Abstract
To investigate the structural and thermodynamic basis of the binding of solvent at internal sites within proteins a number of mutations were constructed in T4 lysozyme. Some of these were designed to introduce new solvent-binding sites. Others were intended to displace solvent from preexisting sites. In one case Val-149 was replaced with alanine, serine, cysteine, threonine, isoleucine, and glycine. Crystallographic analysis shows that, with the exception of isoleucine, each of these substitutions results in the binding of solvent at a polar site that is sterically blocked in the wild-type enzyme. Mutations designed to perturb or displace a solvent molecule present in the native enzyme included the replacement of Thr-152 with alanine, serine, cysteine, valine, and isoleucine. Although the solvent molecule was moved in some cases by up to 1.7Å, in no case was it completely removed from the folded protein. The results suggest that hydrogen bonds from the protein to bound solvent are energy neutral. The binding of solvent to internal sites within proteins also appears to be energy neutral except insofar as the bound solvent may prevent a loss of energy due to potential hydrogen bonding groups that would otherwise be unsatisfied. The introduction of a solvent-binding site appears to require not only a cavity to accommodate the water molecule but also the presence of polar groups to help satisfy its hydrogen-bonding potential. It may be easier to design a site to accommodate two or more water molecules rather than one as the solvent molecules can then hydrogen-bond to each other. For similar reasons it is often difficult to design a point mutation that will displace a single solvent molecule from the core of a protein.
Keywords: Cavities, hydration, water, hydrogen bonds, T4 lysozyme
Although the interiors of proteins are primarily hydrophobic, solvent molecules are occasionally observed within the core (Edsall and McKenzie 1983). Current knowledge of the role of such internal solvent is based in part on surveys of known protein structures (Baker and Hubbard 1984; Rashin et al. 1986; Thanki et al. 1988; Sreenivasan and Axelsen 1992; Levitt and Park 1993; Hubbard et al. 1994; Williams et al. 1994; Baker 1995). The ordering of buried solvent has been studied by NMR (Denisov et al. 1997). Also, theoretical estimates have been made of the entropic cost of localizing a water molecule within a protein (Wade et al. 1990, Wade et al. 1991; Dunitz 1994) as well as the energetics of transferring solvent from the aqueous phase to a more hydrophobic milieu representative of the core of a protein (Wolfenden and Radzicka 1994). Very little is known, however, regarding the structural and energetic changes associated with the binding of solvent at a defined site in a protein. Examples have been reported where the addition or deletion of a water molecule can both decrease (Berndt et al. 1993; Pedersen et al. 1994; Xu et al. 1998) and increase (Das et al. 1989; Hickey et al. 1991; Vriend et al. 1991; Buckle et al. 1996; Lett et al. 1996; Takano et al. 1997) protein stability. The possible functional roles of conserved solvent-binding sites have been examined by mutagenesis and structural analysis (Berghuis et al. 1994; Langhorst et al. 1999).
Here we analyze solvent binding by constructing a number of different mutants of bacteriophage T4 lysozyme. All variants were constructed in the cysteine-free form of the protein (Matsumura and Matthews 1989), here designated as wild-type or WT*. The mutations can be broadly divided into two classes, those that were intended to incorporate additional solvent molecules within the protein structure and those that were intended to remove solvent. The results suggest that new solvent-binding sites can be created by point mutations, although the participation of preexisting polar groups within the protein structure is required. These mutations may also reduce the stability of the protein fairly substantially. In this study there was no point mutation capable of expelling a solvent molecule from the structure of wild-type lysozyme. Evaluation of previously studied mutants suggests that such expulsion can occur, although it is rare.
Results
Mutations that introduce putative solvent-binding sites
Replacements of valine-149
Val-149 is fully buried within the carboxy-terminal domain of T4 lysozyme. It is located such that one methyl group, Cγ1, makes contact with hydrophobic residues while the other methyl group, Cγ2, is directed toward the only buried polar network in T4 lysozyme, including Asp-10, Asn-101, Arg-145, Tyr-161, and a buried water molecule, Sol-208. The local structure is well packed without any cavity detectable by means of a rolling probe of radius 1.2Å (Connolly 1983).
It was previously shown that the replacement Val-149 → Ala (V149A) in T4 lysozyme results in the binding of an internal solvent molecule (Xu et al. 1998). A similar solvent molecule had also been observed in the V149T structure (Blaber et al. 1993). To extend these results, Val-149 was replaced with serine, cysteine, and glycine and, as a further control, with isoleucine. These structures are briefly described below.
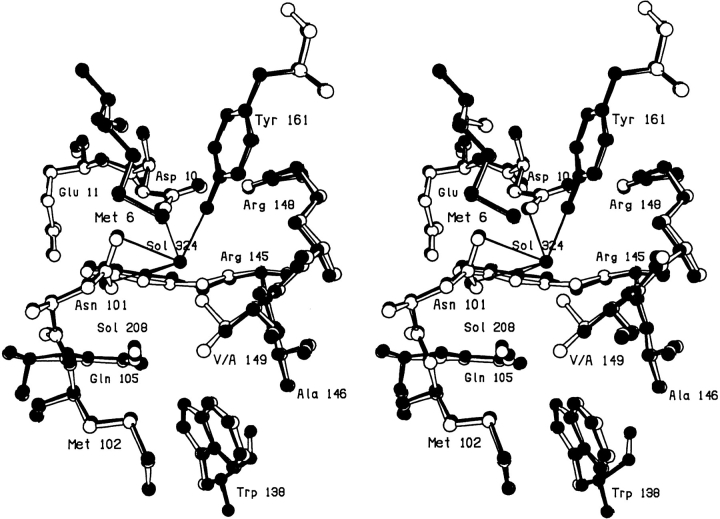
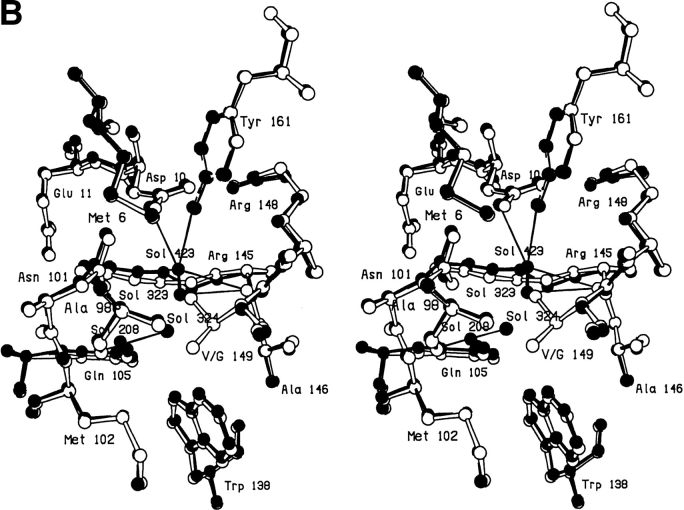
In the V149A mutant the deletion of the two methyl groups causes minimal perturbation in the local structure of the protein (Fig. 1 ▶). A new solvent molecule (Sol-324) is, however, observed to bind close to the site vacated by the γ2 methyl group (Xu et al. 1998). This solvent molecule makes hydrogen bonds to several buried polar atoms (Fig. 1 ▶; Table 1). Another nearby solvent molecule, Sol-208, appears not to participate in hydrogen bonding to Sol-324.
Fig. 1.
Superposition of mutant V149A (solid bonds) (Xu et al. 1998) on WT* (open bonds).
Table 1.
