.2) | 90.4 (95.7) |
| Rmerge† | 0.088 (0.236) | 0.043 (0.097) |
| 〈I/σ(I)〉 | 20.9 (5.6) | 27.3 (7.5) |
R
merge = 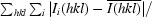
 , where I
i(hkl) is the scaled observed intensity of the ith symmetry-related observation of reflection hkl and
, where I
i(hkl) is the scaled observed intensity of the ith symmetry-related observation of reflection hkl and 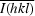 is the average intensity.
is the average intensity.
Figure 2.
Diffraction pattern from a native α-DsbA1 crystal. The X-ray diffraction pattern (a) was recorded from a 0.5° oscillation (Cu Kα wavelength, 1.5418 Å), as described in §2. The enlarged region (b) shows diffraction extending to 2.01 Å resolution.
We then produced SeMet α-DsbA1 crystals with the aim of solving the structure by SAD methods using chromium-wavelength radiation (Cr Kα, 2.29 Å). With chromium radiation, the contribution to the anomalous signal from selenium is doubled (Δf′′ = 2.28 e) compared with that for copper radiation (Δf′′ = 1.14 e). SeMet α-DsbA1 crystals were grown under conditions similar to those used to produce native α-DsbA1 crystals (22% PEG 3350, 0.1 M ammonium citrate pH 5.7 or pH 5.9). Plate-shaped crystals (0.26 × 0.23 × 0.05 mm) grew after 5 d from a 5 µl sitting drop. SeMet α-DsbA1 crystals diffracted to 2.45 Å resolution (Table 1 ▶). The diffraction resolution for the SeMet α-DsbA1 crystal was limited by the experimental setup for chromium-wavelength data measurement, which has a fixed crystal-to-detector distance of 102 mm owing to the use of a helium cone to minimize X-ray absorption. The SeMet α-DsbA1 crystals also belong to space group C2, with unit-cell parameters a = 71.1, b = 49.7, c = 69.2 Å. Assuming the presence of one molecule in the asymmetric unit for both native and SeMet crystals, the Matthews coefficient V M (Matthews, 1968 ▶) and the solvent content were calculated to be 2.2 Å3 Da−1 and 44%, respectively.
Unfortunately, the measured anomalous signal was weak (d′′/σ at 3.4 Å of 1.1); the most likely route for phasing will therefore be MAD or SAD data measurement at a synchrotron.
Acknowledgments
We acknowledge use of the University of Queensland ROCX Diffraction Facility. This work was supported by an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellowship to JLM, Australian Research Council (ARC) grants to JLM and to BH, a University of Queensland Early Career Researcher Grant to BH and an International Postgraduate Research Scholarship (IPRS) to MK.
References
- Breeuwer, J. A. & Jacobs, G. (1996). Exp. Appl. Acarol.20, 421–434. [DOI] [PubMed]
- Brownstein, J. S., Hett, E. & O’Neill, S. L. (2003). J. Invertebr. Pathol.84, 24–29. [DOI] [PubMed]
- Foster, J. et al. (2005). PLoS Biol.3, 599–614.
- Guddat, L. W., Bardwell, J. C., Glockshuber, R., Huber-Wunderlich, M., Zander, T. & Martin, J. L. (1997). Protein Sci.6, 1893–1900. [DOI] [PMC free article] [PubMed]
- Heras, B., Edeling, M. A., Byriel, K. A., Jones, A., Raina, S. & Martin, J. L. (2003). Structure, 11, 139–145. [DOI] [PubMed]
- Iturbe-Ormaetxe, I., Burke, G. R., Riegler, M. & O’Neill, S. L. (2005). J. Bacteriol.187, 5136–5145. [DOI] [PMC free article] [PubMed]
- Jeyaprakash, A. & Hoy, M. A. (2000). Insect Mol. Biol.9, 393–405. [DOI] [PubMed]
- Johnston, K. L. & Taylor, M. J. (2007). Curr. Infect. Dis. Rep.9, 55–59. [DOI] [PubMed]
- Kadokura, H., Katzen, F. & Beckwith, J. (2003). Annu. Rev. Biochem.72, 111–135. [DOI] [PubMed]
- Kurz, M., Iturbe-Ormaetxe, I., Jarrott, R., Cowieson, N. P., Robin, G., Jones, A., King, G. J., O’Neill, S. L., Heras, B. & Martin, J. L. (2008). Submitted.
- Martin, J. L., Waksman, G., Bardwell, J. C., Beckwith, J. & Kuriyan, J. (1993). J. Mol. Biol.230, 1097–1100. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Messens, J. & Collet, J. F. (2006). Int. J. Biochem. Cell B, 38, 1050–1062. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Page, R. & Stevens, R. C. (2004). Methods, 34, 373–389. [DOI] [PubMed]
- Pfarr, K. M. & Hoerauf, A. M. (2006). Mini-Rev. Med. Chem.6, 203–210. [DOI] [PubMed]
- Riegler, M. & O’Neill, S. L. (2007). The Prokaryotes, edited by M. Dworkin, pp. 547–561. New York: Springer-Verlag.
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Taylor, M. J. & Hoerauf, A. (1999). Parasitol. Today, 15, 437–442. [DOI] [PubMed]
- Werren, J. H. & O’Neill, S. L. (1997). Influential Passengers: Inherited Microorganisms and Arthropod Reproduction, edited by S. L. O’Neill, A. A. Hoffmann & J. H. Warren, pp. 1–4. Oxford University Press.
- Wu, M. et al. (2004). PLoS Biol.2, 327–341.
- Zabalou, S., Riegler, M., Theodorakopoulou, M., Stauffer, C., Savakis, C. & Bourtzis, K. (2004). Proc. Natl Acad. Sci. USA, 101, 15042–15045. [DOI] [PMC free article] [PubMed]