The overproduction, purification, crystallization and preliminary crystallographic studies of the native and selenomethionine-labelled P1 domain are reported here as a first step towards the elucidation of the molecular mechanism of YidC as a membrane-protein insertase.
Keywords: YidC/Oxa1/Alb3 protein family, biogenesis of inner membrane proteins
Abstract
In Escherichia coli, the biogenesis of inner membrane proteins (IMPs) requires targeting and insertion factors such as the signal recognition particle (SRP) and the Sec translocon. Recent studies have identified YidC as a novel and essential component involved in membrane insertion of IMPs both in conjunction with the Sec translocon and as a separate entity. E. coli YidC is a member of the YidC (in bacteria)/Oxa1 (in mitochondria)/Alb3 (in chloroplasts) protein family and contains six transmembrane segments and a very large periplasmic domain P1. The overproduction, purification, crystallization and preliminary crystallographic studies of the native and selenomethionine-labelled P1 domain are reported here as a first step towards the elucidation of the molecular mechanism of YidC as a membrane-protein insertase.
1. Introduction
The biogenesis of IMPs is generally accomplished in three distinct steps (Luirink et al., 2005 ▶): the proteins are targeted to the membrane and subsequently inserted into it and the last step comprises the assembly and folding into the final lipid-embedded functional structure. In Escherichia coli, most of the IMPs are targeted and assembled in a co-translational process involving the SRP system, the SRP receptor FtsY (Wild et al., 2004 ▶) and the secretory (Sec) protein translocase complex (Breyton et al., 2002 ▶; Dalbey & Chen, 2004 ▶; van den Berg et al., 2004 ▶). Recently, YidC, a 61 kDa polytopic inner membrane protein, was shown to play an essential but versatile and poorly defined role during the integration, folding and assembly of IMPs both in association with the Sec translocon as well as separately. YidC is homologous to Alb3 and Oxa1, which are involved in the integration of proteins into the chloroplast thylakoid membrane and mitochondrial inner membrane, respectively (Chen et al., 2002 ▶; Serek et al., 2004 ▶; Yi & Dalbey, 2005 ▶). Oxa1 and Alb3 span the membrane five times, whereas E. coli YidC spans the membrane six times and contains a large periplasmic domain, the P1 loop, predicted between TM1 and TM2 (residues 24–342). Previous studies have indicated that five transmembrane segments (TM2–TM6) of the YidC protein which are conserved in evolution contain the region responsible for its insertase function. The domain boundaries and the role of the P1 domain in the function of YidC are still not well understood. The region adjacent to TM2 is of special interest as residues 322–342 have been shown to be essential for cell viability and for YidC insertase function for both Sec-dependent and Sec-independent proteins (Sääf et al., 1998 ▶; Jiang et al., 2002 ▶, 2003 ▶; Xie et al., 2006 ▶). Recently, copurification studies performed with YidC deletion mutants suggested that the P1 domain (or more precisely residues 215–265) is required for the interaction with the SecF protein from the Sec translocase complex (Xie et al., 2006 ▶).
The determination of the three-dimensional structure of the YidC P1 domain will provide new insights into its function and more generally into the YidC protein family. Here, we describe the overproduction, purification, crystallization and preliminary crystallographic analysis of a fragment containing the periplasmic P1 domain of E. coli YidC (referred in this study as P1D) and of its selenomethionine derivative.
2. Materials and methods
2.1. Cloning and protein overproduction
A gene fragment encoding residues 56–329 (P1D) of E. coli YidC was amplified from a cDNA of the full-length yidc by the polymerase chain reaction (PCR) using the Expand High Fidelity PCR system (Roche) with the primers 5′-GGACATGCCATGGGCCAGGGGAAACTGATCTCGGTT-3′ (forward) and 5′-AGGCAGCTCGAGATCAACGGTCAGATCCAGGTGCGG-3′ (reverse). The purified DNA fragment was cloned via the NcoI/XhoI restriction sites into a pET24d vector (Novagen) providing a C-terminal hexahistidine tag. The absence of mutations was checked by DNA sequencing.