Hydrogen-bonding interactions of the introduced solvent molecule(s) near residue 149
| Mutant | Atom | B factor (Å2) | H-bonding partner | Distance (Å) |
| WT* (Val149)a | Asn101 Nδ2 | — | Asp10 Oδ2 | 2.9 |
| Tyr161 Oη | — | Asp10 Oδ2 | 2.6 | |
| Wat208 | — | Asn101 Oδ1 | 2.9 | |
| Arg145 Nɛ | — | Asn101 Oδ1 | 3.2 | |
| Gln105 Oɛ1 | — | Arg145 Nɛ | 3.4 | |
| Wat208 | — | Arg145 Nɛ | 3.5 | |
| V149A | Sol324 | 56 | Asp10 Oδ2 | 2.3 |
| Asn101 Oδ1 | 3.0 | |||
| Asn101 Nδ2 | 3.0 | |||
| Arg145 Nɛ | 2.9 | |||
| Tyr161 Oη | 3.0 | |||
| V149S | Sol323 | 30 | Asp10 Oδ2 | 2.8 |
| Asn101 Oδ1 | 3.4 | |||
| Arg145 O | 2.9 | |||
| Arg145 Nɛ | 3.0 | |||
| Ser149 Oγ | 2.9 | |||
| V149C | Sol323b | 48 | Asp10 Oδ2 | 2.3 |
| Asn101 Oδ1 | 3.2 | |||
| Asn101 Nδ2 | 3.3 | |||
| Arg145 Nɛ | 2.8 | |||
| Cys149 Sδ | 2.9 | |||
| Tyr161 Oη | 3.0 | |||
| V149T | Sol323 | 54 | Asp10 Oδ2 | 2.6 |
| Asn101 Oδ1 | 3.3 | |||
| Arg145 O | 3.1 | |||
| Arg145 Nɛ | 2.7 | |||
| Thr149 Oγ1 | 2.7 | |||
| V149G | Sol323c | 35 | Arg145 O | 3.2 |
| Asn101 Oδ1 | 3.5 | |||
| Sol208 | 3.4 | |||
| Sol324 | 3.3 | |||
| Sol324 | 60 | Ala98 O | 2.9 | |
| Gly149 O | 3.9 | |||
| Sol323 | 3.3 |
a WT* does not bind a solvent molecule at residue 149. In this case the hydrogen bonds shown are those that occur in this region in the WT* structure.
b Occupancy approximately 0.4 (see text).
c Occupancy approximately 0.5 (see text). Hydrogen bonding for the alternative position (Sol423) is not included.
As previously described by Blaber et al. (1993), in the V149T mutant the newly introduced threonine side chain essentially superimposes on the wild-type valyl residue (not shown). The γ-hydroxyl hydrogen bonds to a water molecule that occupies a position very similar to the new water seen in V149A (Table 1). In the V149T structure the distance from the hydroxyl of Thr-149 to the water molecule is 2.7Å (Table 1). This corresponds to a somewhat short, but essentially normal, hydrogen bond. However, 2.7Å is too short for a van der Waals contact between a water molecule and a methyl group (expected distance ∼3.1Å). Thus, when site 149 is occupied by valine, as in WT*, the γ-methyl of the valine sterically excludes the water molecule from the site that it occupies in V149T.
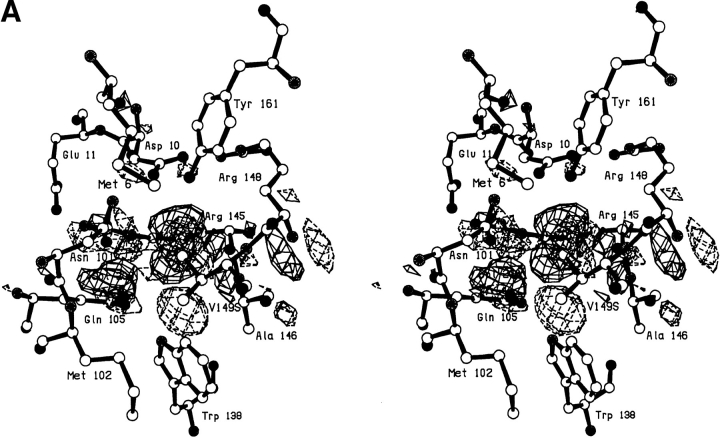
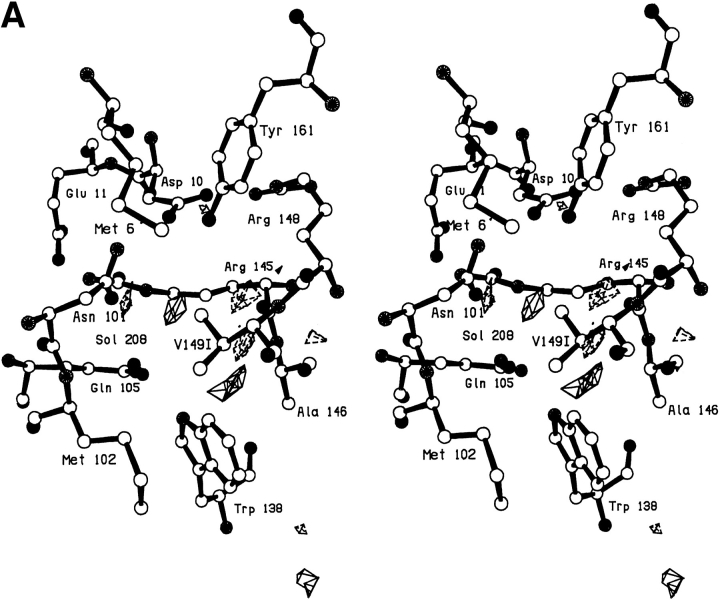
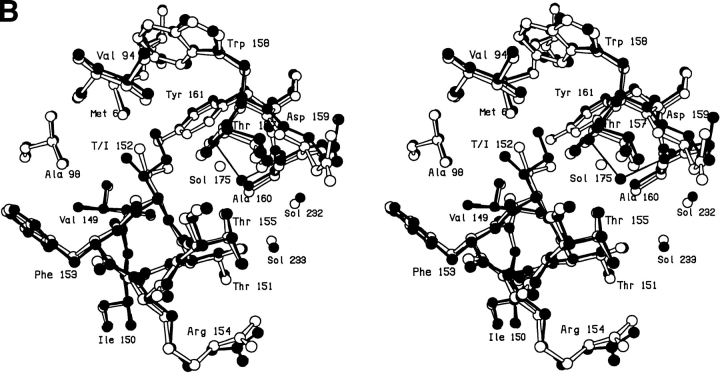
A serine at site 149 behaves very similarly to threonine with the hydroxyl making a hydrogen bond to a solvent molecule (Sol-323) that is bound nearby (Fig. 2A,B ▶; Table 2). The electron density corresponding to this solvent molecule can be clearly seen in Figure 2A ▶. This solvent occupies essentially the same location as in V149A and V149T, and makes multiple hydrogen bonds to nearby polar groups (Table 1). The structure does adjust somewhat relative to WT*. After the deletion of the methyl group, the β-carbon of Ser-149 moves 0.45Å toward the vacated space and, in concert, the main chain atoms move up to 0.35Å (Fig. 2B ▶). The mutation is quite destabilizing (ΔΔG = −4.4 kcal/mol; Table 3).
Fig. 2.
(A) Stereo pair showing the difference in electron density between mutant V149S and wild-type lysozyme. Amplitudes (Fmut,obs − FWT*,obs) where Fmut,obs and FWT*,obs are the observed structure amplitude for the mutant and the wild-type lysozyme, respectively; phases calculated from the refined model structure of the wild-type protein. Positive contours (solid) and negative contours (broken) drawn at +4σ and -4σ, where σ is the rms density throughout the unit cell. (B) Superposition of the refined structure of mutant V149S (solid bonds) on wild-type lysozyme (open bonds). The two sets of coordinates were superimposed based on the main chain atoms within the respective carboxy-terminal domains (i.e., residues 80–162), so as to eliminate possible artifacts due to "hinge-bending."
Table 2.
Hydrogen-bonding of the newly-introduced polar group at sites 149 and 152
| Mutant | Polar atom | Hydrogen-bond partner | Distance (Å) |
| V149S | Oγ | Sol323 | 2.9 |
| V149C | SγA | Oδ1 101 | 3.8 |
| SγB | O 98 | 3.3 | |
| V149T | Oγ1 | Sol323 | 2.8 |
| Thr 152 (WT) | Oγ1 | O 148 | 2.8 |
| Sol175 | 2.7 | ||
| T152S | Oγ | O 148 | 3.0 |
| Sol175 | 2.8 | ||
| T152C | Sγ | O 148 | 3.6 |
| Sol175 | 3.4 |
Table 3.
