A 2.8 Å resolution data set was collected from a crystal of a recombinant plant aminoaldehyde dehydrogenase from P. sativum. This enzyme oxidizes ω-aminoaldehydes arising from polyamine degradation.
Keywords: aminoaldehyde dehydrogenases, Pisum sativum
Abstract
Aminoaldehydes are products of polyamine degradation and are known to be reactive metabolites that are toxic to living cells at high concentrations. These compounds are catabolized by aminoaldehyde dehydrogenases, which are enzymes that contain a nicotinamide adenine dinucleotide coenzyme. Aminoaldehyde dehydrogenase from Pisum sativum was overexpressed in Escherichia coli, purified and crystallized using the hanging-drop method. A complete data set was collected to 2.8 Å resolution at 100 K. Crystals belong to the monoclinic space group P21, with unit-cell parameters a = 86.4, b = 216.6, c = 205.4 Å, β = 98.1°. Molecular replacement was performed and led to the identification of six dimers per asymmetric unit.
1. Introduction
ω-Aminoaldehydes arise from the degradation of polyamines (such as putrescine, spermidine or spermine), which is catalyzed by amine oxidases or polyamine oxidases. The oxidation products (hydrogen peroxide and aminoaldehydes) have been implicated in programmed cell death, induction of cytotoxicity and inhibition of cell division and cause apoptosis in several cell types (Agostinelli et al., 2004 ▶). For instance, 3-aminopropionaldehyde (APAL) is considered to be cytotoxic to mammalian cells and causes apoptotic and necrotic death of both neurons and glial cells (Li et al., 2003 ▶). Aminoaldehyde dehydrogenases (AMADHs) oxidize various primary ω-aminoaldehydes, namely APAL, 4-aminobutyraldehyde (ABAL) and 4-guanidinobutyraldehyde, to the corresponding ω-amino acids β-alanine, 4-aminobutyric acid and 4-guanidinobutyric acid, respectively. To date, AMADHs have been purified from microorganisms, plants and animals. For historical reasons, they are classified separately as 4-aminobutyraldehyde dehydrogenases (EC 1.2.1.19) and 4-guanidinobutyraldehyde dehydrogenases (EC 1.2.1.54) in the enzyme catalogue (Prieto et al., 1987 ▶; Matsuda & Suzuki, 1984 ▶). AMADHs contain a nicotinamide adenine dinucleotide (NAD) coenzyme and a reactive cysteine at the active site. According to amino-acid sequence homology, AMADHs are related to betaine aldehyde dehydrogenases (BADHs; EC 1.2.1.8), which are enzymes that mediate the conversion of betaine aldehydes to betaines (Šebela et al., 2000 ▶).
In pea (Pisum sativum), AMADH is present in two isoforms as demonstrated by activity staining after native gel electrophoresis (Šebela et al., 2001 ▶). This was confirmed when two complete cDNAs (AMADH1 and AMADH2) were obtained by 5′ and 3′ RACE-PCR (Brauner et al., 2003 ▶). The two 503-amino-acid isoenzymes show 81% sequence identity. The subcellular localization of the pea AMADHs is unknown. Although both isoenzymes carry C-terminal peroxisomal targetting signal type 1 (PTS1), analysis of numerous database-deposited amino-acid sequences or ESTs for plant BADHs and AMADHs suggests the coexistence of peroxisomal and nonperoxisomal isoforms in dicots and monocots (Reumann et al., 2005 ▶). In pea root and hypocotyl, most AMADH activity is found in cells belonging to the pericycle and endodermis, while in the epicotyl and shoot apex a major part of the AMADH activity is found in vascular cambium cells (Šebela et al., 2001 ▶). The respective roles and localization of AMADH1 and AMADH2 are not yet known. Native pea AMADH1 shows the highest affinity for APAL and ABAL, while betaine aldehyde is not a substrate. To date, no known three-dimensional structure has been determined of a plant aminoaldehyde dehydrogenase or betaine aldehyde dehydrogenase. Therefore, the structural reason why AMADH1 cannot oxidize betaine aldehyde remains unknown. To gain insight into the substrate specificity, we have purified, crystallized and performed preliminary X-ray diffraction analysis of the recombinant aminoaldehyde dehydrogenase AMADH1 from P. sativum.