The wild-type P1D was overproduced in E. coli strain BL21 pLysS. Cells were grown at 310 K in lysogeny broth medium (LB) complemented with 30 µg ml−1 kanamycin and 34 µg ml−1 chloramphenicol. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at an OD600 of 0.6 and growth was then continued for 4 h at 303 K.
The selenomethionine (SeMet) labelled protein was expressed in methionine-auxotroph strain B834(DE3) with minimal medium supplemented with 17 amino acids, nucleic acid bases, various salts, phosphate, sulfate, vitamins and SeMet (Hendrickson et al., 1990 ▶) and complemented with 30 µg ml−1 kanamycin. Precultures were performed in steps to increase gradually the percentage of minimal medium containing SeMet from 20 to 100%. The last preculture was then used to inoculate the main culture, which was induced with 1 mM IPTG at an OD600 of 0.6 for 12 h at 303 K.
2.2. Protein purification
For purification of the wild-type protein, cell pellets were resuspended in lysis buffer [50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole, 0.02%(v/v) MTG (1-thioglycerol), 2 mM benzamidine (Merck) and protease-inhibitor mix (Serva)], disrupted by sonication and then passed twice through a Microfluidizer M1-10L (Microfluidix). The cell debris and insoluble material were removed by centrifugation (40 000g for 20 min at 277 K). All protein-purification procedures were carried out at 277 K. The supernatant was loaded onto a 1 ml HisTrap Chelating column (GE Healthcare) equilibrated with 50 mM HEPES pH 7.5, 300 mM NaCl and 10 mM imidazole (buffer A). The protein was eluted in buffer B (50 mM HEPES pH 7.5, 300 mM NaCl and 60 mM imidazole). The fractions containing P1D were pooled and subjected to gel-filtration chromatography using a Hi-Load Superdex 75/26-60 column (GE Healthcare) equilibrated in buffer C (10 mM HEPES pH 7.5, 200 mM NaCl). Protein-containing fractions were analyzed by 10% SDS–PAGE and pooled. The SeMet-labelled protein was purified by the same procedure, except that the sonication step was avoided and buffers A and B were supplemented with 0.02%(v/v) MTG and buffer C with 10 mM DTT. The efficiency of SeMet incorporation was assessed by matrix-assisted laser desorption/ionization (MALDI) mass spectroscopy. The molecular-weight difference between the unlabelled (30.903 kDa) and SeMet-labelled (31.091 kDa) proteins indicated 100% incorporation (four sites).
2.3. Size-exclusion chromatography
The apparent molecular weight was analyzed by size-exclusion chromatography using a Superdex 200 HR 10/30 column (GE Healthcare) in buffer C. Standard proteins (γ-globulin, 158.0 kDa; ovalbumin, 43.0 kDa; myoglobin, 17.0 kDa; vitamin B12, 1.3 kDa; Bio-Rad) were used to calibrate the column with the same buffer.
2.4. Circular-dichroism spectroscopy
A Peltier temperature-controlled Jasco 810 spectropolarimeter was used to measure circular-dichroism (CD) spectra. A 2 mm path-length quartz cell was used and the protein concentration was 0.5 mg ml−1 (15 µM). Prior to the experiments, the buffer composition of the protein solution was changed to 10 mM MOPS pH 7.5, 100 mM NaCl using a PD-10 desalting column (GE Healthcare). The CD spectrum was measured at 298 K between 200 and 250 nm and was the average of four scans.
2.5. NMR spectroscopy
An NMR spectrum was acquired at 295 K on a Bruker DRX600 equipped with cyogenic triple-resonance probes and processed with TopSpin. A 500 µl sample at 0.1 mM was used. Prior to the experiments, the buffer composition of the protein solution was changed to 10 mM NaPO4 pH 6.8, 100 mM NaCl using a PD-10 desalting column (GE Healthcare).
2.6. Crystallization
Prior to crystallization trials, the protein was concentrated in Amicon Ultracell-10K concentrators (Millipore). Automated crystallization screening was carried out using sitting-drop vapour diffusion in 96-well plates using a Mosquito nanolitre pipetting robot (TTP Labtec) and sparse-matrix screening. Drops consisting of 100 nl protein solution (20 mg ml−1) and 100 nl reservoir solution were equilibrated over 100 µl reservoir solution. Optimization of the crystallization conditions was performed using the hanging-drop vapour-diffusion method in Greiner 24-well plates with 500 µl reservoir solution and 3 µl drops set up using a 2:1 ratio of protein solution (40 mg ml−1) to reservoir solution.