Thermostabilities of mutant lysozymes
| Proteins | ΔTm (°C) | ΔH (kcal/mol) | ΔΔG (kcal/mol) |
| WT* | 0 | 130 | 0 |
| I58T | −10.1 | 84 | −3.4 |
| N101A | −3.7 | 113 | −1.5 |
| Q105M | −2.7 | 110 | −1.2 |
| V149A | −8.9 | 107 | −3.1 |
| V149S | −13.2 | 94 | −4.4 |
| V149C | −5.5 | 113 | −2.0 |
| V149T | −8.3 | 110 | −3.0 |
| V149I | −0.05 | 123 | −0.1 |
| V149G | −15.3 | 85 | −4.9 |
| T152A | −4.0 | 115 | −1.5 |
| T152S | −5.5 | 117 | −2.0 |
| T152C | −1.4 | 127 | −0.5 |
| T152V | +0.8 | 127 | +0.2 |
| T152I | −1.0 | 125 | −0.4 |
ΔTm is the melting temperature of the mutant minus that of the reference protein, WT*, which unfolds with a Tm of 65.3°C. ΔH is the enthalpy of unfolding determined at the Tm of the given protein. ΔΔG is the difference between the standard free energy of unfolding of the mutant and WT* proteins. The estimated uncertainty in ΔTm is about 0.2°C. For ΔΔG the estimated uncertainty is about 0.15 kcal/mol for most of the variants but increases to about 0.3 kcal/mol for those that are least stable.
Refinement of the structure in which cysteine is substituted at site 149 showed that the sulfur atom takes two conformations corresponding roughly to replacement of one or other methyl groups of the valine. In one conformation the sulfur is directed toward the polar network and has an occupancy of ∼0.6. In the other conformation, directed toward the hydrophobic pocket, the occupancy is ∼0.4. There is a small positive peak (not shown) located near the site at which a new water molecule was observed in the other mutants. This was modeled as a partially occupied solvent molecule. It appears that when the sulfur atom of Cys-149 points toward the polar network there is not enough space to accommodate a water molecule. When the sulfur atom is oriented away from the polar network and toward the hydrophobic pocket, however, a solvent molecule (Sol-323) can bind at a location similar to that in V149A, V149T, and V149S.
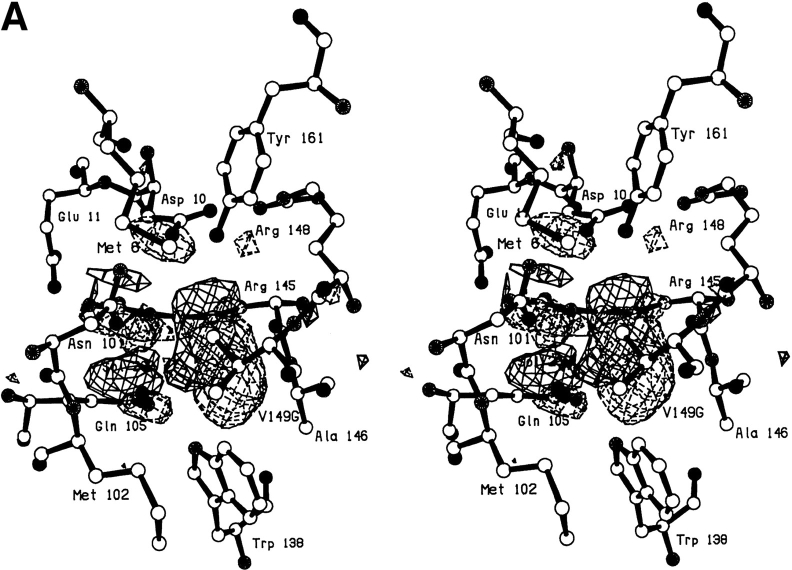
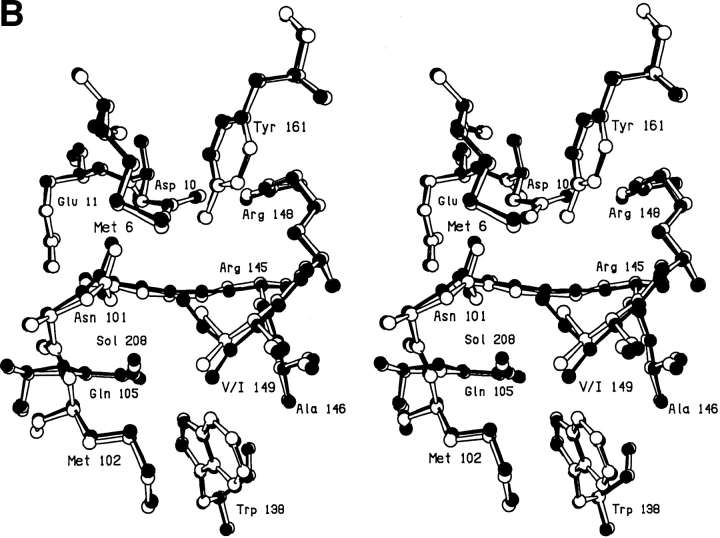
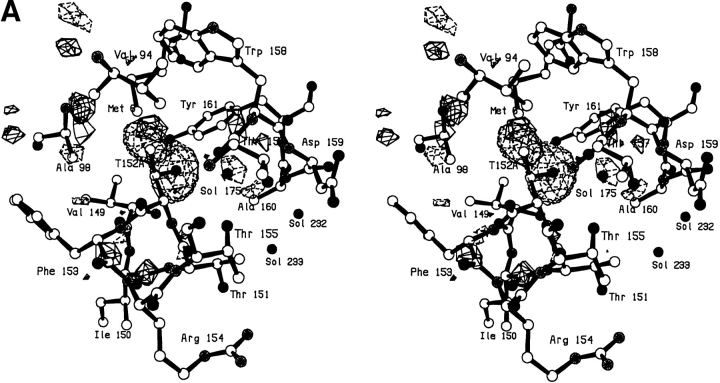
In the V149G mutant, notwithstanding the removal of the side chain, the protein structure changes very little (Fig. 3A,B ▶). The resultant cavity appears to be occupied by two poorly ordered solvent molecules. One of these appears to occupy two alternative sites ∼1.2Å apart (labeled Sol-323 and Sol-432 in Fig. 3B ▶). The second solvent molecule (labeled Sol-324) has a B-factor of 60Å2 indicating that it is also relatively mobile.
Fig. 3.
(A) Difference electron density map for mutant V149G; all conventions as in Fig. 2A ▶. (B) Superposition of mutant V149G (solid bonds) on WT* (open bonds).
As a further control Val-149 was replaced by isoleucine. In this case the local structure "expands" slightly to accommodate the extra methyl group (Fig. 4B ▶). As is apparent from the lack of features in the difference electron density map (Fig. 4A ▶), the overall mutant structure is very similar to the wild-type protein and no additional solvent molecule is bound. Similar behavior was also seen with the V149M protein (Gassner et al. 1999; Protein Data Bank code 1CV6).
Fig. 4.
(A) Difference electron density map for mutant V149I; all conventions as in Fig. 2A ▶. (B) Superposition of mutant V149I (solid bonds) on WT* (open bonds).
Isoleucine-58 to threonine
In WT* lysozyme Ile-58 is completely buried. It had been shown previously that the replacement of Ile-58 with alanine resulted in a cavity of volume 18Å3 that appeared to be empty (Xu et al. 1998). The side chain of the isoleucine is, however, fairly close to an internal site at which two ordered water molecules are bound (Sol-171 and Sol-179) (Weaver and Matthews 1987). The distance from Cγ1 to the closest water is 7.0Å and it was speculated that a threonine replacement, which would substitute a hydroxyl at this position, might create a solvent-binding site. Although the intervening distance was somewhat longer than ideal, the hydroxyl would provide hydrogen bonding functionality and the removal of the δ-methyl group would help to create the requisite space.
In the mutant structure (not shown) no additional water molecules were observed. Rather, the side chains of Leu-15, Ile-27, Ala-63, and Thr-58 collapse into the space left vacant by the δ-methyl group. The mutation is quite destabilizing (ΔΔG = −3.4 kcal/mol), due to the combination of unsatisfied hydrogen bonding potential with the loss of the ethyl group.
Mutations designed to weaken or eliminate solvent-binding sites
Replacements of threonine-152
Threonine-152 is located within the helix that includes residues 143–155 and is fully buried within the carboxy-terminal domain. Its side chain hydroxyl group makes a hydrogen bond with an internal water molecule Sol-175, which in turn hydrogen bonds with the main chain atoms O of Thr-157 and N of Ala-160 (Tables 2 and 4). Sol-175 is one of the four water molecules that naturally bind at internal sites within the T4 lysozyme molecule (Weaver and Matthews 1987). Its B-factor is 20Å2, showing it to be well ordered.