2. Cloning, expression and purification
The AMADH1 ORF (1510 bp, AJ315852) was cloned into a pET28b His-tag vector (Novagen) digested by NdeI and XhoI restriction endonucleases. Thus, the primers used for amplification of AMADH1 cDNA contained NdeI (upstream primer, 5′-GCTGCATATGGCAATCACAGTATCAAGT-3′) and XhoI (downstream primer, 5′-CGTCTCGAGTATCACAGCTTTGAAGGTGG-3′) restriction sites. The construct was then introduced by heat shock into BL21 Star (DE3) chemically competent Escherichia coli cells (Invitrogen) for expression of 6×His-AMADH1. The nucleotide sequence of one selected clone was confirmed by DNA-sequence analysis. For protein expression, transformed BL21 Star (DE3) E. coli cells were precultured in Luria–Bertani (LB) medium containing 1% glucose and kanamycin (30 µg ml−1) at 310 K overnight. Cells were harvested by centrifugation at 4000g for 5 min and then grown in LB medium containing kanamycin (30 µg ml−1) to an OD600 of about 0.6. AMADH1 expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and the culture was incubated at 293 K overnight. After induction, cells were harvested by centrifugation at 5000g for 10 min and stored frozen.
Cells were resuspended in a lysis buffer containing 50 mM Tris–HCl pH 8.0, 10 mM MgCl2 and one tablet of EDTA-free protease inhibitors (Roche) followed by the addition of lysozyme (0.3 mg ml−1). After 1 h incubation at room temperature, 1%(w/v) Triton X-100, RNAse (0.01 µg ml−1) and DNAse (0.04 U µl−1) were added. Finally, after 30 min incubation at 310 K, 100 mM NaCl and 5%(w/v) glycerol were added. The lysate was then centrifuged at 12 000g for 30 min and the supernatant containing expressed 6×His-AMADH1 was recovered.
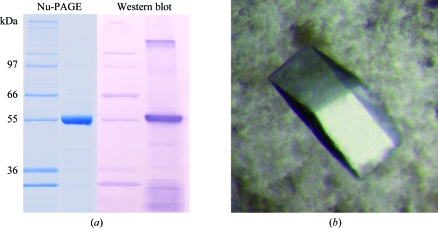
The supernatant was loaded onto a column packed with His-selected cobalt gel (Sigma) equilibrated with 20 mM Tris–HCl buffer pH 8.0 containing 100 mM NaCl, 10 mM imidazole and 5% glycerol. Elution was performed using 250 mM imidazole in the same buffer. Pure AMADH1 was concentrated using 30 kDa cutoff filters (Centricon). The total yield of purified AMADH1 was about 6 mg per litre of culture medium. The enzyme was characterized by a specific activity value of 2.7 µmol s−1 mg−1 measured by monitoring NADH formation at 340 nm using APAL as a substrate (Šebela et al., 2000 ▶). To confirm the purity and correct expression of AMADH1, SDS–PAGE was performed using the Nu-PAGE system from Invitrogen. After electrophoresis, the protein bands were visualized by Coomassie staining (CBB G-250), Western blotted and detected using AMADH antibodies (Fig. 1 ▶ a). The experimental molecular weight of 57 kDa and also the pI of 5.8 (as obtained by isoelectric focusing; not shown) of recombinant AMADH1 were both in agreement with calculated values. MALDI–TOF peptide mass fingerprinting of AMADH1 after in-gel digestion of a 5 pmol sample (Šebela et al., 2006 ▶) allowed unambiguous identification in the MSDB database (accession No. Q8VWZ1). The identified 21 peptides covered 51% of the sequence and matched with an accuracy of 18 p.p.m.; the probability-based MOWSE score was 266.
Figure 1.
(a) Nu-PAGE and Western blot of a recombinant pea aminoaldehyde dehydrogenase (AMADH1). Nu-PAGE was performed using 4–12% bis-tris gel in MOPS buffer. The PVDF membrane was probed with polyclonal anti-AMADH rabbit antibodies, reacted with goat anti-rabbit IgG alkaline phosphatase conjugate and coloured using NBT/BCIP (nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate, Roche). (b) Crystals of AMADH1. The crystal shown (maximum dimension 0.3 mm) is similar to that used for the X-ray diffraction data collection.
3. Crystallization
Prior to crystallization, the enzyme was desalted and concentrated to 18 mg ml−1 by ultrafiltration using a Centricon 30 kDa cutoff device (Amicon). Crystals were grown at 293 K using the hanging-drop vapour-diffusion method. Preliminary crystallization conditions were found using a screening kit (Crystal Screen, Hampton Research). Small crystals appeared within two weeks using condition No. 48 from Crystal Screen 2. Crystallization conditions were then optimized. 2 µl AMADH1 solution with 10 mM NAD (Sigma–Aldrich) was mixed with 2 µl of a reservoir solution containing 0.1 M HEPES pH 7.5, 13%(w/v) PEG 6000 and 5%(v/v) 2-methyl-2,4-pentanediol, leading to both transparent clustered and a few good single crystals (Fig. 1 ▶ b).