Crystals of SeMet-labelled P1D were obtained by mixing 1 µl (40 mg ml−1) protein solution with an equal volume of reservoir solution.
Prior to X-ray analysis, the native crystals were briefly soaked in a cryoprotectant solution consisting of reservoir solution supplemented with 20%(v/v) ethylene glycol.
SeMet crystals were subjected to a post-crystallization treatment (Heras & Martin, 2005 ▶). Crystal dehydration was performed by transferring the crystals into droplets (50 µl) of a dehydrating solution containing the mother liquor with a higher concentration of PEG 3350 [30%(w/v)] and a stepwise increasing concentration [5, 10, 15%(v/v)] of PEG 400, with 5 min incubation in each condition. Crystals were then flash-cooled in liquid nitrogen for data collection.
2.7. Data collection and processing
Diffraction data were collected at beamlines ID14-2 (for the native crystals) and ID29 (for SeMet crystals) at the European Synchrotron Radiation Facility (ESRF) under cryogenic conditions (100 K; Oxford Cryosystems Cryostream). Data were recorded with ADSC Q4R CCD (beamline ID14-2) and ADSC Q315R CCD (beamline ID29) detectors. The fluorescence spectrum, which was recorded from the frozen SeMet crystals prior to data collection, was used to select the wavelength of the Se K absorption edge at the peak (λ1 = 0.9790 Å, maximum of f′′) and at a high-energy remote wavelength (λ2 = 0.9756 Å). Data were processed and scaled with the CCP4-implemented programs MOSFLM and SCALA, respectively (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
The domain boundaries of P1D were not unambiguously defined. P1D was previously predicted to include amino acids 24–342 based on PhoA fusion analysis and hydrophobicity prediction using TOPPREDII (Sääf et al., 1998 ▶; Xie et al., 2006 ▶). However, multiple sequence alignments and secondary-structure prediction (including transmembrane and amphipathic helix prediction) performed with the PredictProtein server (Rost et al., 2004 ▶) and the Network Protein Sequence Analysis server (Combet et al., 2000 ▶) suggested that residues 330–342 are either part of the second transmembrane helix or form an amphipathic helix. Residues 29–50 are predicted to be a low-complexity region.
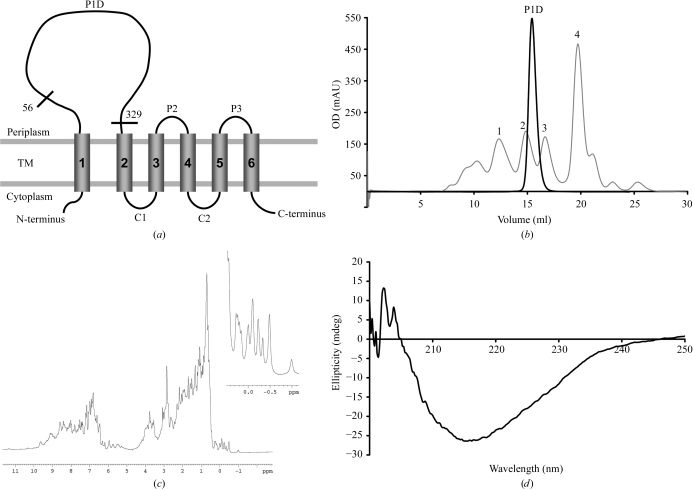
In order to facilitate purification and crystallization, the construct design was based on these predictions and a gene fragment encoding amino acids 56–329 predicted to form a mixed α/β-type domain was cloned (Fig. 1 ▶ a). This P1D was overproduced; typical yields were about 30 mg per litre of culture for the native protein and 10 mg per litre of culture for the SeMet-labelled protein.
Figure 1.