Table 4.
Hydrogen-bonding interactions of the internal water molecule near residue 152
| Mutant | Movement of bound water (Å) | B factor of bound water (Å2) | H-bonding partner | Distance (Å) |
| WT* (Thr152) | — | 20 | Thr152 Oγ | 2.7 |
| Thr157 O | 2.8 | |||
| Ala160 N | 3.1 | |||
| T152A | 0.5 | 26 | Thr157 O | 3.1 |
| Ala160 N | 2.9 | |||
| T152S | 0.3 | 20 | Ser152 Oγ | 2.8 |
| Thr157 O | 2.9 | |||
| Ala160 N | 3.1 | |||
| T152C | 1.6 | 27 | Thr157 O | 2.8 |
| Ala160 N | 3.0 | |||
| Cys152 Sγ | 3.4 | |||
| T152V | 1.7 | 32 | Thr157 O | 2.8 |
| Ala160 N | 3.1 | |||
| T152I | 1.6 | 31 | Thr157 O | 2.7 |
| Ala160 N | 3.1 |
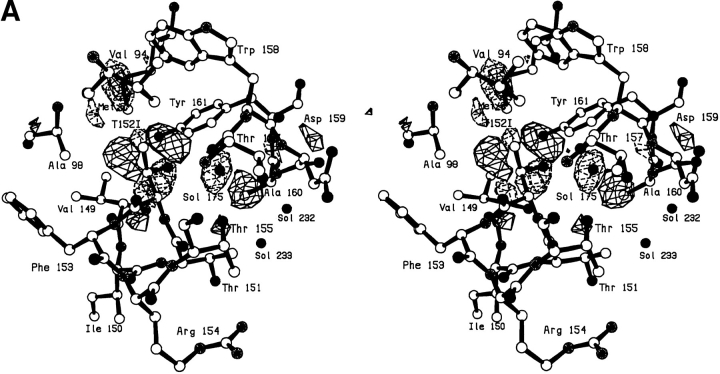
The Thr-152 → Ile mutant was constructed with the idea that it might displace the bound solvent molecule. The replacement of the γ-hydroxyl of the threonine with an ethyl group would not only delete one of the hydrogen bonds to the solvent molecule but would introduce a steric clash were the solvent molecule to remain in place. As can be seen in Figure 5A,B ▶, the solvent molecule moves ∼1.7Å to relieve the steric interference, retains its hydrogen-bonding partners (minus the γ-hydroxyl) (Table 4), but is not displaced from the core of the protein. Positive and negative density showing the movement of the solvent molecule, and also indicating a rotation about the Cα–Cβ bond, can be seen in Figure 5A ▶. There is also some local expansion of the structure to accommodate the larger side chain including backbone shifts up to ∼0.3Å (Fig. 5B ▶). To accommodate the 1.7Å shift in Sol-175, residues 156–160 near the carboxyl terminus of the protein move outward by ∼0.4Å with the side chain atoms of Asp-159 moving up to 0.9Å. Notwithstanding the various changes, mutant T152I is only 0.4 kcal/mol less stable than wild type (Table 3).
Fig. 5.
(A) Difference electron density map for mutant T152I; all conventions as in Fig. 2A ▶. (B) Superposition of mutant T152I (solid bonds) on WT* (open bonds).
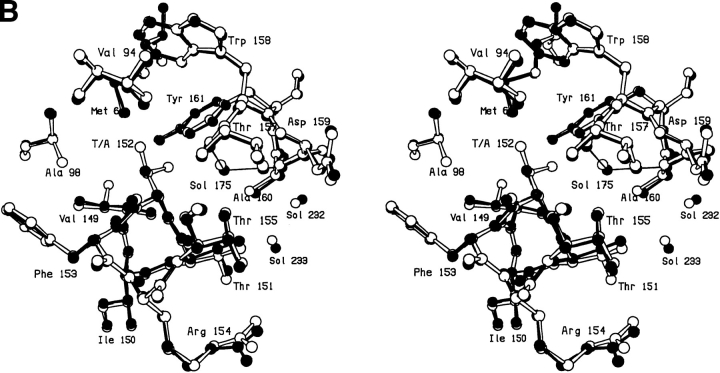
To further probe the structure and energetics at this site, Thr-152 was also replaced with valine, cysteine, serine, and alanine. The changes seen in the T152V mutant are related to those seen for T152I, but are slightly smaller in magnitude (not shown). In this mutant Sol-175 moves 1.6Å compared to 1.7Å in T152I and χ1 decreases by 12° compared to 17°. In this case the mutant protein is marginally less stable than wild type (by 0.4 kcal/mol). In the case of the cysteine mutant the γ-sulfur atom occupies a position similar to the γ-hydroxyl of the threonine, but rotated somewhat toward the site left vacant by the γ-methyl group. The introduced SH group forms an apparent weak hydrogen bond (Gregoret et al. 1991) with Sol-175 and possibly with the carbonyl oxygen of Arg-148 (Table 2). In terms of structure the T152S mutant is very similar to wild type except that Val-94 shifts up to 0.5Å toward the space vacated by the deleted methyl group and the serine hydroxyl appears to be somewhat more mobile than that of the threonine. The mutant is, however, 2 kcal/mol less stable than wild type (Table 3). In general, the structure of the alanine mutant T152A is also very similar to wild type (Fig. 6A,B ▶). Despite the loss of the hydrogen bond to the γ-hydroxyl, Sol-175 maintains a position close to that in wild type (Fig. 6B ▶; Table 4). Val-94 shifts ∼0.4Å so as to somewhat reduce the volume of the cavity created by the loss of the side chain.
Fig. 6.
(A) Difference electron density map for mutant T152A; all conventions as in Fig. 2A ▶. (B) Superposition of mutant T152A (solid bonds) on WT* (open bonds).
Glutamine-105 to methionine and asparagine-101 to alanine
In WT* lysozyme there is another internal water molecule, Sol-208, that appears to be hydrogen bonded most strongly by the side chain amides of Asn-101 and Gln-105 (Table 5). Asn-101 is completely buried but the side chain of Gln-105 is partially exposed to solvent. To eliminate the hydrogen bond from the water molecule to Gln-105, and to maintain approximately the same side chain size, Gln-105 was mutated to methionine. The structure of this mutant reveals that, despite the loss of the hydrogen bond, the water molecule remains bound within the protein. The position of the water molecule does shift 0.3Å relative to its position in WT* lysozyme and its B-factor increases somewhat (Table 5). The hydrogen bond to Asn-101 is preserved (Table 5). In the mutant structure the distance from the solvent atom to Sδ of Met-105 is 3.2Å. This close sulfur–water distance and the orientation of the thioether moiety are suggestive of a sulfur–oxygen hydrogen bond, although the strength of such an interaction is uncertain (Gregoret et al. 1991). The stability of this mutant is only slightly less than that of wild type (ΔΔG = −1.2 kcal/mol).
Table 5.
Hydrogen-bonding interactions of the internal water molecule near residues 101 and 105
| Protein | Movement of bound water (Å) | B factor of bound water (Å2) | H-bonding partner | Distance (Å) |
| WT* (Gln105) | — | 16 | Gln105 Oɛ1 | 2.6 |
| Asn101 Oδ1 | 2.9 | |||
| Asn101 O | 3.7 | |||
| Trp138 Nɛ1 | 3.6 | |||
| Arg145 Nɛ | 3.5 | |||
| Q105M | 0.3 | 24 | Met105 Sδ | 3.2 |
| Asn101 Oδ1 | 2.8 | |||
| Asn101 O | 3.5 | |||
| Trp138 Nɛ1 | 3.8 | |||
| Arg145 Nɛ | 3.5 | |||
| Q105Ea | 0.1 | 18 | Glu105 Oɛ1 | 2.7 |
| Asn101 Oδ1 | 3.0 | |||
| Asn101 O | 3.7 | |||
| Trp138 Nɛ1 | 3.7 | |||
| Arg145 Nɛ | 3.2 | |||
| N101A | 0.8 | 20 | Gln105 Oɛ1 | 2.7 |
| Ala101 O | 2.9 | |||
| Trp138 Nɛ1 | 4.2 | |||
| Arg145 Nɛ | 3.9 | |||
| HOH401 O | 2.8 | |||
| A129F | 0.7 | 53 | Gln105 Oɛ1 | 3.0 |
| Asn101 Oδ1 | 2.8 | |||
| Asn101 O | 2.9 | |||
| Trp138 Nɛ1 | 4.3 | |||
| Arg145 Nɛ | 3.9 |
a Mutant Q105E was constructed in the WT rather than the WT* background (Pjura et al. 1993).