4. Data collection and processing
Crystals were soaked in a cryoprotectant solution (reservoir solution supplemented with 20% glycerol) for 3 min and flash-frozen in liquid nitrogen. Data-collection experiments were carried out at 100 K on the ID14-3 beamline at the European Synchrotron Radiation Facility (Grenoble, France) equipped with an ADSC Q4 CCD detector and tuned to 0.93 Å wavelength. 260° of data were collected in 1° frames, with 5 s exposure per frame. Diffraction intensities were evaluated using the program MOSFLM (Leslie, 1992 ▶). Data were further processed to 2.8 Å resolution using the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). The crystal belongs to the monoclinic space group P21, with unit-cell parameters a = 86.4, b = 216.6, c = 205.4 Å, β = 98,1°. Data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 86.4, b = 216.6, c = 205.4, β = 98.1 |
| Resolution (Å) | 30.0–2.8 (2.95–2.8) |
| No. of observed reflections | 2130930 |
| No. of unique reflections | 178951 |
| Completeness (%) | 97.8 (93.1) |
| Rmerge† (%) | 13.7 (42.9) |
| 〈I〉/〈σ(I)〉 | 7.7 (2.0) |
R
merge = 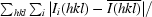
 , where Ii(hkl) is the intensity of an observation and
, where Ii(hkl) is the intensity of an observation and 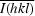 is the mean value of its unique reflection; the summations are over all reflections.
is the mean value of its unique reflection; the summations are over all reflections.
5. Molecular replacement
A search for a sequence-related protein of known crystal structure using the program PHYRE (the successor to 3D-PSSM; Kelley et al., 2000 ▶) indicated several proteins with secondary structure similar to that predicted for AMADH1. All belong to the aldehyde dehydrogenase (ALDH) superfamily and have sequence identities of up to 41% (human and bovine mitochondrial ALDHs and cod liver BADH). Most of the AMADH or ABALDH (4-aminobutyraldehyde dehydrogenase) enzymes have been reported to be homotetramers. The asymmetric unit can contain three or four tetramers, corresponding to Matthews coefficients (Matthews, 1968 ▶) of 2.78 and 2.09 Å3 Da−1, respectively. However, attempts to use molecular replacement to solve the structure of AMADH1 with the program Phaser (Storoni et al., 2004 ▶) gave no solution for three tetramers using the tetrameric structure of human mitochondrial ALDH (PDB code 1cw3) as a model search. When the monomer was used as a model, 12 monomers were placed in the asymmetric unit by the program. Examination of the resulting model showed that the 12 monomers form six dimers, suggesting that the active form of AMADH1 in solution is a dimer, similar to the case for a few ABALDH enzymes. Assuming that the unit cell contains six dimers per asymmetric unit, the calculated Matthews coefficient is 2.78 Å3 Da−1, corresponding to a solvent content of 55.8%. Refinement of the complete AMADH1 structure is under way.
Acknowledgments
This work was supported by grants MSM 6198959215 and MSM 6198959216 from the Ministry of Education, Youth and Sports of the Czech Republic. We thank the staff of ESRF in Grenoble for making station ID14-3 available.
References
- Agostinelli, E., Arancia, G., Dalla Vedova, L., Belli, F., Marra, M., Salvi, M. & Toninello, A. (2004). Amino Acids, 27, 347–358. [DOI] [PubMed]
- Brauner, F., Šebela, M., Snégaroff, J., Peč, P. & Meunier, J. C. (2003). Plant Physiol. Biochem.41, 1–10.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. E. (2000). J. Mol. Biol.299, 499–520. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Li, W., Yuan, X. M., Ivanova, S., Tracey, K. J., Eaton, J. W. & Brunk, U. T. (2003). Biochem. J.371, 429–436. [DOI] [PMC free article] [PubMed]
- Matsuda, H. & Suzuki, Y. (1984). Plant Physiol.76, 654–657. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Prieto, M. I., Martin, J., Balaña-Fouce, R. & Garrido-Pertierra, A. (1987). Biochimie, 69, 1161–1168. [DOI] [PubMed]
- Reumann, S., Ma, C., Lemke, S. & Babujee, L. (2005). Plant Physiol.136, 2587–2608. [DOI] [PMC free article] [PubMed]
- Šebela, M., Brauner, F., Radová, A., Jacobsen, S., Havliš, J., Galuszka, P. & Peč, P. (2000). Biochim. Biophys. Acta, 1480, 329–341. [DOI] [PubMed]
- Šebela, M., Luhová, L., Brauner, F., Galuszka, P., Radová, A. & Peč, P. (2001). Plant Physiol. Biochem.39, 831–839.
- Šebela, M., Štosová, T., Havliš, J., Wielsch, N., Thomas, H., Zdráhal, Z. & Shevchenko, A. (2006). Proteomics, 6, 2959–2963. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]