Characterization of P1D. (a) Schematic representation of the E. coli YidC topology, according to Sääf et al. (1998 ▶). The transmembrane segments (TM), the cytoplasmic (C1 and C2) and periplasmic (P2, P3) loops and the borders of P1D are indicated. (b) Typical size-exclusion chromatography profile (A 280) of P1D on a Superdex 200 HR 10/30 column. The elution positions of the calibration markers (grey line) are indicated (1, γ-globulin; 2, ovalbumin; 3, myoglobin; 4, vitamin B12). (c) One-dimensional 1H NMR spectrum of P1D. Details of the signals upfield of 0.5 p.p.m. are given in the inset. (d) Far-UV CD spectrum of P1D.
The protein was purified by standard protocols as described above. Size-exclusion chromatography revealed a homogeneous monomeric protein solution (Fig. 1 ▶ b). The folding and secondary-structure content of the purified P1D were analysed by 1H NMR and CD spectroscopy. Prior to these experiments, the sample buffer was changed according to the requirements of each method, but this did not affect either the stability or the oligomeric state of P1D. The one-dimensional 1H NMR spectrum showed significant chemical shift dispersions, especially in the amide region downfield of 8.5 p.p.m. and in the methyl group region between 1.0 p.p.m. and −1.0 p.p.m., with a number of signals upfield of 0.5 p.p.m. (Fig. 1 ▶ c). These resonances were well separated sharp lines and their intensities represented the entire population of protein molecules; this is characteristic of a well folded protein. The far-UV CD spectrum exhibited a minimum at 215 nm typical of β-sheet proteins (Fig. 1 ▶ d).
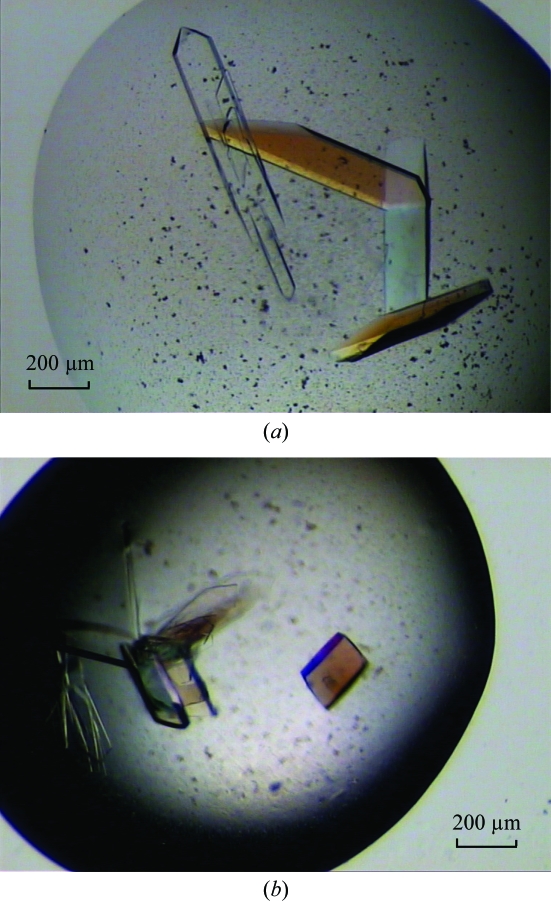
This P1D construct was then subjected to crystallization trials. Initial native crystals appeared within 3 d at 293 K from reservoir solutions containing 0.2 M calcium acetate and 20%(w/v) PEG 3350. After optimization, crystals of dimensions 400 × 200 × 20 µm that were suitable for X-ray analysis were obtained after one week at 293 K from a reservoir solution containing 0.2 M calcium acetate, 20–21%(w/v) PEG 3350 and 10%(v/v) ethylene glycol (Fig. 2 ▶ a). These crystals diffracted to a resolution of 2.2 Å. They belong to the monoclinic space group C2, with unit-cell parameters a = 169.3, b = 56.0, c = 63.6 Å, β = 102.3°. The Matthews coefficient and solvent content, estimated as 2.45 Å3 Da−1 and 50%, respectively (Matthews, 1968 ▶), are consistent with the presence of two monomers in the asymmetric unit. Crystal data and data-collection statistics are shown in Table 1 ▶.
Figure 2.