As noted above, the second side chain that hydrogen bonds the water molecule near Gln-105 is that of Asn-101 (Table 5). When an attempt was made to remove this hydrogen bond by replacing Asn-101 with alanine, an additional solvent molecule (number 401) was observed to bind near the site of the mutated side chain and restore its hydrogen bonding function. At the same time the previously bound water molecule shifts by 0.8Å. This shift permits the formation of a hydrogen bond to the main chain carbonyl of residue 101, in addition to the side chain of Gln-105 (Table 5). Thus, rather than displacing the preexisting solvent molecule, the Asn-101 → Zy Ala mutant actually results in a pair of internal solvent molecules.
It might be noted that several other mutations were previously created at site 105. These include Gln-105 → Gly, Ala, and Glu (Pjura et al. 1993). The structure of the latter mutant is virtually indistinguishable from wild type. The glycine and alanine mutants are different in that the removal of distal atoms of the glutamine side chain result in the exposure of the previously buried solvent molecule to bulk solvent. In the Gln-105 → Zy Ala mutant the water molecule is lost. In contrast, in the Gln-105 → Zy Gly mutant, the water molecule is retained together with a second water molecule (number 568), which adopts a position previously occupied by the side chain of residue 105. This second water molecule is prevented from binding within the Gln-105 → Zy Ala mutant by a close contact with the Cβ atom of Ala-105.
Previously described mutations that affect solvent-binding sites
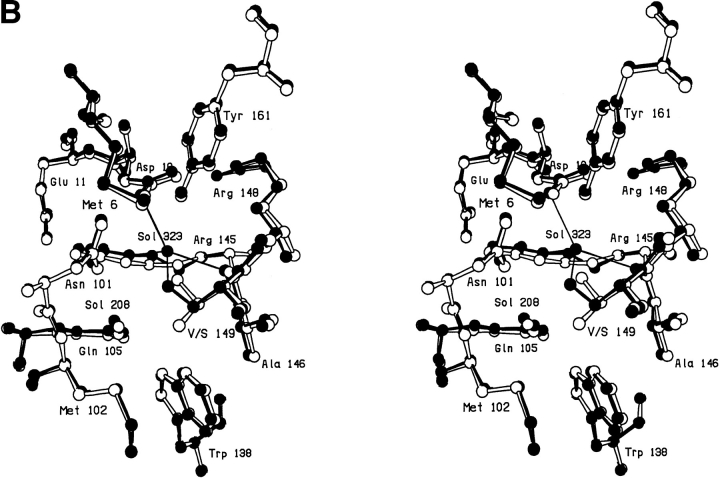
Several mutations introduced into T4 lysozyme in the course of other studies also affect the binding of internal water molecules. As in the case of the Val-149 → Ala mutant described above, a large-to-small mutation that exposes polar groups within the protein core can sometimes generate a hydrated cavity. Another such example is provided by the Met-6 → Ala mutant, where two ordered water molecules fill the void created by the truncation of the methionine side chain (Xu et al. 1998). The first water molecule (number 323) forms hydrogen bonds with the side chain hydroxyls of Thr-152 and Tyr-161 and with the second water molecule (number 324), whereas the second water molecule forms additional hydrogen bonds with the main chain carbonyl of Ala-97 and with the side chain of Asn-101.
Although the mutants described above influence the binding of internal water molecules through direct interactions, other mutants displace water through less obvious mechanisms. In the Ile-3 → Pro mutant, crystal packing forces appear to be responsible for the loss of two internal water molecules (Dixon et al. 1992). This mutant crystallizes in two different crystal forms, a trigonal form that is isomorphous with wild-type lysozyme and an orthorhombic form that has not been seen with any other mutant. In the trigonal form, the positions of the water molecules in the doubly hydrated cavity (numbers 171 and 179) are nearly identical to those observed in wild-type lysozyme. In contrast, only one of the two protein molecules in the asymmetric unit of the orthorhombic form contains water in this cavity. In the protein molecule that does not bind water, the distance between the main chain near position 29 and the central helix is slightly increased, preventing the formation of the network of hydrogen bonds that stabilizes the bound water molecules. Possibly these structural changes may be associated with the large changes in the interdomain hinge-bending angle that are observed in this crystal form.
Forces generated within the protein core by overpacking mutations can also result in the displacement of internal water molecules. Although Ala-129 is over 14Å from the buried water molecule near residues 101 and 105, replacing this residue with large amino acids leads to weaker binding at this position (Liu et al. 2000). In the Ala-129 → Phe mutant, the B-factor of this water increases to 53Å2 compared to 16Å2 in the wild-type structure, whereas in the Ala-129 → Trp mutant binding is completely abolished. Interestingly, the positions of the side chains of Asn-101 and Gln-105, both of which form hydrogen bonds with the buried water molecule in the wild-type protein, do not appear to be affected by the mutations, although the lower resolution of these structures (2.5Å) may obscure subtle changes.
Discussion
There has been a long-standing debate as to whether hydrogen bonds contribute directly to the energy of a folded protein (or to a protein–ligand complex) or whether hydrogen bonds are essentially energy neutral, that is, hydrogen bonds in the folded protein are replaced in the unfolded structure by energetically equivalent hydrogen bonds to solvent (e.g., see Chen et al. 1993; Rose and Wolfenden 1993; Pace et al. 1996).
The present mutants include examples that directly address the energetics of protein–solvent hydrogen bonds. Consider, in particular, the relationship between V149S and V149A. Relative to wild type, each of these mutants lacks the two methyl groups of Val-149. In the case of V149S a hydroxyl group is substituted for one of the methyl groups and makes a hydrogen bond to a bound solvent molecule (Table 3). The structures of V149S and V149A are very similar including, in both cases, the presence of the bound solvent. Notwithstanding the presence of the additional hydrogen bond, the structure of V149S is actually 1.3 kcal/mol less stable than V149A (Table 2). A very similar situation occurs for T152S and T152A. Here again, the structures are very similar including a bound solvent molecule that is present in both, and is hydrogen bonded by the γ-hydroxyl of Ser-152. Here, also, the serine-containing variant is less stable than that containing alanine, in this case by 0.5 kcal/mol (Table 2). It clearly suggests that the hydrogen bonds present in the T152S and the V149S structures do not contribute directly to the stabilities of these mutant proteins. Indeed, the fact that the introduction of the γ-hydroxyl group causes a net reduction in stability suggests that its hydrogen-bonding potential is not fully satisfied. In the unfolded protein, the hydroxyl group presumably forms at least two hydrogen bonds to solvent (Baker and Hubbard 1984). In the folded protein one of these is replaced by the hydrogen bond to the internal solvent molecule and is presumed to be energy neutral. The second hydrogen bond remains unsatisfied in the folded protein and could explain the observed loss in stability relative to the alanine variant.
Following this line of reasoning we can set up a simple system of accounting for the energetics of the various mutants. This scheme ignores all effects due to steric interference and, as such, is only intended for considering mutations that do not significantly perturb the protein structure.
We assume, first, that all hydrogen bonds are energy neutral (in the sense described above). We also assume that solvent molecules only contribute to the stability of the protein insofar as they satisfy potential hydrogen bonding groups on the protein. If such groups are not satisfied, we assume that there is an energy penalty of ∼1 kcal/mol per unsatisfied hydrogen bond to an -OH or -NH group. This is consistent with the estimates of Levitt (1974) and is also essentially equal to the average of the two values quoted above for the serine versus alanine. In the case of a -SH or -S-CH3 group, we assume that a nonsatisfied hydrogen bond costs 0.5 kcal/mol. We assume that the penalty from removing each methyl or methylene group from the core of the protein is 1.5 kcal/mol. In terms of the analysis of Eriksson et al. (1992) and Xu et al. (1998), this term can be considered as the sum of the solvent transfer free energy plus the energy cost of creating the cavity on removal of the methyl group. In the present case, because all the structures are quite similar to wild type (i.e., they do not collapse to fill the putative void created by the mutation), we simply assume a constant value of 1.5 kcal/mol per methyl group. This can be compared with the average value per methyl or methylene group of 1.3 kcal/mol quoted by Pace (1992). Substitution of a buried methyl group by a -SH group was assumed to be energy neutral.
This energy accounting scheme, albeit very qualitative, accounts fairly well for the changes in stability of the different variants. The discrepancies in the cases of V149I and T152I can be attributed to the strain associated with the introduction of a side chain larger than that present in wild type (Figs. 4B and 5B ▶ ▶).
It might be noted that the energy accounting scheme allows for unsatisfied hydrogen-bonding groups on the protein, but does not consider whether the hydrogen bonding potential of the bound solvent is fully satisfied. The solvent molecule Sol-323 that binds to mutants V149A, V149S, V149C, V149T, and V149G, in all cases makes at least four hydrogen bonds (Table 1). In the WT* protein, which lacks the solvent molecule, the polar groups make hydrogen bonds to other partners, typically interacting with each other (Table 1). Thus, in all cases there is no potential hydrogen bonding entity that remains unsatisfied. The solvent molecule near site 152, however, appears to make only three hydrogen bonds in the structures of WT*, T152S and T152C (Table 4). Furthermore, this number is reduced to two in the T152A, T152V, and T152I variants. If we allow for the introduction of this nonsatisfied hydrogen bond by subtracting 1 kcal/mol from the estimated stability (Table 6), the agreement with experiment is improved for T152V and T152I, although not for T152A.
Table 6.
Energetics of mutant lysozymes
| Methyl groups lost relative to WT* | Unsatisfied hydrogen bonds relative to WT* | ||||||
| Protein | Number | Estimated energetic contribution (kcal/mol)a | Number | Estimated energetic contribution (kcal/mol)a | Overall estimated change in stability of mutant relative to WT* (kcal/mol)a | Observed change in stability of mutant, ΔΔG (kcal/mol) | Additional water molecules bound to protein |
| I58T | 2 | −3.0 | 1 | −1 | −4.0 | −3.4 | 0 |
| V149A | 2 | −3.0 | 0 | 0 | −3.0 | −3.1 | 1 |
| V149S | 2 | −3.0 | 1 | −1 | −4.0 | −4.4 | 1 |
| V149C | 2 | −1.5 | 1 | −0.5 | −2.0 | −2.0 | 0.4 |
| C149T | 1 | −1.5 | 1 | −1 | −2.5 | −3.0 | 1 |
| V149I | −1 | 1.5 | 0 | 0 | 1.5 | −0.1 | 0 |
| T152A | 1 | −1.5 | 0 | 0 | −1.5 (−2.5) | −1.5 | 0 |
| T152S | 1 | −1.5 | 0 | 0 | −1.5 | −2.0 | 0 |
| T152C | 1 | 0 | 1 | −0.5 | −0.5 | −0.5 | 0 |
| T152V | −1 | 1.5 | 0 | 0 | 1.5 (0.5) | 0.2 | 0 |
| T152I | −2 | 3.0 | 0 | 0 | 3.0 (2.0) | −0.4 | 0 |
a As discussed in the text the simplest possible system of accounting as sought to reasonably explain the energetic consequences of the different substitutions. It is only intended to apply to substitutions that are at interior sites, that are conservative, and that do not significantly perturb the structure. For this reason, substitutions that involve larger changes such as Asn101 → Ala, Val149 → Gly and Gln105 → Ala are not included. The method used to estimate the energetic contributions arising from the different types of substitution and the gain or loss of hydrogen bonds is described in the text. As noted, the binding of solvent was assumed to be energy neutral. The values in parentheses, however, show the effect of allowing for an increase in unsatisfied hydrogen-bonding of the solvent molecule in the T152A, T152V and T152I structures (see text).
Conclusion
As noted above, one of the principal conclusions of this work is that hydrogen bonds from the protein to bound solvent are energy neutral and do not, per se, contribute to the stability of the folded protein. They do contribute indirectly by helping to avoid unsatisfied hydrogen bond donors and acceptors, each of which destabilizes the protein by ∼1 kcal/mol. A similar conclusion was reached by Langhorst et al. (2000).
A second conclusion is that the creation of a solvent-binding site within a protein with a single mutation requires the presence of other polar groups on the protein. Internal water molecules in proteins are typically found at locations where they can form three or four hydrogen bonds (Rashin et al. 1986; Hubbard et al. 1994; Williams et al. 1994). Therefore, to create a binding site it is necessary to generate a cavity that is large enough to accommodate a water molecule and is quite polar. This may be difficult to accomplish with a point mutation. Consider, for example, the substitution of a serine for a valine. The average volume occupied by a buried valine residue is 142Å3 and that of serine is 99Å3 (Creighton et al. 1993). The difference of 43Å3 substantially exceeds the cavity volumes of 20–30Å3 within which many solvent molecules are observed (Hubbard et al. 1994). Therefore, a Val → Ser substitution should, in principle, create sufficient space to accommodate a water molecule. The problem, however, is to provide the requisite hydrogen bonding partners. Typically a valine will occur in a nonpolar environment. Replacement of the valine with a serine introduces the γ-hydroxyl as a hydrogen bond donor or acceptor. In the absence of additional hydrogen bonding groups, however, this will not be sufficient to create a binding site. The Val-149 → Ser mutant is a case in which the environment happens to be unusually polar. In this situation a water molecule is introduced and makes four or more hydrogen bonds, one of which is to the γ-hydroxyl of the introduced serine (Table 1). The polarity of the site is further demonstrated by the observation that the Val-149 → Ala mutation also results in the introduction of a water molecule with four or more hydrogen bonds (Table 1), although the introduced side chain is incapable of participating in this network. There have been some other examples reported where a solvent-binding site has been created by a single mutation (Blaber et al. 1993; Pedersen et al. 1994; Buckle et al. 1996; Takano et al. 1997). In all such cases the introduced solvent molecule was hydrogen bonded by existing groups on the protein. In cases where a mutation creates a cavity that is large enough to accommodate more than a single water molecule, binding becomes more favorable as two or more water molecules can hydrogen bond with each other. This is the situation that occurs with the I59A and I59G mutants of human lysozyme (Takano et al. 1997), the M6A mutant of T4 lysozyme (Xu et al. 1998), as well as the V149G mutant described here.
The final conclusion is that it is difficult, with a single mutation, to displace a solvent molecule that is bound within the parent structure. We are aware of three cases in which this has been demonstrated experimentally. The first two are the G36S mutant in basic pancreatic trypsin inhibitor (Berndt et al. 1993) and the N52I mutant in Iso-1-cytochrome c (Lett et al. 1996). The third example is provided by the binding pocket of sperm whale deoxymyoglobin that contains an internal water molecule that forms a single hydrogen bond to His-64. Upon mutating this residue to glutamine or leucine, the water molecule is expelled (Quillin et al. 1993). (These examples do not include the Ala-129 → Trp and Ile-3 → Pro mutants of T4 lysozyme, discussed above, in which the reasons for solvent displacement are indirect.) As noted above, an internal water molecule will typically make three or four hydrogen bonds to the protein. Displacement of the solvent will be energetically disfavored because it will tend to leave these hydrogen bonding partners unsatisfied. A single amino acid substitution might successfully replace some of the hydrogen bonding interactions of the solvent molecule, but not all. In the Thr-152 → Ile mutation of T4 lysozyme, for example, the bound water molecule moves 1.7Å but is not displaced from the protein. In the case of Iso-1-cytochrome c the internal solvent molecule is hydrogen bonded by Asn-52, Thr-78, and Tyr-67. In the Asn-52 → Ile mutant the solvent is displaced and Thr-78 and Tyr-67 are able to satisfy their hydrogen-bonding potential by moving closer together and interacting with each other (Lett et al. 1996).