Crystals of P1D. (a) Typical native crystals with approximate dimensions 400 × 200 × 20 µm. (b) SeMet crystals, typical dimensions 200 × 100 × 20 µm.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| SeMet P1D | |||
|---|---|---|---|
| Native P1D | Peak | Remote | |
| Diffraction data statistics | |||
| Synchrotron-radiation source | ID14-2, ESRF | ID29, ESRF | ID29, ESRF |
| Space group | C2 | ||
| Unit-cell parameters (Å, °) | a = 169.26, b = 55.99, c = 63.60, β = 102.29 | a = 161.15 b = 55.64, c = 63.33, β = 101.12 | |
| Resolution range (Å) | 55.73–2.20 (2.32–2.20) | 44.99–2.10 (2.14–2.10) | 44.99–1.80 (1.89–1.80) |
| Completeness (%) | 100 (100) | 83.3 (83.3) | 97.3 (96.4) |
| Multiplicity | 4.8 (4.8) | 3.6 (3.2) | 3.7 (3.8) |
| Total no. of reflections | 142801 | 97276 | 184602 |
| No. of unique reflections | 29819 | 26993 | 49763 |
| Rmerge† (%) | 5.8 (32.6) | 8.3 (37.2) | 6.7 (42.8) |
| Mean I/σ(I) | 21.3 (3.6) | 13.6 (3.1) | 15.3 (3.2) |
| Rano† (%) | 8.4 (28.7) | 5.6 (26.5) | |
| Anomalous completeness (%) | 78.1 (30.6) | 94.6 (94.2) | |
| Phasing statistics | |||
| No. of Se sites | 8 | ||
| Log likelihood‡ | 320028 | ||
| Figure of merit (FOM)‡ (acentric/centric) | 0.392 (0.422/0.133) | ||
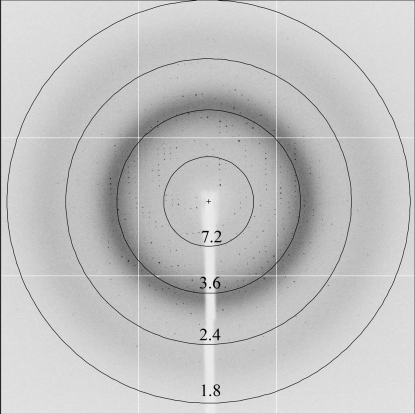
Since no structure of a homologous protein has yet been determined, we produced and crystallized SeMet-labelled protein for phasing by anomalous dispersion using the selenium edge (Fig. 2 ▶ b). SeMet crystals (200 × 100 × 20 µm) appeared after 3–4 d at 293 K from a reservoir solution containing 0.2 M calcium acetate, 21–22%(w/v) PEG 3350 and 10%(v/v) ethylene glycol. When cryoprotected in a similar way as the native crystals, they diffracted to a resolution of 3–3.5 Å using synchrotron radiation. The diffraction quality was improved using crystal dehydration as described above, resulting in an extension of the resolution to 1.8 Å. SeMet crystals belong to the same space group as the native crystals, with unit-cell parameters a = 161.1, b = 55.6, c = 63.3 Å, β = 101.1° (Table 1 ▶). As for the native crystals, the typical Matthews coefficient and solvent content, which were estimated as 2.32 Å3 Da−1 and 47%, respectively (Matthews, 1968 ▶), are consistent with the presence of two monomers in the asymmetric unit. A fluorescence scan was taken around the Se K absorption edge and diffraction data were collected at two wavelengths corresponding to the absorption maximum (λ1 = 0.9790 Å) and remote (λ2 = 0.9756 Å). The first data set collected at the peak with a maximum resolution of 2.1 Å was sufficient to calculate useful single-wavelength anomalous scattering (SAD) phases. A total of eight Se atoms in the asymmetric unit with a mean figure of merit of 0.39 were detected by an automated search using the AUTOSOL routine from the PHENIX program suite (Adams et al., 2002 ▶, 2004 ▶; Table 1 ▶). We therefore expect the structure to be solvable by the SAD method. The second data set (at λ2) subsequently collected at the diffraction limit of the SeMet crystal (1.8 Å; Fig. 3 ▶) will be used in model building and crystallographic refinement.
Figure 3.
Diffraction pattern of P1D. A 0.45° oscillation image is shown, collected from a SeMet crystal at the remote wavelength (λ2 = 0.9756 Å). The resolution ranges specified by the rings are given in Å.
The three-dimensional structure of P1D should provide a molecular basis for understanding its role in the function of YidC during membrane assembly. It should also allow a more rational screening of interaction partners.
Note added in proof: A paper Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC has recently been published (Oliver & Paetzel, 2007 ▶).
Acknowledgments
We thank J. Luirink (VU, Amsterdam) for providing the DNA of the full-length yidc. We thank C. Kohlhaas and U. Dürrwang for their contribution at the beginning of the project and G. Müller and S. Gehrig for excellent technical assistance. We thank B. Simon and M. Sattler from EMBL Heidelberg for the NMR experiments. We acknowledge access to the beamlines at the ESRF in Grenoble and the support of the beamline scientists, especially during the EMBO course EMBO’07 – A Course on Exploiting Anomalous Scattering in Macromolecular Structure Determination. This work was supported by a Marie Curie Intra-European Fellowship (EIF 041951) to SR and by a grant from the Deutsche Forschungsgemeinshaft (SFB638) to IS.
References
- Adams, P. D., Gopal, K., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Pai, R. K., Read, R. J., Romo, T. D., Sacchettini, J. C., Sauter, N. K., Storoni, L. C. & Terwilliger, T. C. (2004). J. Synchrotron Rad.11, 53–55. [DOI] [PubMed]
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Berg, B. van den, Clemons, W. M. Jr, Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C. & Rapoport, T. A. (2004). Nature (London), 427, 36–44. [DOI] [PubMed]
- Breyton, C., Haase, W., Rapoport, T. A., Kuhlbrandt, W. & Collinson, I. (2002). Nature (London), 418, 662–665. [DOI] [PubMed]
- Chen, M., Xie, K., Jiang, F., Yi, L. & Dalbey, R. E. (2002). Biol. Chem.383, 1565–1572. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Combet, C., Blanchet, C., Geourjon, C. & Deleage, G. (2000). Trends Biochem. Sci.25, 147–150. [DOI] [PubMed]
- Dalbey, R. E. & Chen, M. (2004). Biochim. Biophys. Acta, 1694, 37–53. [DOI] [PubMed]
- Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. (1990). EMBO J.9, 1665–1672. [DOI] [PMC free article] [PubMed]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed]
- Jiang, F., Chen, M., Yi, L., de Gier, J. W., Kuhn, A. & Dalbey, R. E. (2003). J. Biol. Chem.278, 48965–48972. [DOI] [PubMed]
- Jiang, F., Yi, L., Moore, M., Chen, M., Rohl, T., Van Wijk, K. J., de Gier, J. W., Henry, R. & Dalbey, R. E. (2002). J. Biol. Chem.277, 19281–19288. [DOI] [PubMed]
- Luirink, J., von Heijne, G., Houben, E. & de Gier, J. W. (2005). Annu. Rev. Microbiol.59, 329–355. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Oliver, D. C. & Paetzel, M. (2007). J. Biol. Chem., doi:10.1074/jbc.M708936200. [DOI] [PubMed]
- Rost, B., Yachdav, G. & Liu, J. (2004). Nucleic Acids Res.32, 321–326. [DOI] [PMC free article] [PubMed]
- Sääf, A., Monne, M., de Gier, J. W. & von Heijne, G. (1998). J. Biol. Chem.273, 30415–30418. [DOI] [PubMed]
- Serek, J., Bauer-Manz, G., Struhalla, G., van den Berg, L., Kiefer, D., Dalbey, R. & Kuhn, A. (2004). EMBO J.23, 294–301. [DOI] [PMC free article] [PubMed]
- Wild, K., Halic, M., Sinning, I. & Beckmann, R. (2004). Nature Struct. Mol. Biol.11, 1049–1053. [DOI] [PubMed]
- Xie, K., Kiefer, D., Nagler, G., Dalbey, R. E. & Kuhn, A. (2006). Biochemistry, 45, 13401–13408. [DOI] [PubMed]
- Yi, L. & Dalbey, R. E. (2005). Mol. Membr. Biol.22, 101–111. [DOI] [PubMed]