Materials and methods
All variants were constructed in the cysteine-free pseudo-wild-type lysozyme (WT*) (Matsumura and Matthews 1989). Mutants were obtained by mutagenizing an M13 clone containing the T4 lysozyme gene by the method of Kunkel et al. (1987) followed by cloning the mutant gene into the expression vector pHN1403, which was then expressed and purified as described (Matsumura and Matthews 1989). Thermal unfolding and two-state van't Hoff analysis were done as described previously (Eriksson et al. 1993) in 0.10 M sodium chloride, 0.010 M sodium acetate at pH 5.4. Standard free energy changes for unfolding (ΔG°) were calculated at 59°C assuming a ΔCp of 2.5 kcal/mol-deg. All data (Table 3) are the average of at least two independent thermal melts.
Mutant proteins were crystallized in space group P3221 using conditions similar to those described by Eriksson et al. (1993). Before crystallographic analysis the crystals were equilibrated with a "standard mother liquor" consisting of 1.05 M K2HPO4, 1.26 M NaH2PO4, 0.23 M NaCl, and 1.4 mM mercaptoethanol at pH 6.7. (In some cases the ratio of sodium and potassium phosphate was changed slightly to adjust to the pH at which the crystals were grown, typically in the range 6.7 to 7.1.) Crystallographic data (Table 7) for the mutant V149I were collected on a Rigaku R-Axis area detector. All other data were measured on a San Diego Multiwire Systems area detector (Hamlin 1985). The program STRAT (Zhang and Matthews 1993) was used to optimize data collection efficiency.
Table 7.
X-ray data collection and refinement statistics
| Mutant | I58T | N101A | Q105M | V149G | V149S | V149C | V149T | V149I | T152A | T152S | T152C | T152V | T152I |
| Data collection | |||||||||||||
| Cell dimensions | |||||||||||||
| a,b (Å) | 60.9 | 61.1 | 61.1 | 60.9 | 60.8 | 60.7 | 61.0 | 60.6 | 60.8 | 60.9 | 60.7 | 60.8 | 60.6 |
| c (Å) | 96.8 | 96.2 | 96.9 | 97.0 | 96.8 | 96.8 | 96.5 | 96.6 | 97.0 | 97.0 | 96.6 | 96.8 | 96.8 |
| Resolution (Å) | 1.9 | 1.9 | 1.8 | 1.8 | 1.85 | 1.7 | 1.8 | 1.8 | 1.9 | 1.8 | 1.85 | 1.8 | 1.7 |
| Data completeness (%) | 96 | 93 | 89 | 92 | 91 | 89 | 87 | 90 | 93 | 92 | 87 | 90 | 85 |
| Rmerge (%) | 5.8 | 4.0 | 7.2 | 3.3 | 3.0 | 3.2 | 3.3 | 5.0 | 5.6 | 4.6 | 4.8 | 2.9 | 5.0 |
| Refinement statistics | |||||||||||||
| R (%) | 16.4 | 17.4 | 17.3 | 16.1 | 15.6 | 16.7 | 15.5 | 15.0 | 15.4 | 16.1 | 15.5 | 15.4 | 16.1 |
| Δbond (Å) | 0.020 | 0.014 | 0.019 | 0.016 | 0.016 | 0.017 | 0.015 | 0.015 | 0.016 | 0.017 | 0.015 | 0.015 | 0.015 |
| Δangle (°) | 2.9 | 2.1 | 2.6 | 1.9 | 2.0 | 2.0 | 1.9 | 1.9 | 2.0 | 2.1 | 1.9 | 2.0 | 2.0 |
| PDB code | 1G1V | 1I6S | 1G1W | 1G0P | 1G06 | 1G07 | 126L | 1G0Q | 1G0G | 1G0J | 1G0K | 1G0L | 1G0M |
The cell dimensions of WT* are a = b = 60.9Å, c = 96.9Å. All crystals have space group P3221, the same as that of wild-type lysozyme.
Initial electron density maps with coefficients Fobs(mutant) −Fobs(WT*) and 2Fobs(mutant) − Fobs(WT*) phased with the WT* model (Weaver and Matthews 1987; Eriksson et al. 1993) were used to verify the mutation and to establish the initial atomic positions of the side chain of the mutated residue (e.g., Figs. 2A and 3A ▶ ▶). Refinement was carried out with TNT (Tronrud et al. 1987) using the procedure described by Baldwin et al. (1996). Putative water molecules were initially identified and located from the maps with coefficients Fobs(mutant) − Fcalc(mutant) where putative solvent sites were omitted from the calculation of Fcalc. To be identified as a water molecule an electron density peak had to be roughly spherical in shape and with height at least three standard deviations higher than the root-mean-square density of the map. Also, during subsequent refinement the crystallographic B-factor was required to remain within the approximate range 10Å2 to 75Å2. Although not used as a primary criterion for identification, all of the presumed water molecules were found to have both hydrogen bond donors and acceptors ∼2.7–3.2Å away (Tables 1, 4, and 5). Such an environment is consistent with a bound water molecule but would not be expected if, for example, the electron density in the binding site was due to a partially occupied metal ion.
Cavity volumes were calculated by the method of Connolly (1983) and were analyzed using the procedure described by Xu et al. (1998). PDB codes are given in Table 7.
Acknowledgments
We are most grateful to Sheila Snow and Leslie Gay for help with protein purification, Kane Anderson for help with the construction of mutants I58T and Q105M, Joel Lindstrom for help with the thermodynamic analyses, and Rhett Kovall for help with data collection. This work was supported in part by National Institutes of Health grant GM21967 to B.W.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.02101.
References
- Baker, E.N. 1995. Solvent interactions with proteins as revealed by X-ray crystallographic studies. In Protein-solvent interactions (ed. R.B. Gregory), pp. 143–189. Marcel Dekker, Inc., New York.
- Baker, E.N. and Hubbard, R.E. 1984. Hydrogen bonding in globular proteins. Prog. Biophys. Molec. Biol. 44 97–179. [DOI] [PubMed] [Google Scholar]
- Baldwin, E., Xu, J., Hajiseyedjavadi, O., Baase, W.A., and Matthews, B.W. 1996. Thermodynamic and structural compensation in size-switch core repacking variants of bacteriophage T4 lysozyme. J. Mol. Biol. 259 542–559. [DOI] [PubMed] [Google Scholar]
- Berghuis, A.M., Guillemette, J.G., McMendon, G., Sherman, F., Smith, M., and Brayer, G.D. 1994. The role of a conserved internal water molecule and its associated hydrogen bond network in cytochrome c. J. Mol. Biol. 236 786–799. [DOI] [PubMed] [Google Scholar]
- Berndt, K.D., Beunink, J., Schröder, W., and Wüthrich, K. 1993. Designed replacement of an internal hydration water molecule in BPTI: Structural and functional implications of a glycine-to-serine mutation. Biochemistry 32 4564–4570. [DOI] [PubMed] [Google Scholar]
- Blaber, M., Lindstrom, J.D., Gassner, N., Xu, J., Heinz, D.W., and Matthews, B.W. 1993. Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala → Ser and Val → Thr substitutions in T4 lysozyme. Biochemistry 32 11363–11373. [DOI] [PubMed] [Google Scholar]
- Buckle, A.M., Cramer, P., and Fersht, A.R. 1996. Structural and energetic responses to cavity-creating mutations in hydrophobic cores: Observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities. Biochemistry 35 4298–4305. [DOI] [PubMed] [Google Scholar]
- Chen, Y.W., Fersht, A.R., and Henrick, K. 1993. Contribution of buried hydrogen bonds to protein stability. The crystal structures of two barnase mutants. J. Mol. Biol. 234 1158–1170. [DOI] [PubMed] [Google Scholar]
- Connolly, M.L. 1983. Solvent-accessible surfaces of proteins and nucleic acids. Science 221 709–713. [DOI] [PubMed] [Google Scholar]
- Creighton, T.E., Bagley, C.J., Cooper, L., Darby, N.J., Freedman, R.B., Kemmink, J., and Sheikh, A. 1993. On the biosynthesis of bovine pancreatic trypsin inhibitor (BPTI). Structure, processing, folding and disulphide bond formation of the precursor in vitro and in microsomes. J. Mol. Biol. 232 1176–1196. [DOI] [PubMed] [Google Scholar]
- Das, G., Hickey, D.R., McLendon, D., McLendon, G., and Sherman, F. 1989. Dramatic thermostabilization of yeast iso-1-cytochrome c by an asparagine–isoleucine replacement at position 57. Proc. Natl. Acad. Sci. 86 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov, V.P., Venu, K., Peters, J., Hörlein, H.D., and Halle, B. 1997. Orientational disorder and entropy of water in protein cavities. J. Phys. Chem. 101 9380–9389. [Google Scholar]
- Dixon, M.M., Nicholson, N., Shewchuk, L., Baase, W.A., and Matthews, B.W. 1992. Structure of a hinge-bending bacteriophage T4 lysozyme mutant, Ile 3 → Pro. J. Mol. Biol. 227 917–933. [DOI] [PubMed] [Google Scholar]
- Dunitz, J.D. 1994. The entropic cost of bound water in crystals and biomolecules. Science 264 670. [DOI] [PubMed] [Google Scholar]
- Edsall, J.T. and McKenzie, H.A. 1983. Water and proteins. II. The location and dynamics of water in protein systems and its relation to their stability and properties. Adv. Biophys. 16 53–183. [DOI] [PubMed] [Google Scholar]
- Eriksson, A.E., Baase, W.A., Zhang, X-J., Heinz, D.W., Blaber, M., Baldwin, E.P., and Matthews, B.W. 1992. The response of a protein structure to cavity-creating mutations and its relationship to the hydrophobic effect. Science 255 178–183. [DOI] [PubMed] [Google Scholar]
- Eriksson, A.E., Baase, W.A., and Matthews, B.W. 1993. Similar hydrophobic replacements of Leu 99 and Phe 153 within the core of T4 lysozyme have different structural and thermodynamic consequences. J. Mol. Biol. 229 747–769. [DOI] [PubMed] [Google Scholar]
- Gassner, N.C., Baase, W.A., Lindstrom, J.D., Lu, J., Dahlquist, F.W., and Matthews, B.W. 1999. Methionine and alanine substitutions show that the formation of wildtype-like structure in the carboxy-terminal domain of T4 lysozyme is the rate-limiting step in folding. Biochemistry 38 14451–14460. [DOI] [PubMed] [Google Scholar]
- Gregoret, L.M., Rader, S.D., Fletterick, R.J., and Cohen, F.E. 1991. Hydrogen bonds involving sulfur atoms in proteins. Proteins 9 99–107. [DOI] [PubMed] [Google Scholar]
- Hamlin, R. 1985. Multiwire area X-ray diffractometers. Method Enzymol. 114 416–452. [DOI] [PubMed] [Google Scholar]
- Hickey, D.R., Berghuis, A.M., Lafond, G., Jaeger, J.A., Cardillo, T.S., McLendon, D., Das, G., Sherman, F., Brayer, G.D., and McLendon, G. 1991. Enhanced thermodynamic stabilities of yeast iso-1-cytochromes c with amino acid replacements at positions 52 and 102. J. Biol. Chem. 266 11686–11694. [PubMed] [Google Scholar]
- Hubbard, S.J., Gross, K-H., and Argos, P. 1994. Intramolecular cavities in globular proteins. Prot. Engin. 7 613–626. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A., Roberts, J.D., and Zakour, R.A. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Method Enzymol. 154 367–382. [DOI] [PubMed] [Google Scholar]
- Langhorst, U., Loris, R., Denisov, V.P., Doumen, J., Roose, P., Maes, D., Halle, B., and Steyaert, J. 1999. Dissection of the structural and functional role of a conserved hydration site in RNase T1. Protein Sci. 8 722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhorst, U., Backmann, J., Loris, R., and Steyaert, J. 2000. Analysis of a water mediated protein–protein interactions within RNase T1. Biochemistry 39 6586–6593. [DOI] [PubMed] [Google Scholar]
- Lett, C.M., Berghuis, A.M., Frey, H.E., Lepock, J.R., and Guillemette, J.G. 1996. The role of a conserved water molecule in the redox-dependent thermal stability of Iso-1-cytochrome c. J. Biol. Chem. 271 29088–29093. [DOI] [PubMed] [Google Scholar]
- Levitt, M. 1974. Energy refinement of hen egg-white lysozyme. J. Mol. Biol. 82 393–420. [DOI] [PubMed] [Google Scholar]
- Levitt, M. and Park, B.H. 1993. Water: Now you see it, now you don't. Structure 1 223–226. [DOI] [PubMed] [Google Scholar]
- Liu, R., Baase, W.A., and Matthews, B.W. 2000. The introduction of strain and its effects on the structure and stability of T4 lysozyme. J. Mol. Biol. 295 127–145. [DOI] [PubMed] [Google Scholar]
- Matsumura, M. and Matthews, B.W. 1989. Control of enzyme activity by an engineered disulfide bond. Science 243 792–794. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. 1992. Contribution of the hydrophobic effect to globular protein stability. J. Mol. Biol. 226 29–35. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Shirley, B.A., McNutt, M., and Gajiwala, K. 1996. Forces contributing to the conformational stability of proteins. FASEB J. 10 75–83. [DOI] [PubMed] [Google Scholar]
- Pedersen, J.T., Olsen, O.H., Betzel, C., Eschenburg, S., Branner, S., and Hastrup, S. 1994. Cavity mutants of savinase. Crystal structures and differential scanning calorimetry experiments give hints of the function of the buried water molecules in subtilisins. J. Mol. Biol. 242 193–202. [DOI] [PubMed] [Google Scholar]
- Pjura, P., McIntosh, L.P., Wozniak, J.A., and Matthews, B.W. 1993. Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties. Proteins 15 401–412. [DOI] [PubMed] [Google Scholar]
- Quillin, M.L., Arduini, R.M., Olson, J.S., and Phillips, G.N. Jr. 1993. High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J. Mol. Biol. 234 140–155. [DOI] [PubMed] [Google Scholar]
- Rashin, A.A., Iofin, M., and Honig, B. 1986. Internal cavities and buried waters in globular proteins. Biochemistry 25 3619–3625. [DOI] [PubMed] [Google Scholar]
- Rose, G.D. and Wolfenden, R. 1993. Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu. Rev. Biophys. Biomol. Struct. 22 381–415. [DOI] [PubMed] [Google Scholar]
- Sreenivasan, U. and Axelsen, P.H. 1992. Buried water in homologous serine proteases. Biochemistry 31 12785–12791. [DOI] [PubMed] [Google Scholar]
- Takano, K., Funahashi, J., Yamagata, Y., Fuji, S., and Yutani, K. 1997. Contribution of water molecules in the interior of a protein to the conformational stability. J. Mol. Biol. 274 132–142. [DOI] [PubMed] [Google Scholar]
- Thanki, N., Thornton, J.M., and Goodfellow, J.M. 1988. Distributions of water around amino acid residues in proteins. J. Mol. Biol. 202 637–657. [DOI] [PubMed] [Google Scholar]
- Tronrud, D.E., Ten Eyck, L.F., and Matthews, B.W. 1987. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Cryst A43 489–503. [Google Scholar]
- Vriend, G., Berendsen, H.J.C., van der Zee, J.R., van den Burg, B., Venema, G., and Eijsink, V.G.H. 1991. Stabilization of the neutral protease of Bacillus stearothermophilus by removal of a buried water molecule. Prot. Engin. 4 941–945. [DOI] [PubMed] [Google Scholar]
- Wade, R.C., Mazor, M.H., McCammon, J.A., and Quiocho, F.A. 1990. Hydration of cavities in proteins: A molecular dynamics approach. J. Am. Chem. Soc. 112 7057–7059. [Google Scholar]
- ———. 1991. A molecular dynamics study of thermodynamic and structural aspects of the hydration of cavities in proteins. Biopolymers 31 919–931. [DOI] [PubMed] [Google Scholar]
- Weaver, L.H. and Matthews, B.W. 1987. Structure of bacteriophage T4 lysozyme refined at 1.7Å resolution. J. Mol. Biol. 193 189–199. [DOI] [PubMed] [Google Scholar]
- Williams, M.A., Goodfellow, J.M., and Thornton, J.M. 1994. Buried waters and internal cavities in monomeric proteins. Protein Sci. 3 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden, R. and Radzicka, A. 1994. On the probability of finding a water molecule in a nonpolar cavity. Science 265 936–937. [DOI] [PubMed] [Google Scholar]
- Xu, J., Baase, W.A., Baldwin, E., and Matthews, B.W. 1998. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 7 158–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X-J. and Matthews, B.W. 1993. STRAT: A program to optimize X-ray data collection on an area detector system. J. Appl. Cryst. 26 457–462. [Google Scholar]